Abstract
Bone biomechanical performance is a complex trait or, more properly, an ensemble of complex traits. Biomechanical performance incorporates flexibility under loading, yield and failure load, and energy to failure; all are important measures of bone function. To date, the vast majority of work has focused on yield and failure load and its surrogate, bone mineral density. We performed a reciprocal intercross of the mouse strains HcB-8 and HcB-23 to map and ultimately identify genes that contribute to differences in biomechanical performance. Mechanical testing was performed by 3-point bending of the femora. We measured femoral diaphysis cross-sectional anatomy from photographs of the fracture surfaces. We used beam equations to calculate material level mechanical properties. We performed a principal component (PC) analysis of normalized whole bone phenotypes (17 input traits). We measured distances separating mandibular landmarks from calibrated digital photographs and performed linkage analysis. Experiment-wide α = 0.05 significance thresholds were established by permutation testing. Three quantitative trait loci (QTLs) identified in these studies illustrate the advantages of the comprehensive phenotyping approach. A pleiotropic QTL on chromosome 4 affected multiple whole bone phenotypes with LOD scores as large as 17.5, encompassing size, cross-sectional ellipticity, stiffness, yield and failure load, and bone mineral density. This locus was linked to 3 of the PCs but unlinked to any of the tissue level phenotypes. From this pattern, we infer that the QTL operates by modulating the proliferative response to mechanical loading. On this basis, we successfully predicted that this locus also affects the length of a specific region of the mandible. A pleiotropic locus on chromosome 10 with LOD scores displays opposite effects on failure load and toughness with LOD scores of 4.5 and 5.5, respectively, so that the allele that increases failure load decreases toughness. A chromosome 19 QTL for PC2 with an LOD score of 4.8 was not detected with either the whole bone or tissue level phenotypes. We conclude that first, comprehensive, system-oriented phenotyping provides much information that could not be obtained by focusing on bone mineral density alone. Second, mechanical performance includes inherent trade-offs between strength and brittleness. Third, considering the aggregate phenotypic data allows prediction of novel QTLs.
Key Words: Biomechanics, Bone modeling, Quantitative trait loci, Linkage, Pleiotropy, Principal components
Introduction
Bone biomechanical performance is complex and encompasses several different interrelated properties, each of which is subject to both genetic and environmental variability. Studies of bone genetics have addressed an array of related phenotypes, including various measures of biomechanical performance, bone size, bone shape, volumetric bone mineral density (BMD), and areal BMD. Covariation among these phenotypes is well established [Jepsen et al., 2007; Jepsen et al., 2009], leading some to assert that the biomechanical phenotypes of energy to failure, yield load, and maximum load are essentially equivalent [Ritchie et al., 2008].
When a quantitative trait locus (QTL) appears to control multiple traits, it is said to be pleiotropic. However, invoking pleiotropy as an explanation for the observed biology is really begging the question. Rather, pleiotropy is indicative of the existence of a ‘physiological gap’ between the pleiotropic gene's primary function and the studied phenotypes. Once a QTL has been identified, the goals of follow-up work are to identify the responsible gene and to fill the physiological gap, thus advancing the mechanistic understanding of how a particular genotype contributes to the observed phenotypes.
Our laboratory has performed linkage mapping of multiple bone phenotypes in a reciprocal intercross of the recombinant congenic strains HcB-8 and HcB-23 [Demant and Hart, 1986]. Here, we use the results of this work to illustrate the advantages of using multiple phenotypes to detect and study the pleiotropy of bone QTLs. While some of the data presented here have been published previously [Saless et al., 2009; Saless et al., 2010a, 2010b], several novel results and interpretations are reported here for the first time. These include the mandibular linkage data and the effect size plots for the chromosome 4 QTL. Furthermore, the emphasis here is on the integrated interpretation of the full data, which was not featured in the prior literature. Our primary interest is to understand biomechanical performance. Therefore, the majority of the individual traits we have studied are either outcomes of biomechanical tests or indices of bone size, which is related to biomechanical performance.
Materials and Methods
Detailed methods have been described elsewhere [Saless et al., 2009; Saless et al., 2010a, 2010b]. Briefly, methods were as follows. Mechanical testing was performed by 3-point bending of the femora. We measured femoral diaphysis cross-sectional anatomy from photographs of the fracture surfaces. We used beam equations to calculate material level mechanical properties. We performed a principal component (PC) analysis of normalized whole bone phenotypes using the R function prcomp. We measured distances separating mandibular landmarks from calibrated digital photographs. We performed linkage analysis using R/qtl and QTL Cartographer. Experiment-wide α = 0.05 significance thresholds were established by permutation testing and ranged between LOD scores 2.7 and 3.0. All other statistical analyses were performed with SigmaStat (SPSS) and data are shown as means ± SEM. All animal studies were approved by the William S. Middleton Memorial Veterans Hospital and University of Wisconsin IACUCs.
Results
Biomechanical properties of HcB-8 and HcB-23 have been summarized in detail [Saless et al., 2010b]. Briefly, HcB-8 bones are smaller and more cylindrical, and they have lower yield and maximum loads but absorb more energy prior to fracture and have a higher Young's modulus. Therefore, it is not the case that one strain has a better biomechanical performance than the other but rather that their mechanical performances differ, with HcB-23 bones being stronger but achieving greater strength at the expense of greater brittleness.
We chose these strains for the genetic experiments in part because of their contrasting biomechanical behavior but also because, as members of the same recombinant congenic strain series, they share alleles over approximately 75% of the genome [Groot et al., 1992]. This impacts the genetic analysis in 2 important ways. First, it reduces the significance threshold from an LOD score of approximately 4.3 for a genome-wide intercross to one of approximately 2.8. This is simply a reflection of the shared genetic information, resulting in the performance of fewer statistical tests. Second, the strains’ construction allowed crossovers to occur between the ancestral strains. Recombination breakpoints serve to constrain the possible locations of QTLs mapped in a cross between HcB strains in a way that is not possible in an intercross between ordinary inbred strains.
Chromosome 4 contains only a short informative region between HcB-8 and HcB-23, spanning approximately 6 Mb of physical length and about 1 cM of genetic length. It harbors the single most robust QTL found in the entire experiment (fig. 1). It encompasses 10 whole bone level phenotypes and displays LOD scores as high as 17.5 (shape factor), as well as double digit LOD scores for maximum load, outer major axis length, and cross-sectional area [Saless et al., 2009]. It is also linked to 3 of the 4 PC we constructed from the raw data, which together account for 80% of the overall phenotypic variance observed in the experiment [Saless et al., 2010a]. However, it is not linked to any material property phenotypes [Saless et al., 2010b]. Thus, this locus is pleiotropic at the level of whole bone structure and mechanical performance and also at the level of PCs, which by definition are mutually orthogonal.
Fig. 1.
Chromosome 4 linkage. The x-axis shows the map position and the y-axis shows the LOD score. The map includes only 1 cM. The longer line shows the α = 0.05 significance threshold. CSA = Cross-sectional area; OutMajAx = outer major axis; InMinAx = inner minor axis; / = cross-sectional moment of inertia. Reprinted with permission of the FASEB Journal.
On the basis of these data, particularly the negative result of the material property mapping, the primary effect of the QTL was on bone size. Further, because the linkage signal was stronger for the shape factor than for any other included trait, we further inferred that the QTL affected not size per se but rather modeling in response to mechanical loading. Notably, prior work has demonstrated that at least one gene mediating responsiveness to mechanical loading is present on chromosome 4 [Robling et al., 2003].
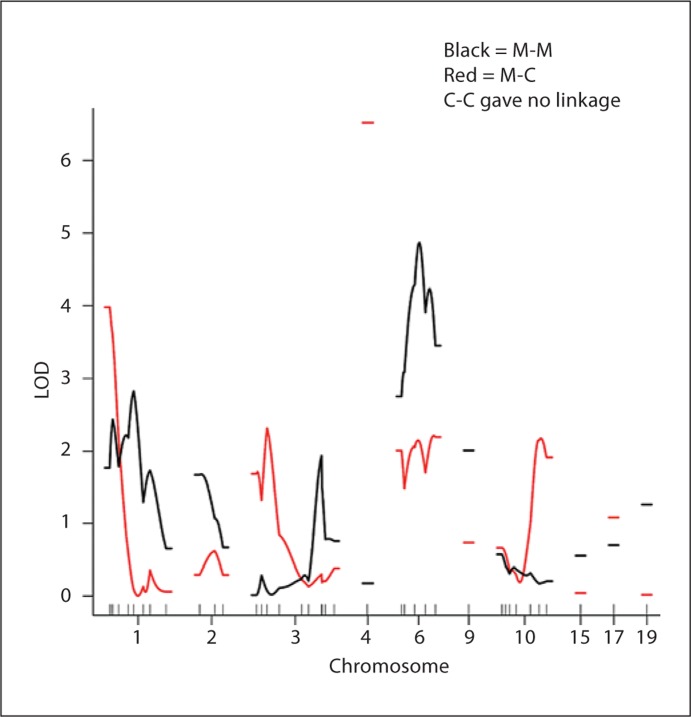
To test this interpretation, we performed linkage analysis of mandible length dividing the mandible into 3 segments separated by anatomical landmarks. The anterior segment extends from the mandibular symphysis to the first molar (M-M), the middle segment extends from the first molar to the coronoid process (M-C), and the posterior segment extends from the coronoid process to the condylar process. The M-M segment does not contain any insertions for muscles of mastication, while the M-C segment contains the insertion of the masseter. We predicted that M-C length would be linked to chromosome 4, while M-M length would not be. This is exactly what we found, as shown in figure 2. M-M length is linked to chromosome 6, and a second QTL for M-C length is also present on chromosome 1. We did not map any QTLs for C-C length.
Fig. 2.
Linkage mapping of mandibular length. The x-axis shows the location and the y-axis shows the LOD score. The α = 0.05 significance levels are LOD = 2.8 for M-M and 2.9 for M-C.
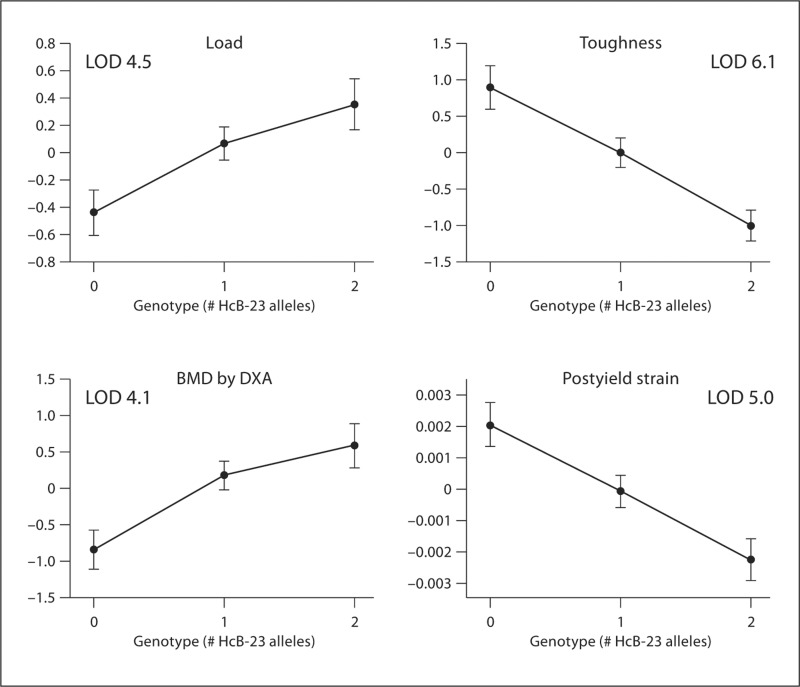
The parental strains display opposite behavior with regard to strength, as expressed by maximum load, and ductility, as expressed by energy to failure or toughness. A QTL on chromosome 10 displays the same behavior at the level of a single pleiotropic QTL as illustrated in figure 3. For this QTL, the HcB-8 allele increases toughness and postyield strain while simultaneously decreasing maximum load and BMD [Saless et al., 2009, 2010b]. This behavior is precisely that which was previously shown to maximize the statistical power of multiple phenotype mapping [Allison et al., 1998].
Fig. 3.
Effect plots for D10Mit106. Top left = Maximum load; top right = toughness; bottom left = BMD; bottom right = postyield strain.
We performed a PC analysis of the whole femur phenotypes and performed linkage mapping of the 4 whose eigenvalues exceeded 1 [Saless et al., 2010a]. We found a QTL for PC2 on chromosome 19 that was undetected in either of the whole bone properties. Inspection of the eigenvector suggests that PC2 represents size without mechanical performance, including large negative contributions from yield load, maximum load, stiffness, energy to failure, and BMD and large positive contributions from inner minor axis, inner major axis, outer major axis, and shape factor. The QTL has an LOD score of 4.7 and accounts for 3.5% of the phenotypic variance, and HcB-8 contributes the high phenotype allele. The informative segment of chromosome 19 is short, spanning approximately 10 Mb. Among the genes in the interval, Ostf1, encoding osteoclast stimulating factor 1, is an obvious positional candidate gene. Ostf1, an intracellular signaling protein that promotes osteoclast maturation and activity [Reddy et al., 1998] was originally isolated from mouse embryo cDNA. This gene is an attractive candidate because the loading of PC2 is heavily weighted by inner minor and inner major axis lengths, phenotypes that reflect the size of the marrow space.
Discussion
We have presented several examples of the advantages resulting from the simultaneous investigation of multiple phenotypes. On chromosome 4, we used the pattern of pleiotropy to infer the mechanism by which the QTL acts and to use the inference to predict an additional phenotype. The findings on chromosome 10 demonstrate a limit to the ability to simultaneously increase strength and ductility. In this case, the same QTL has opposite effects on these properties. Moreover, it supports the premise that bone responses to load and energy are distinct in spite of claims that they convey similar biological information [Ritchie et al., 2008]. Previously, we used FTIR to show that HcB-8 bones have more mature collagen cross-links and more crystalline apatite than do HcB-23 bones [Blank et al., 2003]. These findings provide a chemical basis for the differences in biomechanical performance between HcB-8 and HcB-23. On chromosome 19, use of PC allowed us to map a QTL that could not be detected by any single phenotype. PC analysis depends on the use of multiple phenotypes, extracting their shared information to reduce the dimensionality of the data. None of the insights described here could have been obtained had we studied more limited data.
In the case of chromosome 4, the lack of a tissue level phenotype allowed us to infer that the primary effect of the QTL is on growth. In the case of the chromosome 10 QTL, we cannot distinguish whether the primary genetic effect is on strength or on ductility as these traits have not been dissociated genetically. On chromosomes 4 and 19, we have also exploited the short length of the informative chromosome segments to identify candidate genes – Ece1 on chromosome 4 and Ostf1 on chromosome 19 [Saless et al., 2009, 2010a, 2010b]. On chromosome 10, the informative region is larger, and we have not yet identified a candidate gene within the interval.
Our studies have some important limitations. Using recombinant congenic strains as parental strains made it impossible for us to detect bone QTLs in genomic regions where HcB-8 and HcB-23 harbor the same allele. The end points of the informative chromosome regions have not been precisely localized. We have analyzed the bone properties of the F2 progeny only at a single age, so our results are unable to address developmental phenotypes either during growth or following maturity. The biological interpretation of the PC is also potentially problematic as they are synthetic phenotypes. We had hoped that the limited power of biomechanical testing [Leppanen et al., 2008], particularly in the context of measuring plasticity, energy absorption, and their material equivalents, i.e. strain and toughness, might be mitigated by the use of PC analysis. However, PC3, which corresponds best to these properties, proved to be unsuitable for linkage analysis.
The data presented here reveal some of the complexity regarding the establishment and maintenance of skeletal size, shape, and tissue quality. Pleiotropy is one manifestation of that complexity, and its existence serves as both a tool and a spur to elucidate the mechanisms that separate gene function from measured phenotypes. We have illustrated how considering the functional relationship among phenotypes allows inference of the unifying mechanisms by which those phenotypes arise. Further progress will depend on embracing the complexity of bone biomechanical performance and developing a greater understanding of its chemical and structural basis.
Acknowledgements
This material is based on work supported by the Office of Research and Development, Biomedical Laboratory R&D Service, Department of Veterans Affairs (R.D.B.), and this work was performed in part at the Geriatrics Research, Education, and Clinical Center of the William S. Middleton Memorial Veterans Hospital. This is Madison GRECC manuscript 10-23. This study was also supported by NIH R01 AR054753 (R.D.B.).
Glossary
Abbreviations used in this paper
| cAMP | cyclic adenosine monophosphate |
| CAP1 | cyclase-associated protein 1 |
| C/EBPα | CAAT enhancer binding protein alpha |
| C/EBPβ | CAAT enhancer binding protein beta |
| Glut4 | glucose transporter 4 |
| IGF1R | insulin-like growth factor 1 receptor |
| IR | insulin receptor |
| IRS | insulin receptor substrate |
| LDLR | low-density lipoprotein receptor |
| MAPK | mitogen-activated protein kinase |
| PI3K | phosphoinositide 3-kinase |
| PPARγ | peroxisome proliferator-activated receptor gamma |
| Srebf1 | sterol regulatory element binding factor 1 |
References
- Allison D.B., et al. Multiple phenotype modeling in gene-mapping studies of quantitative traits: power advantages. Am J Hum Genet. 1998;63:1190–1201. doi: 10.1086/302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank R.D., et al. Spectroscopically determined collagen Pyr/deH-DHLNL cross-link ratio and crystallinity indices differ markedly in recombinant congenic mice with divergent calculated bone tissue strength. Connect Tissue Res. 2003;44:134–142. doi: 10.1080/03008200390223918. [DOI] [PubMed] [Google Scholar]
- Demant P., Hart A.A. Recombinant congenic strains – a new tool for analyzing genetic traits determined by more than one gene. Immunogenetics. 1986;24:416–422. doi: 10.1007/BF00377961. [DOI] [PubMed] [Google Scholar]
- Groot P.C., et al. The recombinant congenic strains for analysis of multigenic traits: genetic composition. FASEB J. 1992;6:2826–2835. doi: 10.1096/fasebj.6.10.1634045. [DOI] [PubMed] [Google Scholar]
- Jepsen K.J., et al. Genetic randomization reveals functional relationships among morphologic and tissue-quality traits that contribute to bone strength and fragility. Mamm Genome. 2007;18:492–507. doi: 10.1007/s00335-007-9017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K.J., et al. Phenotypic integration of skeletal traits during growth buffers genetic variants affecting the slenderness of femora in inbred mouse strains. Mamm Genome. 2009;20:21–33. doi: 10.1007/s00335-008-9158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppanen O.V., Sievanen H., Jarvinen T.L. Biomechanical testing in experimental bone interventions – may the power be with you. J Biomech. 2008;41:1623–1631. doi: 10.1016/j.jbiomech.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Reddy S., et al. Isolation and characterization of a cDNA clone encoding a novel peptide (OSF) that enhances osteoclast formation and bone resorption. J Cell Physiol. 1998;177:636–45. doi: 10.1002/(SICI)1097-4652(199812)177:4<636::AID-JCP14>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Ritchie R.O., et al. Measurement of the toughness of bone: a tutorial with special reference to small animal studies. Bone. 2008;43:798–812. doi: 10.1016/j.bone.2008.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robling A.G., et al. Evidence for a skeletal mechanosensitivity gene on mouse chromosome 4. FASEB J. 2003;17:324–326. doi: 10.1096/fj.02-0393fje. [DOI] [PubMed] [Google Scholar]
- Saless N., et al. Quantitative trait loci for biomechanical performance and femoral geometry in an intercross of recombinant congenic mice: restriction of the Bmd7 candidate interval. FASEB J. 2009;23:2142–2154. doi: 10.1096/fj.08-118679. [DOI] [PubMed] [Google Scholar]
- Saless N., et al. Linkage mapping of principal components for femoral biomechanical performance in a reciprocal HCB-8xHCB-23 intercross. Bone. 2010a;48:647–653. doi: 10.1016/j.bone.2010.10.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saless N., et al. Linkage mapping of femoral material properties in a reciprocal intercross of HcB-8 and HcB-23 recombinant mouse strains. Bone. 2010b;46:1251–1259. doi: 10.1016/j.bone.2010.01.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks A.B., et al. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]