Abstract
Ubiquitination is a widely-studied regulatory modification involved in protein degradation, DNA damage repair and the immune response. Conjugation of ubiquitin to a substrate lysine occurs in an enzymatic cascade involving an E1 ubiquitin activating enzyme, and E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase. Assays for ubiquitin conjugation include electrophoretic mobility shift assays and detection of epitope-tagged or radiolabeled ubiquitin, which are difficult to quantitate accurately and are not amenable to high throughput screening. We have developed a colorimetric assay that quantifies ubiquitin conjugation by monitoring pyrophosphate released in the first enzymatic step in ubiquitin transfer, the ATP-dependent charging of the E1 enzyme. The assay is rapid, does not rely on radioactive labeling, and requires only a spectrophotometer for detection of pyrophosphate formation. We show that pyrophosphate production by E1 is dependent on ubiquitin transfer and describe how to optimize assay conditions to measure E1, E2, or E3 activity. The kinetics of polyubiquitin chain formation by Ubc13-Mms2 measured by this assay are similar to those determined by gel assays, indicating that the data produced by this method are comparable to methods that measure ubiquitin transfer directly. This assay is adaptable to high-throughput screening of ubiquitin and ubiquitin-like conjugating enzymes.
Keywords: Ubiquitin Conjugation, Ubiquitin-like proteins, Ubc13-Mms2, ubiquitin-activating enzyme
Introduction
Protein ubiquitination is a reversible post-translational modification that regulates diverse pathways in eukaryotic cells by controlling protein stability, activity, binding and intracellular targeting [1; 2]. Biological processes that are regulated by ubiquitination include proteasomal protein degradation, DNA damage signaling, the inflammatory response and transcription [2; 3; 4]. Ubiquitin, and ubiquitin-like modifiers (Ubl2) such as SUMO and NEDD8, are small proteins that are attached to substrates via an isopeptide linkage between the C-terminus of ubiquitin (or the Ubl) and a substrate lysine [1; 2]. Substrates can be modified by a single ubiquitin or by a polyubiquitin chain, in which one ubiquitin is covalently linked to the next via one of its seven surface lysines [5; 6; 7]. Ubiquitin and Ubls are conjugated to substrate lysines by the sequential activity of three enzymes: an E1 activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ligase [2; 8]. The human genome encodes several E1 enzymes, 35 E2 enzymes, and more than 500 E3 enzymes [3; 4; 9; 10]. Our understanding of which E2 and E3 enzymes work in concert to conjugate ubiquitin to specific substrates is incomplete and remains an active area of research [4; 9; 11].
The first step in the ubiquitination cascade is the ATP-dependent charging of the E1 activating enzyme (Figure 1). The E1 uses ATP to adenylate the C-terminus of ubiquitin, forming ubiquitin-AMP and pyrophosphate. The ubiquitin-AMP ester bond is then cleaved by an active site cysteine within E1, resulting in a thioester bond between the C-terminus of ubiquitin and the sulfhydryl of cysteine [12; 13; 14]. Ubiquitin is then transferred from the active site cysteine of the E1 to an active site cysteine in the E2 [2; 4]. The E3 ligase then binds to both the E2 and the substrate, promoting isopeptide bond formation with a substrate lysine [3]. Members of the HECT family of E3 ligases first transfer the ubiquitin from the E2 to an active site cysteine in the E3 before conjugating the ubiquitin to the substrate, while the RING domain E3 ligases simply target E2 to the substrate [3; 8]. Substrate specificity is thought to be intrinsic to the specific combination of E2 and E3, although the number of functional combinations of E2s and E3s is not known. Studies of the mechanism and specificity of ubiquitin and Ubl transfer enzymes are important for understanding ubiquitin and Ubl signaling in the cell, and for identifying targets for therapeutic intervention.
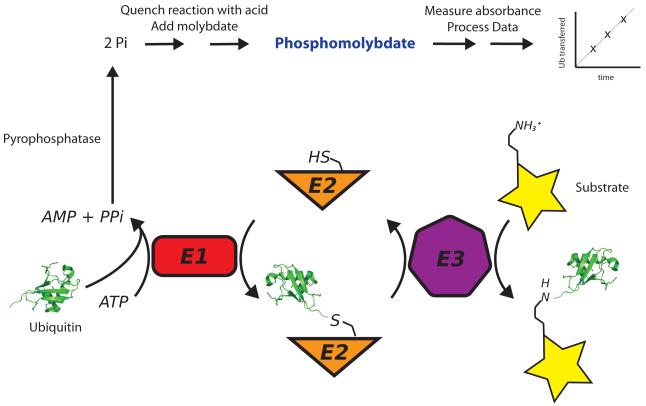
Figure 1. Schematic of Molybdenum Blue Assay.
The ubiquitination cascade (bottom) involves the sequential action of E1, E2 and E3 enzymes, leading to ubiquitination of the substrate lysine. In the first step, the ubiquitin activating enzyme, E1, adenylates ubiquitin, forming ubiquitin-AMP and releasing pyrophosphate. To assay ATP consumption (top), the pyrophosphate product is broken down by pyrophosphatase into two phosphate molecules. Addition of ascorbic acid and ammonium molybdate in hydrochloric acid halts the reaction progress and initiates phosphomolybdate formation. The solution is then developed by addition citric acid and sodium arsenite and the phosphomolybdate formed measured by visible spectroscopy.
The last ten years have seen growing interest in the ubiquitin conjugation machinery and in the proteasome as targets for drug discovery. The efficacy of the proteasome inhibitor, Bortezomib, in treating multiple myeloma and mantle lymphoma has spurred efforts to target upstream events in the ubiquitin pathway by inhibiting E1, E2 and E3 enzymes [15]. Nutlin, an inhibitor of the RING E3, Mdm2, appears to be effective in the treatment of multiple myeloma [16]. Recently, an inhibitor of the E1 activating enzyme for the ubiquitin-like molecule, Nedd8, entered Phase I trials for the treatment of cancer [9]. These two examples offer the promise of success in targeting the ubiquitin conjugation pathway by therapeutics. However, progress in developing and screening small molecule inhibitors of E1, E2 and E3 has been limited by the lack of a simple, high throughput assay for ubiquitination using native substrates [16].
The most widely used activity assay for ubiquitin or Ubl transfer is the band shift assay, in which ubiquitinated and unmodified proteins are separated by gel electrophoresis and detected by one of a variety of methods, including staining or Western blot [17; 18; 19]. However, these assays make it difficult to quantitate protein modification accurately because of variability or non-linearity in the signal from stained or antibody-bound proteins. The attachment of epitopes or fluorescent tags to ubiquitin or substrates permits accurate quantification and enhances sensitivity; however, adding an epitope tag such as hexahistidine, glutathione-S-transferase (GST), green fluorescent protein (GFP), Myc or HA to ubiquitin or to the substrate has the potential to disrupt interactions between the substrate and the transfer enzymes [20; 21; 22]. Ubiquitin radiolabeled with 125[I] or 32[P] is also used [23; 24; 25], but requires special handling due to the hazards of working with radioactivity, particularly gamma emitters like 125[I]. In addition, radiolabeling must be performed regularly because of the short half-lives of these isotopes. None of the gel shift assays are amenable to high throughput formats, both because of the time needed to perform the assay and the variability in signal between gels. Ubiquitination and Ubl transfer assays that rely upon Förster resonance energy transfer (FRET) between GFP or fluorescently labeled ubiquitin and Ubls have also been developed for real-time, quantitative measurement of ubiquitination [22; 26; 27; 28; 29; 30]. However, the FRET assay requires attaching a fluorophore to ubiquitin or to the substrate, which may interfere with the function of ubiquitin transfer enzymes and may preclude the use of native substrates [21]. In addition to methods that directly measure the ubiquitinated product, coupled enzyme methods that monitor ATP cleavage by E1 have used radiolabeled ATP and quantitated the phosphate released [31; 32]. However, none of the above methods are amenable to large-scale or high-throughput screening.
We have developed a spectrophotometric method for measuring ubiquitin and Ubl transfer that does not require tagged substrates or gel electrophoresis and can be used to quantitate each step within the ubiquitin transfer cascade. The assay couples ubiquitin transfer by E2 and E3 to the formation by E1 of pyrophosphate, a product of the charging reaction, which is broken down to phosphate by pyrophosphatase. The inorganic phosphate is quantitated by measuring the absorbance of molybdenum blue, a reduced complex of phosphate and molybdate that absorbs visible light. All reagents required for the assay are easily obtainable and the assay only requires a spectrophotometer capable of measuring absorbance between 600 nm and 850 nm. With the use of a plate reader, the assay can be readily applied to high-throughput screening of compounds that target ubiquitin or Ubl transfer enzymes.
Materials and Methods
Chemicals and reagents
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) or Fisher Scientific (Pittsburgh, PA, US) and used without further purification. Inorganic pyrophosphatase from E. coli (I5907) was purchased from Sigma-Aldrich and resuspended in 20 mM HEPES, pH 7.5, 100 mM NaCl then aliquoted, frozen and stored at −20 °C. We note that, while pyrophosphatase from S. cerevisiae can be substituted for the bacterial enzyme, the yeast enzyme has measurable ATPase activity and produces higher background absorbance values in the molybdenum blue assay compared to the bacterial enzyme (data not shown). We therefore recommend using the bacterial enzyme.
Protein Expression and Purification
S. cerevisiae Ubc13 and Mms2 were expressed in E. coli and purified as previously described by Eddins, et al. [33]. Hexahistidine tagged, human E1 enzyme was expressed in E. coli using a pET32 vector. Cells were grown at 37°C to an O.D. of 0.3, shifted to 16°C and induced with IPTG overnight. Cells were lysed and then loaded onto a nickel affinity column and eluted with imidazole. Peak fractions were pooled, dialyzed into buffer containing 50 mM NaCl, loaded onto a mono Q column (GE Healthcare) and eluted with a 0 – 400 mM Nacl gradient. Peak fractions were pooled, concentrated and purified by size exclusion chromatography on a Superdex 200 column to 20 mM Tris, pH 7.8, 100 mM NaCl, and 7.5 mM β-mercaptoethanol (GE Healthcare). Fractions containing E1 were pooled, concentrated and stored at −80 °C until use. A gel showing the purity of recombinant E1 is shown in Supplemental Figure 1
Wild type and mutant human ubiquitin were expressed in E. coli and purified following the protocol of Pickart and Raasi [20; 34]. Ubiquitin mutants are denoted in the text with superscripts; UbK63R or UbΔG75,ΔG76, for example, indicate mutation of lysine 63 to arginine or the deletion of glycine 75 and glycine 76 from ubiquitin.
S. cerevisiae Rad5 RING domain (residues 846 - 1196) was expressed in E. coli from a pMalc2xHV plasmid containing an N-terminal hexahistidine tag followed by a tobacco etch virus (TEV) protease cleavage site. The RING domain was purified by nickel affinity chromatography then dialyzed with 1 mg of TEV protease overnight to remove the affinity tag. Dialyzed protein was then concentrated and further purified by size exclusion chromatography on a Superdex 75 column in 20 mM Tris, pH 7.5, 100mM NaCl, and 5 mM β-mercaptoethanol. Fractions containing the Rad5 RING domain were pooled, concentrated, and stored at −80 °C until use.
Solutions for Molybdenum Blue Assay
The quench and developer solutions were modified from conditions described in Gawronski and Benson [35] and McCain and Zhang [36]. The quench solution contains 8% L-ascorbic acid and 0.67% ammonium molybdate in 0.67 N HCl. This solution is made fresh from a 2:3 dilution of a solution of 12% ascorbic acid in 1 N HCl with 2% ammonium molybdate in water. The ammonium molybdate/ascorbic acid solution turns yellow upon mixing with ammonium molybdate and is stable for only a few days in the dark at 4 °C (the solution will eventually turn green or brown and signal will deteriorate significantly). The developer solution contains 2% sodium citrate-trihydrate, 2% sodium (meta)arsenite in 2% acetic acid. Acetic acid is added last. This solution is stable for about a week at room temperature.
Generating a Standard Curve
A 1 mM sodium phosphate stock solution used for standards was made in buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl. This stock is diluted to a final phosphate concentration between 0 and 200 μM in 20 mM HEPES, pH 7.5, 100 mM NaCl in a final volume of 50 μL. To each standard, 150 μL of the quench solution is added and the quenched reactions are incubated at room temperature. After 5 minutes, 150 μL of the developer solution is added and the absorbance of the sample is measured at 850 nm. The absorbance of each standard is then plotted as absorbance versus the concentration of phosphate in mM or μM. The data are then fitted to a linear equation to obtain the slope in units of abs • mM−1 or abs • μM−1. Absorbance values from ubiquitination reactions are then converted to μM phosphate using the slope of the phosphate standard curve (Eq. 1).
| (Eq.1) |
Since there are two phosphates generated from every pyrophosphate molecule, the concentration of pyrophosphate formed (usually in μM) is then divided by 2 (Eq. 2).
| (Eq. 2) |
It is important to generate the standard curve using the same instrument as that used to conduct the experiments, since instrument-to-instrument variations, measurement path length, and other factors can affect the absolute absorbance measurements.
Molybdenum Blue Assay of ubiquitin conjugation
For the assays described in this paper, reactions contained 50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM DTT, 10 mM MgCl2, 500 U inorganic pyrophosphatase, 0.05 to 0.6 μM E1 enzyme, 0.1 to 4 μM Ubc13-Mms2 enzyme, 1 μM Rad5 RING domain, 80 μM UbK63R and 0 to 1 mM UbΔG75, ΔG76 or Ubd77. Reactions were initiated by the addition of 2 mM ATP. All reactions were performed at 37 °C. At the designated time points, a 50 μL aliquot of the reaction was removed and quenched with 150 μL of the quench solution. After 5 minutes at room temperature (23 °C to 26 °C), 150 μL of the developer solution was added and the absorbance of the solution was then measured at 850 nm in a Cary 50 Bio UV-Vis spectrometer (Agilent Technologies, Santa Clara, CA) or by recording a spectrum from 500 nm to 900 nm in a POLARStar Omega plate reader (BMG LABTECH GmbH, Offenburg, Germany). If necessary, the volume of the reaction aliquot can be decreased and the ratios of the aliquot to quench and developer proportionally reduced. The absorbance at 850 nm was used to quantitate activity; however any wavelength between 600 nm and 875 nm can be used for measurement. The amount of pyrophosphate formed should be equivalent to the amount of ubiquitin transferred by E1 to E2. The quantity of pyrophosphate formed is also equivalent to the amount of ubiquitin conjugated to substrate if either E2 or E3 is the rate limiting enzyme (see Results and Discussion). Each new E2 or E3 enzyme to be assayed will require optimization of conditions (see Results and Discussion for more details).
Reaction rates in μM per minute were determined from linear fits of μM pyrophosphate formed versus time. The rate of pyrophosphate formation was then plotted as a function of ubiquitin concentration and fit to the Michaelis-Menten equation to determine kinetic constants (Eq. 3):
| (Eq. 3) |
where v is the initial velocity at a given substrate concentration, kcat is the apparent first order rate constant of the enzyme, [E]t = the total concentration of enzyme, [S] is the total concentration of substrate, KM is the concentration of substrate required to reach one-half the maximum velocity of the enzyme.
Fluorescent-Ubiquitin assays
UbiquitinK63C was labeled with fluorescein-5-maleimide (Thermo Scientific, Pittsburgh, PA) in PBS buffer for 2 hours at room temperature in the dark. Labeling reactions were dialyzed overnight into 2 L of 20 mM HEPES pH 7.5, 1 mM EDTA. Ubiquitin was further purified by cation exchange chromatography as described [20]. Fractions that contained ubiquitin were pooled and dialyzed overnight into 2 L of 20 mM HEPES pH 7.5. The concentration of ubiquitin was determined using the BCA assay kit (Thermo Scientific, Pittsburgh, PA) following manufacturer’s protocol. Labeled ubiquitin was stored at 4 °C wrapped in foil.
Band shift assays using 10 μM fluorescently labeled ubiquitin (labeled at cysteine 63) with an additional 70 μM ubiquitinK63R (total donor ubiquitin concentration = 80 μM) were performed using the assay conditions and reagents described above for the molybdenum blue assay. At designated time points, reactions were stopped by the addition of 4X NuPAGE SDS-PAGE loading buffer with dye (NP0007 Invitrogen, Carlsbad, CA). Reactions were separated by SDS-PAGE and the bands visualized on a Typhoon 9410 phosphorimager (GE Healthcare, Waukesha, WI). Band intensity was measured using Quantity One version 4.6.7 (Bio-Rad, Hercules, CA). Fluorescence intensity was converted to concentration by dividing the band intensity by the slope obtained from a calibration curve of fluorescein ubiquitin standards of known concentration on the same gel. Data were plotted and fitted as described above to determine kinetic constants.
Results and Discussion
Assay Description
We set out to develop a quantitative assay that could be used with native substrates for both general activity screening assays and mechanistic studies. Activation of ubiquitin or an Ubl by E1, the first step in the ubiquitin transfer cascade (Figure 1), involves formation of ubiquitin-AMP and pyrophosphate from ubiquitin and ATP. The ubiquitin adenylate is then cleaved by the active site cysteine of the E1, resulting a thioester bond between E1 and the C-terminus of ubiquitin and AMP. The ubiquitin is then transferred to the active site cysteine of the E2 in a transthiolation reaction (Figure 1). The E3 ligase binds to the E2~ubiquitin conjugate and promotes transfer of the ubiquitin or Ubl to the substrate lysine. The release of pyrophosphate is therefore, in principle, directly related to the amount of ubiquitin transferred because E1 must transfer ubiquitin to E2 before it can undergo another round of ATP cleavage. It is therefore possible to use the rate of pyrophosphate production by E1 as a measure of ubiquitin transfer. Pyrophosphate is measured by first hydrolyzing it with the enzyme, pyrophosphatase, to produce inorganic phosphate, which is then complexed with molybdate in the presence of ascorbic acid to produce a blue color, sometimes referred to as molybdenum blue [34; 35; 36; 37]. The relative absorbance of visible or infrared light by phosphomolybdate is proportional to the amount of phosphate generated, and hence to the number of times the E1 enzyme is charged with ubiquitin. The absorbance of phosphomolybdate compounds has been used to assay the activity of phosphatases, synthetases, and other enzymes that release inorganic phosphate [35; 36]. We therefore investigated whether this method could be adapted to measuring substrate ubiquitination by monitoring pyrophosphate release by E1.
To assay ubiquitination using the phosphomolybdate assay, all proteins are first mixed in the absence of ATP and equilibrated at 37 °C. The ubiquitination reaction is then initiated with the addition of ATP. At designated times, an aliquot of each reaction is removed and diluted into a solution of ammonium molybdate, ascorbic acid and hydrochloric acid as described in Methods. Hydrochloric acid lowers the pH of the solution, which decreases the catalytic activity of the ubiquitin conjugation enzymes and effectively quenches the reaction [38; 39]. Molybdate forms a complex with phosphate, which is reduced by ascorbic acid and gives rise to the blue color [35; 40]. Phosphomolybdate formation is then stopped after 5 minutes by the addition of a citrate/arsenite solution, which competes for free molybdate and enhances the color stability of the phosphomolybdate [41]. The phosphomolybdate formed at each time point was then quantified by measuring the absorbance at 850 nm. The absorbance is divided by two, since two phosphates are produced for every ubiquitin transferred because ATP is cleaved to AMP and pyrophosphate. These numbers are corrected for background ATP hydrolysis that is not dependent on the activity of E1 or E2. The concentration of phosphate is then determined from the corrected absorbance by comparison with the standard curve (see Methods).
We developed the assay using the enzymes that synthesize K63-linked polyubiquitin, which is synthesized in yeast by the E2, Ubc13, in complex with Mms2, a non-catalytic subunit that confers the specificity for K63 of the acceptor ubiquitin [21; 33; 42; 43]. Ubc13-Mms2 can synthesize free polyubiquitin chains in the absence of the RING E3 ligase, Rad5 [42; 44; 45]. When added to the in vitro reaction in the absence of other substrates, Rad5 stimulates the rate of free chain synthesis by Ubc13-Mms2 [45]. It is therefore possible to monitor the rate of the unstimulated E1/E2 reaction, as well as the E3-stimulated reaction.
We first determined whether the rate of ATP cleavage by E1 was increased during free chain synthesis by Ubc13-Mms2, which would confirm that ATP cleavage by E1 is coupled to transfer of ubiquitin to E2. We carried out the reactions under conditions that would lead to formation of diubiquitin only by using 80 μM of UbK63R, which can only serve as a donor ubiquitin, and saturating amounts of UbD77, which can only serve as an acceptor because D77 prevents the ubiquitin from binding to and reacting with the E1 [20]. Figure 2 shows the time course of pyrophosphate produced during diubiquitin formation by Ubc13-Mms2, which is repeatedly charged by E1 for each ubiquitin transferred. A control reaction containing the E1, ubiquitin, pyrophosphatase, and ATP, which corrects for background ATP hydrolysis that is not due to ubiquitin transfer to E2, is nearly 100-fold lower than with Ubc13-Mms2 (1.2 μM/min vs. 0.014 μM/min). We verified that pyrophosphatase was saturating by assaying the rate of pyrophosphate formation over a broad range of pyrophosphatase concentrations and found that it did not change significantly at pyrophosphatase concentrations greater than 20 Units (Supplemental Figure 2). We note that there is significant ATPase activity at pyrophosphatase concentrations greater than 500 Units (data not shown) and recommend controlling for this activity by assaying phosphate formation in the absence of E1.
Figure 2. Pyrophosphate production during diubiquitin synthesis.
The plot shows pyrophosphate formation during diubiquitin synthesis by Ubc13-Mms2 in the absence (open circles) and presence (black circles) of the RING domain of the Rad5 E3. The control (black triangles) shows the background signal in the absence of Ubc13-Mms2. Reactions contained 0.6 μM Ubc13-Mms2, 1 μM Rad5 RING domain, 0.3 μM E1, 80 μM ubiquitinK63R (the donor ubiquitin), and 800 μM ubiquitinD77 (the acceptor ubiquitin. -Data points are an average of three independent measurements and the error bars are the standard deviation of the experiments. Lower panel shows a gel of diubquitin synthesis using fluorescently labeled ubiquitin.
We next measured the rate of pyrophosphate production in the presence of a RING domain E3 ligase to show that pyrophosphate production can reflect the activity of an E3. In the presence of the RING domain of yeast Rad5, we measured a rate of 2.9 μM/min, a 3-fold stimulation in the rate of pyrophosphate production (Figure 2). This is comparable to a previously reported 4-fold stimulation in the rate of ubiquitin chain formation by Ubc13/Mms2 and intact Rad5, which was assayed using band shift assays [45]. These data indicate that measuring phosphate production is a viable assay of E2 and RING E3 activity.
Different sets of enzymes involved in ubiquitin and Ubl modification require distinct conditions for assaying activity. In order to determine whether some of these conditions might affect molybdenum blue color formation, we determined how different assay buffers, salts and additives affect assay results, thereby establishing the range of conditions under which the assay can be used. Absorbance of a phosphomolybdate standard was compared in the presence of a variety of buffers, salts, reducing agents and reaction additives such as BSA and EDTA, and the absorbance was compared over a broad range of concentrations (Supplemental Figure 3). Phosphomolybdate absorbance was unaffected by most reaction components tested except for phosphate buffers, high concentrations of DTT (>20 mM), HEPES buffer (>100 mM), and bovine serum albumin (>3 mg/mL; this also showed significant precipitation). For best results, phosphate standards should be obtained using buffers that are identical to reaction conditions.
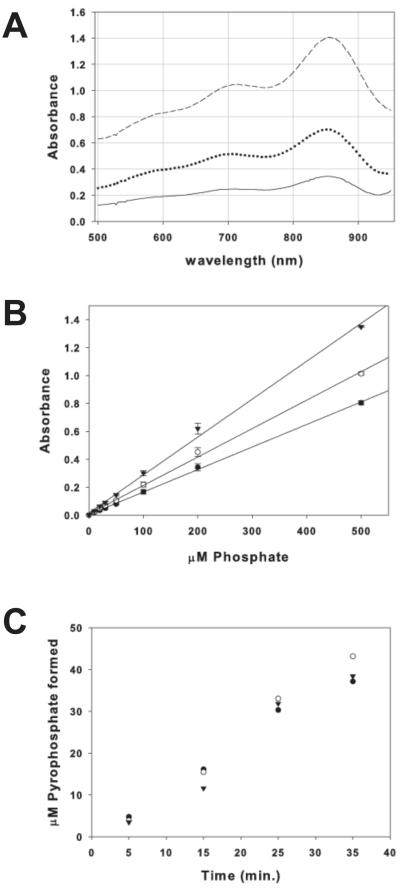
Phosphomolybdate absorbs most strongly between 600 nm and 900 nm; therefore any spectrophotometer or plate reader that can measure absorbance between these wavelengths can be used for this assay (Figure 3a). We find that the absorbance of phosphomolybdate at 600 nm, 710 nm, and 850 nm is linear and highly reproducible from 5 μM to 200 μM phosphate (Figure 3b). Phosphomolybdate absorbs most strongly around 850 nm, but any wavelength between 600 nm and 875 nm is suitable for assaying ubiquitin transfer and produces similar results (Figure 3c). The blue color is stable at all three wavelengths for greater than four hours in the presence of ubiquitin, with a ~3% loss in signal (data not shown).
Figure 3. Absorbance of phosphomolybdate.
(A) Absorbance spectrum of phosphomolybdate from 100 μM (solid line), 200 μM (dotted line), and 500 μM (dashed line) sodium phosphate in 20 mM HEPES, pH 7.5 and 100 mM NaCl. Spectra were taken of 200 μL solutions in a 96-well plate using a POLARStar Omega plate reader (B) Linearity of phosphomolybdate absorbance at 600 nm (filled circles), 710 nm (open circles), and 850 nm (filled triangles). (C) Molybdenum blue assay of time course of Ubc13-Mms2 ubiquitin chain formation measured at 600 nm (filled circles), 710 nm (filled triangles), and 850 nm (open circles).
Application of the molybdenum blue assay to high-throughput investigations of ubiquitin conjugation
The molybdenum blue assay is easily adapted to a multi-well plate format for high-throughput measurement of ubiquitin conjugation using a plate reader. Possible applications include screening for activity by different E2-E3 pairs, potential substrates or the effect of small molecule inhibitors of E1, E2, or E3 enzyme activity. The assay is particularly well suited to screening for E2-E3 pairs and ubiquitin/Ubl targets because it can be used with native enzymes and substrates.
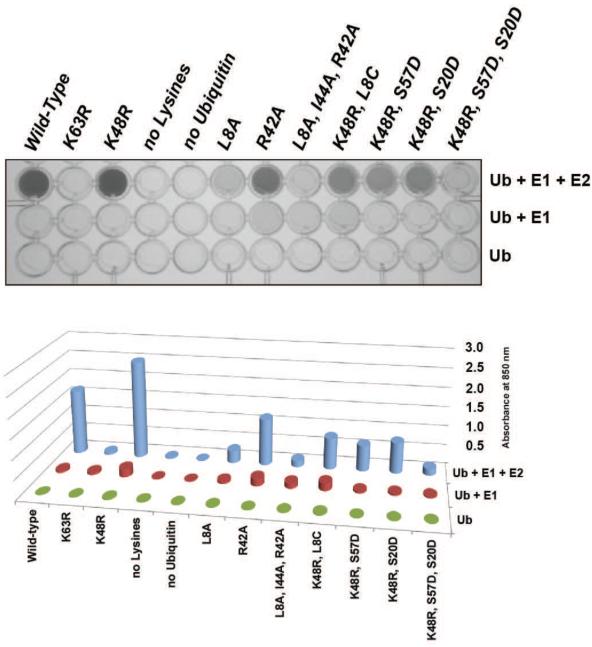
To demonstrate the application of the molybdenum blue assay to high throughput screening, we used the assay in a plate format to measure the effects of several different ubiquitin mutations on K63-linked polyubiquitin formation by Ubc13-Mms2. Assays containing 0.5 μM E1, 2 μM E2 (Ubc13-Mms2) and 100 or 500 μM of each mutant ubiquitin were incubated at 37°C for 60 minutes (Figure 4). For each mutant ubiquitin, control reactions measured pyrophosphate production by E1 plus ubiquitin and ubiquitin alone in separate wells in the plate. Reactions were quenched after 60 minutes and the absorbance was measured in a plate reader as described in Methods. The results in Figure 4 show that Ubc13-Mms2 formed ubiquitin chains with all ubiquitin mutants tested except the K63R substitution, which cannot serve as an acceptor ubiquitin for K63-linked polyubiquitin chain synthesis [21]. Substitutions at residues 8, 20, 42, 44, or 57 of ubiquitin decreased chain formation by factors ranging from 3 to 10-fold as compared to wild-type ubiquitin. Importantly, the relative effects of each ubiquitin mutation as assayed by the phosphomolybdate plate assay agree well with band shift assays of ubiquitin chain formation (Supplemental Figure 4). The effects of mutations at residues 8, 42, and 44 of ubiquitin agree with structural studies showing that these residues are important for acceptor ubiquitin binding to Mms2 [33; 46]. Residues 20 and 57 of ubiquitin are proximal to the acceptor ubiquitin binding surface in Ubc13 suggesting that these substitutions disrupt this interaction [33]. These data show that it is possible to rapidly screen the effects of ubiquitin mutations on ubiquitin transfer using the molybdenum blue assay.
Figure 4. High throughput screen for the effect of ubiquitin mutations on polyubiquitin chain formation.
The effect of ubiquitin mutations on polyubiquitin chain formation by Ubc13-Mms2. Wells in row 1 contain E1, Ubc13-Mms2 and ubiquitin, row 2 contains E1 and ubiquitin, and row 3 contains ubiquitin alone. Protein concentrations used were 2 μM Ubc13-Mms2, 0.5 μM E1, 500 μM ubiquitin (except for no-lysine ubiquitin which was at 100 μM). Reactions were incubated for 1 hour at 37 °C, quenched and the absorbance measured. Assay was performed in duplicate, with representative data shown.
Establishment of the rate-limiting enzyme
For quantitative investigations using the molybdenum blue assay, it is essential to carry out the assay under conditions in which the enzyme of interest (E1, E2 or E3) is rate-limiting. Under these conditions, the rate of pyrophosphate formation will reflect the rate-limiting step in the enzymatic cascade. To address whether isopeptide bond formation by Ubc13-Mms2 was the rate-limiting step in our assays, we determined how the rate of pyrophosphate formation changed with increasing concentration of E1 and E2. Reaction rates are dependent both on the concentration of substrate and the concentration of enzyme [47]. Therefore, if E2 is rate limiting, increasing the concentration of E2 two-fold will increase the rate of pyrophosphate formation by two-fold as well. A smaller change would indicate that E1 or, in the molybdenum blue assay, pyrophosphatase is rate limiting. Since the rates of catalysis will likely be different for each combination of E1, E2, and E3 enzymes, it is therefore important to determine the concentration range where the activity of the enzyme of interest is rate-limiting if the molybdenum blue assay is to be used to determine kinetic constants. Once the concentrations at which E2 is the rate-limiting step have been established, the steady-state kinetics of ubiquitin transfer can be measured with the confidence that the measured rates of phosphate production correlate with isopeptide bond formation catalyzed by E2. To maximize signal-to-noise and, hence, accuracy, assays should utilize the maximum enzyme concentration that is still rate-limiting.
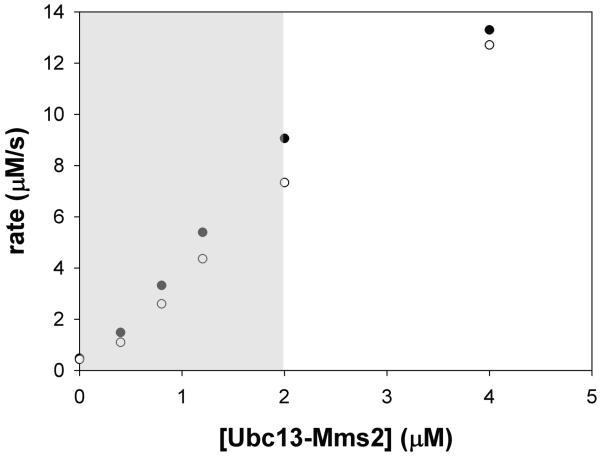
As an example of how the rate-limiting concentration of E2 is determined, we measured the rate of pyrophosphate formation at concentrations of E2 ranging from 0 to 4 μM and at two different concentrations of E1. Figure 5 shows a plot of the rate of pyrophosphate formation as a function of Ubc13/Mms2 concentration. The rate of pyrophosphate formation varies linearly with E2 concentration between 0 and 2 μM Ubc13-Mms2 at both 0.3 μM and 0.6 μM E1 (shown by the gray box in Figure 5), indicating that ubiquitin conjugation by Ubc13-Mms2 is the rate-limiting step under these conditions. At concentrations of Ubc13-Mms2 greater than 2 μM, however, this relationship no longer strictly holds, suggesting that the E2 is no longer entirely rate limiting.
Figure 5. Rate-limiting concentration range of Ubc13-Mms2.
Rate of pyrophosphate formation in μM/sec at 0, 0.4, 0.8, 1.2, 2, and 4 μM Ubc13-Mms2 at 0.3 μM E1 (filled circles) and 0.6 μM E1 (open circles). Grey box indicates concentrations where ubiquitin transfer by Ubc13-Mms2 is directly correlated with pyrophosphate production by E1.
Steady-State Kinetics of Diubiquitin Formation by Ubc13-Mms2
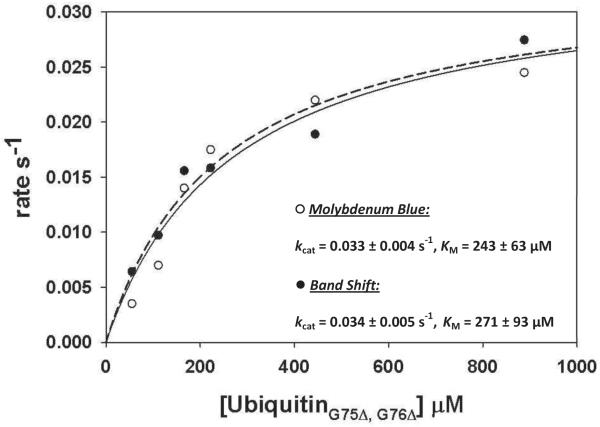
In order to compare the results of the molybdenum blue assay with a conventional ubiquitin conjugation assay, we compared the steady-state kinetics of diubiquitin formation by Ubc13-Mms2 as measured using a band shift assay and the molybdenum blue assay. Formation of diubiquitin only was ensured by utilizing a donor ubiquitin with a K63R substitution, which blocks chain formation by Ubc13-Mms2, and an acceptor ubiquitin lacking the two C-terminal glycine residues, which cannot be activated by E1. The rate of diubiquitin formation was assayed by the two methods at several concentrations of the acceptor ubiquitin. In the molybdenum blue assay, absorbance measurements were corrected for background ATP hydrolysis by subtracting from each measurement the absorbance in a matched reaction lacking E2. Data were plotted as the rate (in s−1) versus concentration of ubiquitin and fit to the Michaelis-Menten equation to calculate kinetic constants. The saturation plot shown in Figure 6 displays the curves from the molybdenum blue and gel assays along with the calculated rate constants. The turnover number for E2 and Michaelis constant determined from the molybdenum blue (kcat = 0.033 ± 0.004 s−1 and KM = 243 ± 63 μM, R2 = 0.95) and gel shift assay (kcat = 0.034 ± 0.005 s−1 and KM = 271 ± 93 μM, R2 = 0.94) are in very good agreement. These values are comparable to those previously reported by Carlile, et al. (kcat = 0.02 s−1 and KM = 212 μM) using a gel assay with 125[I]-ubiquitin [45]. Taken together, our results indicate that kinetic data obtained using the molybdenum blue assay are comparable to kinetic data obtained by other methods.
Figure 6. UbiquitinΔG75,ΔG76 saturation curves for Ubc13-Mms2.
Reactions contained 0.3 μM Ubc13-Mms2, 0.2 μM E1, 0 to 950 μM ubiquitinΔG75,ΔG76, and 80 μM ubiquitin K63R. For molybdenum blue assay reactions (black circles), 500 Units of pyrophosphatase was present and for the band shift assay (open circles), 10 μM ubiquitin labeled with fluorescein at position 63 and with 70 μM unlabeled ubiquitinK63R. Each rate is an average of 3 to 5 separate experiments. Error bars are not shown for clarity.
Assay Controls and Caveats
The key to successfully measuring ubiquitin conjugation in a molybdenum blue assay is to control for background ATPase activity that could artificially inflate or deflate the apparent rate of ubiquitin transfer. While we have explained the importance of determining the rate-limiting enzyme when using the molybdenum blue assay, good laboratory practice and additional controls may be necessary to increase accuracy. The background level of phosphate released can be affected by the purity of the enzymes, substrates, and reagents used in the assay. In general, a background value or rate reflecting ATP cleavage by solvent, pyrophosphatase and E1 (if E1 is not the enzyme under investigation) should be subtracted from all measurements. This value can be from a single reaction which measures the combined enzymatic and non-enzymatic ATP cleavage rates or from separate reactions independently measuring the rate of ATP cleavage by solvent, pyrophosphatase, and E1. Fresh ATP stocks should be used to reduce the background phosphate signal that is due to the slow hydrolysis of ATP during prolonged storage. Signs of ATP degradation will initially appear as elevated background and can lead to lowered rates because E1 is inhibited by AMP [12]. We typically make a large 100 mM stock of ATP, which is aliquoted and stored at −20 °C and discarded after six months. For highly quantitative measurements, the E1, E2 and E3 enzymes and ubiquitin should be >95% pure to reduce the likelihood of contaminating ATPase activity. The background rate of phosphate formation in the absence of substrate due to a contaminate can be subtracted from the overall rate of phosphate formation or this activity can be removed through further purification. For a more qualitative activity screen, for example in high-throughput screens of different E2-E3 pairs, it is possible to use partially purified proteins provided that the appropriate controls for the ATPase activity of the contaminates are performed.
The molybdenum blue assay correlates pyrophosphate production with total ubiquitin transferred. Therefore, any transfer of ubiquitin, including auto-modification of E3 enzymes, modification of multiple sites within the same substrate and ubiquitin chain formation, will be counted in the final absorbance measurement. If measurement of heterogeneous modifications is not desired, it is important to establish the order of modification (i.e. substrate > E3 > chains) and the timescale in which the desired modification occurs. In the simplest case, in which the substrate is the first protein modified, the assay length can be kept sufficiently short such that nearly all the ubiquitin conjugation events involve the product of interest. In cases where the substrate is not preferentially modified first or where mixed modification is unavoidable, we recommend combining the molybdenum blue assay with a band shift assay. Aliquots of the reaction should be taken before initiation and after the last time point for separation by SDS-PAGE. The gel should be stained for total protein or blotted for the ubiquitin or Ubl to identify the modified proteins. For each protein, the percent of the product of interest is determined and used to partition the total rate of ubiquitin transfer measured in the molybdenum blue assay into the component rates of modification.
In addition to background formation of ATP, any reagents added to the reaction must not act by altering the activity of pyrophosphatase or affecting the chemical reaction to form phosphomolybdate. This is especially important when screening chemical libraries for inhibitors or activators of ubiquitin conjugation. Since this interference with the assay is not a likely event, follow-up controls can be carried out only on compounds that show an effect in the initial high-throughput screen.
Conclusions
We have developed a straightforward, colorimetric assay for ubiquitin conjugation that can be used in kinetic studies as well as in high-throughput screens for the effects of mutations, small molecules, different E2-E3 pairs and the presence of different potential substrates. The assay couples release of pyrophosphate by E1 to the transfer of ubiquitin by E2 and/or E3. The pyrophosphate is quantitated by breaking it down with pyrophosphatase into phosphate, which is combined with molybdate to form a blue phosphomolybdate complex, sometimes referred to as molybdenum blue. The assay is rapid, simple, safe and as inexpensive to perform as gel shift assays, while as quantitative as radioactive and FRET assays that have been previously used to investigate ubiquitin conjugation [22; 23; 26; 27; 28; 29; 45; 48]. Additional advantages of the assay are the ability to use native, untagged ubiquitin as well as native substrates. Since ATP cleavage by an E1 is universal for activation of ubiquitin and ubiquitin-like proteins such as SUMO and NEDD8, the molybdenum blue assay is applicable to these systems, as well. This assay will open the door to in-depth screening of therapeutics targeting ubiquitin conjugation and mechanistic studies of E2 and E3 enzymes.
Supplementary Material
Acknowledgements
We thank Dr. Reuven Wiener for the generous gift of the mutant ubiquitin proteins and the RING domain of Rad5. We also thank Dr. Tao Wang for additional ubiquitin mutants and Anthony DiBello for purifying human E1.
Footnotes
This work was supported by the Howard Hughes Medical Institute. C.E.B. was supported by a Ruth Kirchstein Fellowship from the National Institute of General Medical Science (F32GM089037).
Abbreviations used: Ub, ubiquitin; Ubl, ubiquitin-like protein; FRET, Forster Resonance Energy Transfer; DTT, Dithiothreitol; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide electrophoresis; ATP, Adenosine Triphosphate; AMP, adenosine monophosphate; BSA, Bovine serum albumen; EDTA, ethylenediaminetetraacetic acid; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–9. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- [3].Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- [4].Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2010;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- [6].Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- [7].Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007;17:726–35. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–31. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- [12].Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem. 1982;257:10329–37. [PubMed] [Google Scholar]
- [13].Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982;257:2543–8. [PubMed] [Google Scholar]
- [14].Haas AL, Warms JV, Rose IA. Ubiquitin adenylate: structure and role in ubiquitin activation. Biochemistry. 1983;22:4388–94. doi: 10.1021/bi00288a007. [DOI] [PubMed] [Google Scholar]
- [15].Petroski MD. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008;9(Suppl 1):S7. doi: 10.1186/1471-2091-9-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Eldridge AG, O’Brien T. Therapeutic strategies within the ubiquitin proteasome system. Cell Death Differ. 2010;17:4–13. doi: 10.1038/cdd.2009.82. [DOI] [PubMed] [Google Scholar]
- [17].Yunus AA, Lima CD. Purification and activity assays for Ubc9, the ubiquitin-conjugating enzyme for the small ubiquitin-like modifier SUMO. Methods Enzymol. 2005;398:74–87. doi: 10.1016/S0076-6879(05)98008-7. [DOI] [PubMed] [Google Scholar]
- [18].Yunus AA, Lima CD. Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods Mol Biol. 2009;497:167–86. doi: 10.1007/978-1-59745-566-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hershko A, Eytan E, Ciechanover A, Haas AL. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982;257:13964–70. [PubMed] [Google Scholar]
- [20].Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- [21].Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–43. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- [22].Boisclair MD, McClure C, Josiah S, Glass S, Bottomley S, Kamerkar S, Hemmila I. Development of a ubiquitin transfer assay for high throughput screening by fluorescence resonance energy transfer. J Biomol Screen. 2000;5:319–28. doi: 10.1177/108705710000500503. [DOI] [PubMed] [Google Scholar]
- [23].Haas AL. Purification of E1 and E1-like enzymes. Methods Mol Biol. 2005;301:23–35. doi: 10.1385/1-59259-895-1:023. [DOI] [PubMed] [Google Scholar]
- [24].Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol Cell. 2010;39:784–96. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I [kappa] B [alpha] Molecular Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- [26].Gururaja TL, Pray TR, Lowe R, Dong G, Huang J, Daniel-Issakani S, Payan DG. A homogeneous FRET assay system for multiubiquitin chain assembly and disassembly. Methods Enzymol. 2005;399:663–82. doi: 10.1016/S0076-6879(05)99044-7. [DOI] [PubMed] [Google Scholar]
- [27].Martin SF, Tatham MH, Hay RT, Samuel ID. Quantitative analysis of multi-protein interactions using FRET: application to the SUMO pathway. Protein Sci. 2008;17:777–84. doi: 10.1110/ps.073369608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kao MW, Yang LL, Lin JC, Lim TS, Fann W, Chen RP. Strategy for efficient site-specific FRET-dye labeling of ubiquitin. Bioconjug Chem. 2008;19:1124–6. doi: 10.1021/bc700480j. [DOI] [PubMed] [Google Scholar]
- [29].Carlson CB, Horton RA, Vogel KW. A toolbox approach to high-throughput TR-FRET-based SUMOylation and DeSUMOylation assays. Assay Drug Dev Technol. 2009;7:348–55. doi: 10.1089/adt.2008.0188. [DOI] [PubMed] [Google Scholar]
- [30].Wee KE, Lai Z, Auger KR, Ma J, Horiuchi KY, Dowling RL, Dougherty CS, Corman JI, Wynn R, Copeland RA. Steady-state kinetic analysis of human ubiquitin-activating enzyme (E1) using a fluorescently labeled ubiquitin substrate. J Protein Chem. 2000;19:489–98. doi: 10.1023/a:1026501515450. [DOI] [PubMed] [Google Scholar]
- [31].Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. Prog Clin Biol Res. 1985;180:215. [PubMed] [Google Scholar]
- [32].Rose IA, Warms JV. A specific endpoint assay for ubiquitin. Proc Natl Acad Sci U S A. 1987;84:1477–81. doi: 10.1073/pnas.84.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–20. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- [34].Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–24. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gawronski JD, Benson DR. Microtiter assay for glutamine synthetase biosynthetic activity using inorganic phosphate detection. Anal Biochem. 2004;327:114–8. doi: 10.1016/j.ab.2003.12.024. [DOI] [PubMed] [Google Scholar]
- [36].McCain DF, Zhang ZY. Assays for protein-tyrosine phosphatases. Methods Enzymol. 2002;345:507–18. doi: 10.1016/s0076-6879(02)45042-2. [DOI] [PubMed] [Google Scholar]
- [37].Heinonen JK, Lahti RJ. A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphatase. Anal Biochem. 1981;113:313–7. doi: 10.1016/0003-2697(81)90082-8. [DOI] [PubMed] [Google Scholar]
- [38].Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J Mol Biol. 2007;369:608–18. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2010;39:548–59. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lowry OH, Lopez JA. The determination of inorganic phosphate in the presence of labile phosphate esters. J Biol Chem. 1946;162:421–8. [PubMed] [Google Scholar]
- [41].Baginski ES, Foa PP, Zak B. Determination of Phosphate - Study of Labile Organic Phosphate Interference. Clinica Chimica Acta. 1967;15:155. &. [Google Scholar]
- [42].Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–53. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- [43].Tsui C, Raguraj A, Pickart CM. Ubiquitin binding site of the ubiquitin E2 variant (UEV) protein Mms2 is required for DNA damage tolerance in the yeast RAD6 pathway. J Biol Chem. 2005;280:19829–35. doi: 10.1074/jbc.M414060200. [DOI] [PubMed] [Google Scholar]
- [44].Petroski MD, Zhou X, Dong G, Daniel-Issakani S, Payan DG, Huang J. Substrate modification with lysine 63-linked ubiquitin chains through the UBC13-UEV1A ubiquitin-conjugating enzyme. J Biol Chem. 2007;282:29936–45. doi: 10.1074/jbc.M703911200. [DOI] [PubMed] [Google Scholar]
- [45].Carlile CM, Pickart CM, Matunis MJ, Cohen RE. Synthesis of free and proliferating cell nuclear antigen-bound polyubiquitin chains by the RING E3 ubiquitin ligase Rad5. J Biol Chem. 2009;284:29326–34. doi: 10.1074/jbc.M109.043885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lewis MJ, Saltibus LF, Hau DD, Xiao W, Spyracopoulos L. Structural basis for non-covalent interaction between ubiquitin and the ubiquitin conjugating enzyme variant human MMS2. J Biomol NMR. 2006;34:89–100. doi: 10.1007/s10858-005-5583-6. [DOI] [PubMed] [Google Scholar]
- [47].Fersht A. Structure and mechanism in protein science : a guide to enzyme catalysis and protein folding. W.H. Freeman; New York: 1999. [Google Scholar]
- [48].Kim YP, Jin Z, Kim E, Park S, Oh YH, Kim HS. Analysis of in vitro SUMOylation using bioluminescence resonance energy transfer (BRET) Biochem Biophys Res Commun. 2009;382:530–4. doi: 10.1016/j.bbrc.2009.03.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.