Abstract
Two point mutations (T21I and P40T) within amelogenin have been identified from human DNA sequences in 2 instances of amelogenesis imperfecta. We studied the folding and self-assembly of recombinant amelogenin (rM180) compared to the T21I and P40T mutants analogs. At pH 5.8 and 25°C, rM180 and the P41T mutant existed as monomers, whereas the T21I mutant formed small oligomers. At pH 8 and 25°C, all of the amelogenin samples formed nanospheres with hydrodynamic radii (RH) of around 15–16 nm. Upon heating to 37°C, particles of P41T increased in size (RH = 18 nm). During thermal denaturation at pH 5.8, both of the mutant proteins refolded more slowly than the wild-type (WT) rM180. Variable temperature tryptophan fluorescence and dynamic light scattering studies showed that the WT transformed to a partially folded conformation upon heating and remained stable. Thermal denaturation and refolding studies indicated that the mutants were less stable and exhibit a greater ability to prematurely aggregate compared to the WT. Our data suggest that in the case of P41T, alterations in the self-assembly of amelogenin are a consequence of destabilization of the secondary structure, while in the case of T21I they are a consequence of change in the overall hydrophobicity at the N-terminal region. We propose that alterations in the assembly (i.e. premature aggregation) of mutant amelogenins may have a profound effect on intra- and extracellular processes such as amelogenin secretion, proteolysis, and its interactions with nonamelogenins as well as with the forming mineral.
Key Words: Enamel, Amelogenin, Amelogenesis imperfecta, Dynamic light scattering, Thermal denaturation, Fluorescence spectroscopy, Protein aggregation
Introduction
The formation of an amelogenin matrix that serves to template the growth of enamel crystals is a key event that occurs during tooth enamel development [Simmer and Fincham, 1995; Moradian-Oldak, 2001]. Disruptions to the amelogenin sequence can disrupt the entire enamel-forming process, resulting in teeth that can be either hyper- or hypomineralized. Amelogenesis imperfecta is a disorder in which enamel is abnormally formed, often because of the malfunction of amelogenin or nonamelogenin proteins during enamel development. Two mutations in amelogenin that have been observed in humans are T21I and P40T [Lench and Winter, 1995; Collier et al., 1997; Wright, 2006], both of which cause the enamel to be hypomineralized and to have a brown discoloration. In most cases, the molecular mechanisms of defective enamel formation have still not been elucidated, although a possible molecular mechanism of the induced Y34H mutant in mice has been described recently [Barron et al., 2010].
The current report is an expansion of our recent study as we follow up in more depth the folding and self-assembly of a recombinant wild-type amelogenin (rM180) and the T21I and P41T mutants [Moradian-Oldak et al., 2000; Lakshminarayanan et al., 2010]. It is important to note that as a recombinant mouse mutant was used in this work an extra methionine is present at the 29th residue, meaning human P40T becomes mouse P41T. Here, we directly visualized the self-associated forms of the mutated amelogenins by transmission electron microscopy (TEM) and further performed dynamic light scattering (DLS) and fluorescence on the nanospheres formed at pH 8 by utilizing a more powerful instrument. We determined how each mutation affects the intra- and intermolecular interactions of amelogenin and hypothesized how such mutations affect amelogenin interactions with other components of the extracellular matrix, affecting physiological events such as enzymatic processing of the amelogenin matrix.
Material and Methods
Protein Expression, Purification, and Preparation
Preparation of the recombinant His-tagged rM180 and the mutants has been described previously [Moradian-Oldak et al., 2000]. The proteins were further purified on a Jupiter C4 semi-preparative reversed phase column (10 × 250 mm, 5 μm) Varian Prostar HPLC system (Varian, Palo Alto, Calif., USA). All of the proteins contain ‘RGHHHHHHGS’ residues at the N-terminus and lack a phosphorylation on Ser-16 [Fincham et al., 1994; Moradian-Oldak et al., 2000] in contrast to the native forms. The amelogenin concentration was set at 0.4 mg·ml–1. Sodium acetate (25 mM) buffer was used for experiments at pH 5.8 while Tris buffer (25 mM) was used for experiments at pH 8. Isothermal experiments were performed at 25°C.
Circular Dichroism Spectropolarimetry
Circular dichroism spectropolarimetry (CD) was performed using a JASCO J-815 spectropolarimeter (JASCO, Easton, Md., USA). The spectra were recorded between 190 and 240 nm with a step size of 0.5 nm and a scan rate of 50 nm/min. Four scans were averaged and no smoothing was required. All experiments were performed under a constant nitrogen flush using a 1-mm path length quartz cuvette. Variable temperature (VT) experiments were performed between 10 and 70°C, pH 5.8, using a ramp rate of 1°C/min.
Fluorescence Spectroscopy
Intrinsic tryptophan steady-state fluorescence spectroscopy was performed using a PTI QuantaMaster QM-4SE spectrofluorometer (PTI, Birmingham, N.J., USA). The amelogenin solutions were excited at 290 nm. The emission spectra were monitored between 300 and 400 nm with a step size of 1 nm using a 10-mm path length cell. A Peltier controller accurate to 0.1°C was used for VT measurements. Samples at pH 5.8 were heated from 25 to 70°C and the emission spectra recorded at intervals of 5°C. Each measurement was made after 5 min of equilibration time at the designated temperature.
Dynamic Light Scattering
DLS studies were performed using a Wyatt DynaPro Nanostar DLS instrument (Wyatt Technology, Santa Barbara, Calif., USA). Amelogenin samples were prepared by dissolving the lyophilized protein in cold deionized water to a concentration of 0.8 mg/ml. The stock solutions were kept at 4°C for 2 days prior to the addition of buffer. Isothermal experiments were carried out on amelogenin samples of 0.4 mg/ml at 25 and 37°C. Sodium acetate buffer (25 mM) was used for the samples at pH 5.5, while Tris buffer (25 mM) was used for the samples at pH 8. As Tris has a high temperature coefficient, the Tris buffers were prepared at the relevant temperature in each case. VT experiments were performed between 20 and 60°C at a ramp rate of 0.25°C/min. The data were analyzed using Dynamics 7.0 software as previously described [Lakshminarayanan et al., 2010]. The hydrodynamic radii (RH) that are presented in the text were calculated from regularization histograms with %mass set as the y-axis value.
Transmission Electron Microscopy
T21I and P41T amelogenin samples (0.4 mg·ml–1, pH 5.8) were heated to 37°C in a drying oven for 30 min. For each sample, a 2-μl droplet was placed on a 3-mm carbon-coated copper grid with the temperature maintained at 37°C. The droplet was wicked away after 30 s. A 2-μl droplet of uranyl acetate (2% w/w) was then placed on the grid and wicked away after 15 s. TEM was performed using a JEOL 1200EX TEM (JEOL, Tokyo, Japan).
Results
Isothermal Spectroscopic Studies at pH 8 and 5.8
CD, spectrofluorometry, and DLS were performed on samples of rM180 and the 2 mutants at room temperature (25°C) at both pH 5.8 and pH 8. We performed the majority of the experiments at pH 5.8 as rM180 existed as monomers. We analyzed the proteins at pH 8 to observe the effect of mutations on assembly and to understand the effect of assembly on the fluorescence emission maximum. DLS and TEM analyses were also performed at 37°C.
Analysis of Assembly at 25°C
At pH 5.8, DLS revealed that both rM180 and P41T had RH of around 3.4 and 3.5 nm, respectively (table 1). For proteins with an equivalent chain length, a monomer with a globular conformation would have an RH of 2.2 nm, while a protein monomer with an extended conformation would have an RH of 4.2 nm [Uversky, 2002]. That rM180 and the P41T mutant were measured to have an RH of about 3.5 nm implies that they exist as partially folded monomers at pH 5.8. Surprisingly, the T21I mutant had an RH of 7 nm, indicating the presence of small oligomers. Intrinsic tryptophan fluorescence spectroscopy was performed on the same samples to probe the environment around the tryptophan residues. rM180 and P41T had emission maxima at 346 and 345.5 nm, respectively, but T21I differed once again, having a blue-shifted emission maximum at 340.5 nm.CD at pH 5.8 revealed that rM180 and P41T were essentially identical in terms of folding (data not shown) as both had minima at 202 nm with virtually identical ellipticity values. Although T21I also had a minimum at 202 nm, the ellipticity value was much lower.
Table 1.
A comparison of the RH of each amelogenin at pH 5.8 and pH 8 and at 25° C and 37° C
| pH | Protein | RH (25° C), nm | RH (37° C), nm |
|---|---|---|---|
| 5.8 | rM180 | 3.418±015 | 3.748±0.23 |
| T21I | 7.078±0.23 | 7.388±0.20 | |
| P41T | 3.528±0.11 | aggregated | |
| 8 | rM180 | 15.018±0.38 | 14.718±0.19 |
| T21I | 14.998±0.47 | 15.308±0.41 | |
| P41T | 16.418±0.71 | 18.288±0.29 |
At pH 8, rM180 and the T21I mutant formed nanospheres with RH of around 15 nm, whereas P41T formed slightly larger nanospheres with RH of just over 16 nm (table 1). The change was small but reproducible (n = 4). Fluorescence spectroscopy revealed that each of the amelogenins had blue shifted to 337–338 nm upon nanosphere formation, but no significant difference could be observed between the wild-type and each mutant.
Analysis of Assembly at 37°C
When rM180 and T21I were heated to 37°C at pH 5.8, no significant change in RH was observed from their respective RH values at 25°C. However, P41T at pH 5.8 showed significant signs of aggregation at 37°C (table 1). TEM was performed in order to visualize the mutant amelogenins at 37°C and pH 5.8. At this temperature, light scattering indicated that T21I existed primarily in the form of small oligomers, whereas P41T had already begun to aggregate. The T21I oligomers (n = 51) appeared to be relatively homogeneous with an average diameter of 18.3 ± 2.4 nm (fig. 1a). The P41T aggregates (n = 38) were heterogeneous and nonspherical with diameters ranging from 130 to 560 nm with an average of 306.7 ± 110.5 nm (fig. 1b).
Fig. 1.
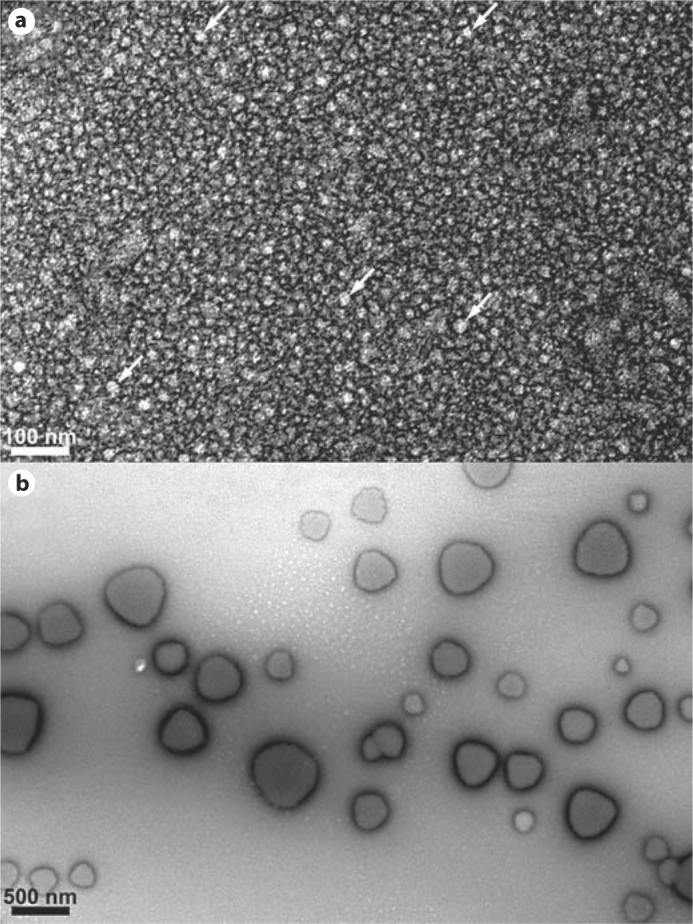
TEM images showing the T21I mutant at pH 5.8, 0.4 mg/ml, and 37°C (a). The particles were 18.2 nm in diameter on average, which is in approximate agreement with DLS. White arrows highlight 5 of the most clearly apparent oligomers. b The P41T mutant formed large aggregates at 37°C with diameters ranging from 100 to 500 nm, again in agreement with DLS. Scale bars = 100 and 500 nm, respectively.
At pH 8 and 37°C, rM180 and T21I formed nanospheres of a similar size to those observed at 25°C. On the other hand, P41T nanospheres increased in RH by around 2 nm when heated to 37°C (table 1).
VT Studies
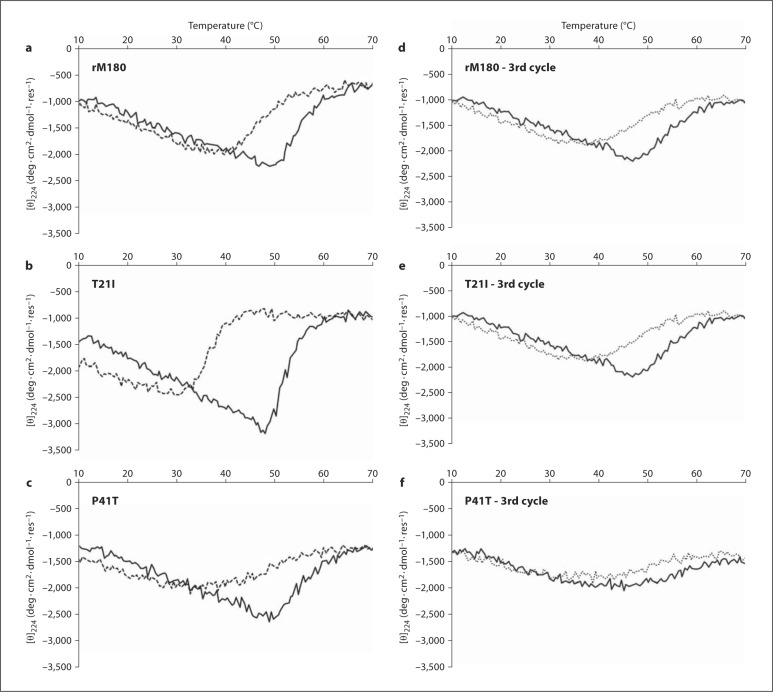
VT studies were performed at pH 5.8 to minimize aggregation and maximize the mass of monomers in the solution. VT-CD on rM180 revealed a biphasic denaturation curve with a midpoint (Tm) at 50°C (fig. 2a). The cooling profile (refolding curve) showed some hysteresis but ultimately returned to its original folding state by 40°C (full spectra not shown). T21I also exhibited a biphasic denaturation profile, but the turning point was sharper and occurred just below 50°C (fig. 2b), whereas its cooling profile was markedly different, exhibiting significant hysteresis. Refolding began at 45°C and the cooling profile crossed the heating profile at 33°C. The heating profile of P41T looked similar to that of rM180 with an initial turning point at just under 50°C (fig. 2c). Upon cooling, however, P41T refolded more slowly than rM180, returning to its original state at 30°C. Subsequent heating and cooling cycles on rM180 were extremely similar in appearance to the original heat/cool cycle (fig. 2d). When T21I and P41T were subjected to subsequent heat/cool cycles, the denaturation and refolding curves broadened and weakened (fig. 2e, f). In addition, the midpoint of the P41T denaturation curve shifted from 49 to 43°C.
Fig. 2.
First heating (solid lines) and cooling (dashed lines) CD profiles performed at pH 5.8 (monitoring at 224 nm) of rM180 (a), T21I (b), and P41T (c) measured at pH 5.8. Third heating (solid lines) and cooling (dashed lines) profiles of rM180 (d), T21I (e), and P41T (f).
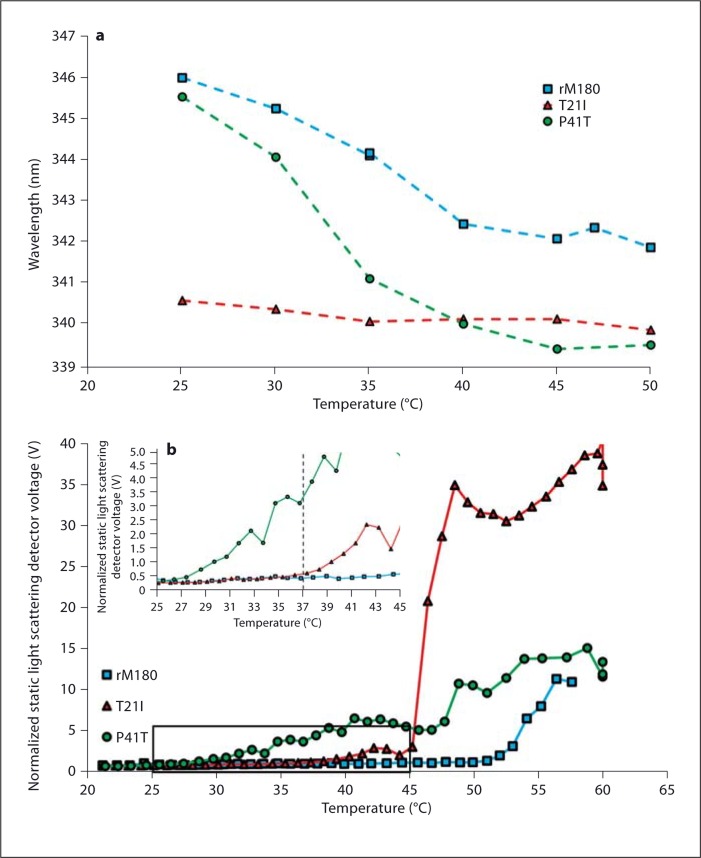
VT fluorescence spectroscopy of the 3 proteins showed that the emission from both rM180 and the P41T mutant shifted to lower wavelengths upon heating from 25 to 50°C (fig. 3a). Between 25 and 40°C, P41T exhibited a shift from 345.5 to 340 nm (5.5-nm blue shift), and rM180 exhibited a shift from 346 to 342.5 nm (3.5-nm blue shift). Further heating to 50°C did not significantly alter the emission wavelength in either protein. T21I already exhibited a lower wavelength emission of 340.5 nm at 25°C. This emission wavelength did not vary significantly when the temperature was increased to 50°C.
Fig. 3.
a Fluorescence heating profiles of rM180, T21I, and P41T measured at pH 5.8. The maximum emission wavelength was plotted as the samples were heated. b VT light scattering plots of rM180, T21I, and P41T. The inset provides a close-up view of the area identified by the box, giving a clearer view of the onset of aggregation of T21I and P41T.
VT light scattering was performed in order to track the amount of aggregation occurring in the samples during heating. By observing the total amount of light scattered by the sample, it was possible to qualitatively measure the level of aggregation as the samples were heated (fig. 3b). Upon heating, rM180 did not display significant aggregation until just above 50°C. Both of the mutant samples aggregated at lower temperatures. T21I remained relatively stable until signs of a small amount of aggregation were detected at 38°C. At 45°C, a sudden increase in aggregation was observed that occurred within a range of 1°C. P41T began to show signs of aggregation as low as 28°C (fig. 3b inset) and continued to gradually aggregate with increasing temperature. No sudden increase in scattering was observed during heating.
Discussion
Two N-terminal mutations on amelogenin (T21I and P40T) cause a hypomineralized enamel phenotype in human subjects [Lench and Winter, 1995; Collier et al., 1997; Wright, 2006], resulting in soft and discolored teeth. Considering the notion that amelogenin assembly is critical for normal enamel biomineralization we applied spectroscopic and imaging approaches to analyze the folding, self-assembly, and aggregation properties of the His-tagged recombinant full-length murine amelogenin (rM180) compared to the T21I and P41T mutants. None of the 3 spectroscopic techniques performed at pH 5.8 and 25°C could identify any differences between rM180 and P41T, each existing as ≈3.5-nm monomers and displaying a disordered secondary structure. Both of these properties are typical of premolten globule (PMG) type proteins [Uversky, 2002]. Although the T21I mutant also appeared to have a PMG conformation, the lower ellipticity at 200 nm revealed that it was slightly more ordered than both rM180 and P41T. More significantly, the fluorescence emission maximum of T21I was blue shifted by 5 nm and DLS showed that the RH was 7 nm, which was double the radius of the monomeric rM180 and P41T (table 1).
Isothermal experiments at 25°C could not identify any differences between the wild-type rM180 amelogenin and the amelogenesis imperfecta-causing mutant P41T amelogenin. However, as soon as P41T was heated above 28°C it began to aggregate. Aggregation continued gradually over a range of 15°C. That the PMG nature of the amelogenin was maintained in P41T despite the mutation indicates that P41 is not critical to preserving the conformation at room temperature [Lakshminarayanan et al., 2010]. However, upon heating above 28°C, P41T began to form heterogeneous aggregates indicating that P41 is necessary to maintain the thermodynamic stability of a more ordered intermediate conformation that is formed upon heating. That amelogenin forms an intermediate conformation upon heating was confirmed by a combination of fluorescence, CD, and DLS. Firstly, CD showed that rM180 increased its amount of conformational order with increasing temperature. Secondly, fluorescence showed that the emission wavelength of rM180 blue shifted when heating from 25 to 50°C, revealing that some or all of the tryptophans in the molecules were moving into more hydrophobic environments. Thirdly, DLS was necessary to show that rM180 did not undergo any self-assembly interactions when the structural rearrangements were occurring.
At pH 8, recombinant mouse amelogenin formed nanospheres with an RH of around 15 nm at 25 and 37°C (table 1). T21I exhibited identical behavior in maintaining an RH of around 15 nm despite the temperature increase. Interestingly, P41T did not aggregate upon heating as it did at pH 5.8. Instead, the nanospheres increased in size from just over 16 nm to just over 18 nm in RH (table 1).
The replacement of threonine by isoleucine adds more hydrophobic character to the molecule in the N-terminal tyrosine-rich amelogenin peptide (TRAP) region or the self-assembly A domain, a region that has been previously postulated to be partially responsible for self-assembly in amelogenin [Paine and Snead, 1997]. It was therefore not surprising that the T-to-I replacement might facilitate self-association even under conditions that favor the monomeric form of amelogenin. The observation that T21I existed as oligomers under these conditions suggested that the ‘self-binding’ constant of T21I is greater than rM180 (i.e. the dissociation constant is lower). Using surface plasmon resonance (SPR) spectroscopy, it was found that rM180-rM180 dissociation occurred more rapidly than either T21I-rM180 dissociation or P41T-rM180 dissociation [Paine et al., 2002]. Reinterpretation of the published SPR data indicates that binding affinities occur in the order T21I-T21I > T21I-rM180 > rM180-rM180. The same order applies for the P41T mutant. Recent work on ‘amelogenin-to-MMP20’ interactions [Tanimoto et al., 2008] also used SPR to calculate relative binding constants. Amelogenin solutions were passed over an immobilized layer of deactivated MMP20 and it was found that P41T amelogenin exhibited a lower k(on) signal than did the wild-type rH174 (recombinant human amelogenin). Using the same logic described earlier, this decrease can be attributed to a larger P41T-P41T binding interaction occurring in solution. Importantly, the k(off) values were identical, revealing that both rH174 and P41T had the same dissociation constant with MMP20.
In summary, both mutations caused amelogenin molecules to interact more tightly under conditions where the wild-type amelogenin was monomeric. The T21I mutant existed as oligomers at pH 5.8 and 25°C. That it existed in an oligomeric form indicates that important self-assembling interactions have been stabilized, presumably as the TRAP region has been made more hydrophobic by the mutation. The P41T mutant displayed characteristics identical to those of the wild type at pH 5.8 and 25°C, but as soon as it was heated it began to aggregate. As the conformation of the wild-type amelogenin has been observed to become more ordered with an increase in temperature, we could infer that P41T misfolded as it attempted to adopt the more ordered conformation. In addition, this misfold may have caused the nanospheres to increase in size upon heating to 37°C at pH 8. We propose that each mutation described here increases the self-association of amelogenin at 37°C, albeit via different mechanisms affecting amelogenesis in vivo. Intracellular aggregation as a result of increased self-affinity (case of T21I) or misfolding (case of P41T) could hinder amelogenin secretion. Such a mechanism was recently elucidated for the Y34H mutant in mice [Barron et al. 2010]. A mutation in the tri-tyrosyl region can also affect interactions between amelogenin and other proteins (e.g. enamelin or ameloblastin). Increased self-affinity could also impede proteolytic processing in the extracellular domain and affect protein-mineral interactions.
Acknowledgements
We would like to thank Professor Ralf Langen for use of the TEM and Nickolas Chelyapov at the Nanobiophysics Core Facility at USC for CD and fluorescence spectroscopy. The funding was provided by NIH-NIDCR grants DE-020099, DE-013414, and DE-015644.
Glossary
Abbreviations used in this paper
| CD | circular dichroism spectropolarimetry |
| DLS | dynamic light scattering |
| P41T | His-tagged recombinant full-length murine amelogenin with a threonine replacing a proline at residue 41 |
| PMG | premolten globule |
| RH | hydrodynamic radii |
| rM180 | His-tagged recombinant full-length murine amelogenin |
| SPR | surface plasmon resonance |
| T21I | His-tagged recombinant full-length murine amelogenin with an isoleucine replacing a threonine at residue 21 |
| TEM | transmission electron microscopy |
| TRAP | tyrosine-rich amelogenin peptide |
| VT | variable temperature |
References
- Barron M.J., Brookes S.J., Kirkham J., Shore R.C., Hunt C., Mironov A., Kingswell N.J., Maycock J., Shuttleworth C.A., Dixon M.J. A mutation in the mouse Amelx tri-tyrosyl domain results in impaired secretion of amelogenin and phenocopies human X-linked amelogenesis imperfecta. Hum Mol Genet. 2010;19:1230–1247. doi: 10.1093/hmg/ddq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier P.M., Sauk J.J., Rosenbloom J., Yuan Z.A., Gibson C.W. An amelogenin gene defect associated with human X-linked amelogenesis imperfecta. Arch Oral Biol. 1997;42:235–242. doi: 10.1016/s0003-9969(96)00099-4. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Sarte J. Moradianoldak. P.E. Mass-spectrographic analysis of a porcine amelogenin identifies a single phosphorylated locus. Calcif Tissue Int. 1994;55:398–400. doi: 10.1007/BF00299322. [DOI] [PubMed] [Google Scholar]
- Lakshminarayanan R., Bromley K.M., Lei Y.P., Snead M.L., Moradian-Oldak J. Perturbed amelogenin secondary structure leads to uncontrolled aggregation in amelogenesis imperfecta mutant proteins. J Biol Chem. 2010;285:40593–40603. doi: 10.1074/jbc.M110.131136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lench N.J., Winter G.B. Characterization Of molecular defects in X-Linked amelogenesis imperfecta (Aih1) Hum Mutat. 1995;5:251–259. doi: 10.1002/humu.1380050310. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J. Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol. 2001;20:293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Paine M.L., Lei Y.P., Fincham A.G., Snead M.L. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J Struct Biol. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- Paine M. L., Lei Y.P., Dickerson K., Snead M.L. Altered amelogenin self-assembly based on mutations observed in human X-linked amelogenesis imperfecta (AIH1) J Biol Chem. 2002;277:17112–17116. doi: 10.1074/jbc.M110473200. [DOI] [PubMed] [Google Scholar]
- Paine M.L., Snead M.L. Protein interactions during assembly of the enamel organic extracellular matrix. J Bone Miner Res. 1997;12:221–227. doi: 10.1359/jbmr.1997.12.2.221. [DOI] [PubMed] [Google Scholar]
- Simmer J.P., Fincham A.G. Molecular mechanisms of dental enamel formation. Crit Rev Oral Biol Med. 1995;6:84–108. doi: 10.1177/10454411950060020701. [DOI] [PubMed] [Google Scholar]
- Tanimoto K., Le T., Zhu L., Witkowska H.E., Robinson S., Hall S., Hwang P., DenBesten P., Li W. Reduced amelogenin-MMP20 interactions in amelogenesis imperfecta. J Dent Res. 2008;87:451–455. doi: 10.1177/154405910808700516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- Wright J.T. The molecular etiologies and associated phenotypes of amelogenesis imperfecta. Am J Med Genet A. 2006;140A:2547–2555. doi: 10.1002/ajmg.a.31358. [DOI] [PMC free article] [PubMed] [Google Scholar]