Abstract
The ER-mitochondrial junction provides a local calcium signaling domain that is critical for both matching energy production with demand and the control of apoptosis. Here, we visualize ER-mitochondrial contact sites and monitor the localized [Ca2+] changes ([Ca2+]ER-mt) using drug-inducible fluorescent interorganelle linkers. We show that all mitochondria have contacts with the ER but plasma membrane-mitochondrial contacts are less frequent because of interleaving ER stacks in both RBL-2H3 and H9c2 cells. Single mitochondria display discrete patches of ER contacts and show heterogeneity in the ER-mitochondrial Ca2+ transfer. Pericam-tagged linkers revealed IP3-induced [Ca2+]ER-mt signals that exceeded 9μM and endured buffering bulk cytoplasmic [Ca2+] increases. Altering linker length to modify the space available for the Ca2+ transfer machinery had a biphasic effect on [Ca2+]ER-mt signals. These studies provide direct evidence for the existence of high Ca2+ microdomains between the ER and mitochondria, and suggest an optimal gap width for efficient Ca2+ transfer.
Keywords: calcium signaling, mitochondria, ER, physical coupling, local Ca2+ regulation
Introduction
ER and mitochondria form junctions supported by sturdy protein tethers (Csordas et al., 2006; Mannella et al., 1998; Shore and Tata, 1977). The tethers show various lengths and seem to include the Mmm1/Mdm10/Mdm12/Mdm34 complex in yeast (Kornmann et al., 2009) and IP3 receptor (IP3R)-containing complexes (Szabadkai et al., 2006), Mfn2 (de Brito and Scorrano, 2008), and several other proteins (Csordas et al., 2006; Merkwirth and Langer, 2008) in mammalian cells. The ER-mitochondrial local coupling is essential for survival in yeast (Kornmann et al., 2009) and is required for many aspects of cell function in mammalian cells, including lipid biosynthesis, protein trafficking and calcium signaling (Goetz and Nabi, 2006; Hayashi et al., 2009; Vance, 2003; Voelker, 2009; Wiedemann et al., 2009). Regarding calcium, the junctions provide a site where the IP3R/RyR-mediated calcium-oscillations are propagated locally to the mitochondria to control energy metabolism, mitochondrial apoptosis and to exert feedback effects on cytoplasmic calcium signaling (Breckenridge et al., 2003; Hajnoczky et al., 1999; Hajnoczky et al., 1995; Jouaville et al., 1995; Jouaville et al., 1999; Pinton et al., 2001; Szalai et al., 1999). Indirect evidence supports that at the ER-mitochondrial junctions, the mitochondrial membrane can be exposed to a high-calcium microdomain generated by the open IP3Rs or ryanodine receptors (RyRs). This microdomain would attain activation of the low-affinity mitochondrial Ca2+ uniporter (Csordas et al., 1999; Rizzuto et al., 1993; Rizzuto et al., 1998; Rizzuto and Pozzan, 2006; Szalai et al., 2000). Despite the broad significance of the ER-mitochondrial junctions and the emerging research efforts to further explore them, several fundamental aspects of the coupling remain elusive. Localization and dynamic monitoring of the ER-mitochondrial contacts in live cells have been prevented by the lack of a specific marker. Visualization and quantitation of the [Ca2+]ER-mt changes were also precluded by the lack of an ER-mitochondrial interface targeted probe. Here we describe a genetic approach to address these problems and provide direct evaluation of the [Ca2+]ER-mt microdomain and the underlying ER-mitochondrial structure in live cells.
Results
Drug-inducible ER-mitochondrial and PM-mitochondrial linkers
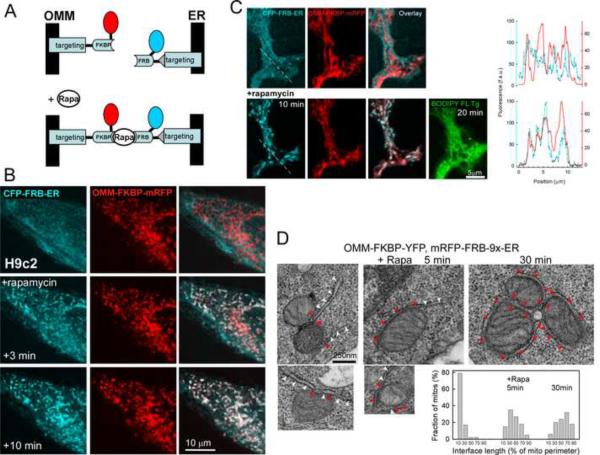
To study the structure and function of ER-mitochondrial associations, we and others have generated genetically encoded bifunctional linkers that upon expression resulted in both tightening of the interorganellar contacts and expansion of the interface area (Csordas et al., 2006; Kornmann et al., 2009). These constitutive linkers consisted of ER and outer mitochondrial membrane (OMM) targeting sequences (targeting sequences of Sac1 or yUBC6 and mAKAP1, respectively) connected through a fluorescent protein. To adapt this approach for labeling and functional characterization of the preexisting, physiological ER-mitochondrial interface, we coupled the OMM and ER targeting sequences with the two components of the FKBP-FRB heterodimerization system (Inoue et al., 2005), respectively (Fig1A). Addition of rapamycin causes heterodimerization between adjacent FKBP and FRB domains to rapidly connect the ER-and OMM targeted anchors (Fig1A). Thus, induction of the bridge formation would be initially confined to the areas where the ER and OMM were naturally close. These areas include the contacts where ER and mitochondria are coupled by direct physical links and some random associations. Rapamycin is applied only for a short time (in most cases ≤10min) to minimize the broad effects initiated by its endogenous receptors.
Fig 1. Inducible tethering of the OMM to the ER.
(A) Scheme illustrating the rapamycin-inducible bridge-forming modules. OMM targeting is established via the mAKAP1(34–63) presequence that is fused to an FKBP12 protein tagged with mRFP1. For ER-targeting, the C-terminal localization sequence (521–587) of the human Sac1 phosphatase is added to the C-terminus of an FRB fragment that is also tagged with CFP (Varnai and Balla, 2007). A covalent linkage between the FKBP and FRB domains is established via rapamycin that is administered to the linker-expressing cells.
(B) Confocal images of an individual H9c2 cell expressing OMM-ER linker pairs show the distribution of the respective fluorophores before and after (3 and 10 min) rapamycin (100nM) treatment. Note the rapamycin-induced marked conglomeration of CFP fluorescence to the mitochondria appearing in white on the cyan/red overlay images.
(C) High-resolution confocal images of a projection of an RBL-2H3 cell overexpressing CFP-labeled ER-targeted and mRFP-labeled OMM-targeted inducible linker modules. Effect of 10 min rapamycin is shown. To further illustrate the redistribution of the ER-targeted module to the mitochondria, the corresponding line profiles of the cyan and red fluorescence (along the dashed line in the first images on the left) are drawn on the right. As a reference, the ER is also visualized using BODIPY FL Tg applied ~20 min after rapamycin addition. BODIPY FL Tg was added in the end of the rapamycin time course to avoid any bleedthrough from fluorescein to the CFP channel and to avoid SERCA pump inhibition.
(D) TEM imaging of RBL-2H3 cells overexpressing OMM-ER linker pairs and incubated without (left) or with rapamycin (100nM) for 5 min (middle) and 30 min (right), respectively. Images are representative of 12, 5 and 5 cells, and a total of 91, 63 and 70 mitochondria for control, 5 min and 30 min treatment conditions, respectively. Bar charts: The length of close associations (interface length) was calculated for each mitochondrion as % of the perimeter of the mitochondrial cross-section. The interface length values were binned by 20 % (x axis: 10 corresponds to 0–20%, 30 corresponds to 20–40% and so on) and their distribution is shown for each condition.
For information on localization and lateral mobility of the linkers see also FigS1.
Visualization of ER-mitochondrial contacts in live cells
H9c2 and RBL-2H3 cells, expressing CFP-FRB-ER and OMM-FKBP-RFP showed respective ER and mitochondrial fluorescence distribution (Fig1B and C upper). The organellar distribution of the respective constructs was confirmed by double labeling with ER and mitochondrial markers (ER Tracker, ER lumen targeted FP and TMRE and mitochondrial matrix targeted FP, respectively (FigS1A and not shown)). Addition of rapamycin (100 nM) led to rapid redistribution of the majority of the CFP fluorescence to the mitochondria (Fig1B middle and lower). Effectively, every mitochondrion showed enrichment in CFP fluorescence in 3 min and further accumulation by 10 min (Fig1B). The rapamycin-induced CFP redistribution was also resolved in 3D in RBL-2H3 cells (FigS1B). In some experiments, the ER was labeled with an independent marker, BODIPY FL thapsigargin (Tg) after rapamycin treatment (Fig1C). BODIPY FL Tg showed broad ER distribution, similar to that of CFP-FRB-ER prior to rapamycin treatment, indicating that the FRB redistribution within the ER toward the mitochondria occurred without a change in the gross ER structure. Further along this line, fluorescence photobleaching-recovery studies have shown great mobility of the FRB-ER in the ER network and the OMM-FKBP within individual mitochondria (FigS1C). Thus, rapamycin induces binding of CFP-FRB-ER with the OMM-FKBP12-mRFP on the surface of adjacent mitochondria where the two organelles are juxtaposed and lateral movement of the constructs in their respective membranes allows rapid accumulation of CFP and mRFP in the interface areas.
To evaluate the effect of CFP-FRB-ER and OMM-FKBP-RFP and their rapamycin-induced linkage on the ER-mitochondrial interface transmission electron microscopy (TEM) studies were performed in RBL-2H3 cells (Fig1D). In these cells, we have previously quantitated the effect of a constitutive ER-OMM linker on the ultrastructure (Csordas et al., 2006). Before rapamycin addition, the ER-mitochondrial associations involved <20% of the total mitochondrial surface (Fig1D left), a value similar to that described in cells expressing no linker construct (Csordas et al., 2006). Five min treatment with rapamycin resulted in some increase in the contact area for the majority of the mitochondria (Fig1D middle), whereas 30 min treatment led to massive expansion of the associations (Fig1D right) similar to what has been documented previously in the constitutive ER-OMM linker expressing cells (Csordas et al., 2006). Importantly, colocalization of CFP and mRFP fluorescence occurred before expansion of the interface (Fig1B 3min and Fig3CD vs Fig1D 5min). Based on these results, the initial site of the linker accumulation corresponds to the natural site of the ER-mitochondrial contacts and this process is followed by a progressive increase in the area of associations.
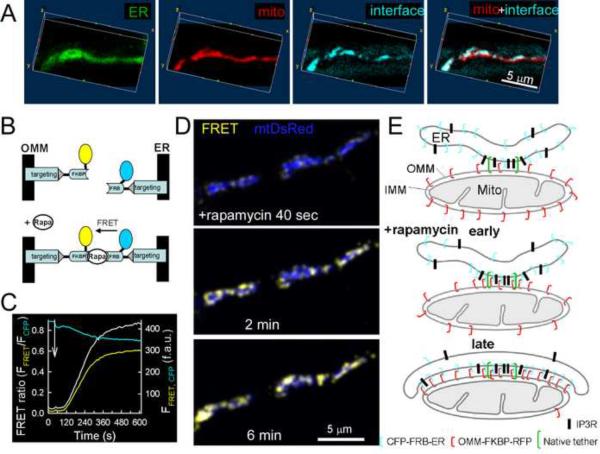
Fig 3. Visualization of ER-mitochondrial focal contacts using inducible linkers in processes of RBL-2H3 cells.
(A) 3D reconstruction of the ER (BODIPY FL Tg, green), mitochondria (RFP-labeled OMM-targeted linker module, red) and their close interfaces (delineated by the CFP-labeled ER-targeted linker module, cyan) from a z-stack of a cell process after (~15 min) rapamycin treatment.
(B) The close ER-mitochondrial contacts are depicted as the rapamycin-induced FRET increase between CFP (ER-targeted module) and YFP (OMM-targeted module).
(C) Time course of CFP (cyan) and FRET fluorescence (yellow, corrected to fluorescence bleed-through from CFP and YFP) during induction of the linkage by rapamycin (addition is marked by an arrow). Traces also show the FRET ratio (FRET/CFP, white). Calculations were made for whole cell areas (n=10 experiments, 7–15 cells in each).
(D) Spatial-temporal progression of the ER-OMM linkage formation followed as the change in the distribution and magnitude of FRET increase (yellow) over time. The blue image shows the mitochondrial distribution (mtDsRed, recorded simultaneously with FRET) (representative images, n=6 cells). As a background correction, approx. 5% of the maximal signal was subtracted from the images, which subtraction practically did not change the heterogeneity observed.
(E) Schematics showing the progress of the rapamycin-induced ER-OMM linkage formation. In the early phase of rapamycin treatment, the ER and OMM-targeted modules are linked and retained at the ER-mitochondrial contact areas (middle). Longer incubation with rapamycin leads to expansion of the interface (lower). Putative distribution of the IP3Rs is also shown. Dependent on the linker length, narrowing of the ER-OMM gap also occurs at the area of linkage (not shown).
Visualization of PM-mitochondrial contacts in live cells
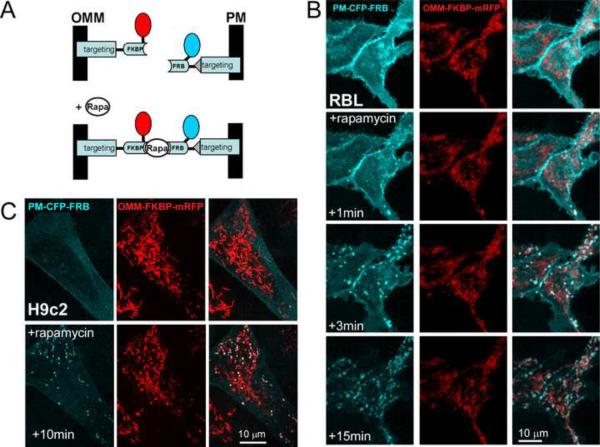
To test if mitochondria can also be linked to the plasma membrane (PM), we created an FRB-construct that was directed to the PM using the targeting sequence of Lyn (Varnai and Balla, 2007) (Fig2A). Fluorescent proteins coupled to the targeting sequence of Lyn showed colocalization with another PM-targeted domain (CAAX domain from K-ras) and did not coincide with ER-targeted signals (yUBC6, Sac1, not shown). In PM-FRB-CFP expressing RBL-2H3 (Fig2B) and H9c2 cells (Fig2C), x–y confocal sections acquired at the level of the adherent surface showed homogenous distribution of the CFP fluorescence and brighter labeling of the cells' edges. Upon addition of rapamycin, the CFP fluorescence rapidly concentrated in spots coinciding with some of the OMM-FKBP-RFP fluorescence. However, no CFP accumulation was observed for many mitochondria, indicating that a large fraction of the peri-PM mitochondria lacks contact with the PM.
Fig 2. Visualization of OMM-PM associations.
(A) Scheme illustrating the rapamycin-inducible OMM-PM bridge-forming modules. For PM targeting the N-terminal palmitoylation/myristoylation signal of the Lyn protein is fused to the N-terminus of the FRB.
(B) Confocal images of individual RBL-2H3 cells overexpressing OMM-PM linker pairs show the distribution of the respective fluorophores before and after (1, 3 and 15 min) rapamycin (100nM) treatment. Note the rapamycin-induced marked conglomeration of CFP fluorescence to the mitochondria appearing in white on the cyan/red overlay images.
(C) Confocal images of an H9c2 cell overexpressing OMM-PM linker pairs show the distribution of the respective fluorophores before and after 10 min rapamycin (100nM) treatment.
Visualization of ER-mitochondrial contact zones on individual mitochondria
To further study the ER interaction sites at the level of individual mitochondria, we obtained high magnification CFP and RFP images in protrusions of RBL-2H3 cells, which frequently contain elongated mitochondria. First, 3D reconstruction of the CFP and RFP images was used (Fig3A). This approach has revealed distinct areas of the ER linkage on single mitochondria. Since a weak fluorescence of the adherent surface was detected in the CFP channel and the ER appeared at the PM side of the mitochondria, these images also indicated that a stack of ER was inserted between the mitochondrion and the plasma membrane (6 out of 6 z-series). This provides an explanation why some mitochondria close to the PM could not participate in PM-mitochondrial linkage (Fig2). Secondly, we created CFP and YFP-tagged ER-mitochondrial linker pairs that allowed visualization of the rapamycin-induced interactions by FRET (Fig3B). An advantage of this approach is that FRET offers higher spatial resolution than fluorescence colocalization. When fluorescence was monitored over whole cell areas, rapamycin induced an increase in FRET fluorescence and a simultaneous decrease in CFP fluorescence, confirming the interaction between ER-targeted CFP and the OMM-targeted YFP (Fig3C). The FRET ratio showed a rapid initial rise (first 3min) followed by a continuous prolonged increase (Fig3C). In the light of the TEM data (Fig1D), the rapid rising phase might represent the linkage formation at the natural contact sites, whereas the slower phase might reflect the expansion of the contact areas. Therefore, we attempted to resolve the early phase of the FRET increase at the level of single mitochondria (Fig3D). The FRET fluorescence visualized distinct spots of ER interaction sites on the OMM (200–300 nm diameter), which became apparent in the first 2min of the rapamycin treatment and then expanded (Fig3D). FRET spots were apparent on every mitochondrion that was inspected (79 out of 79 in 6 cells). Thus, these studies provide evidence for organization of the contacts as focal interaction sites and for heterogeneity among individual mitochondria in ER-mitochondrial interface area. Collectively, the results of the fluorescence colocalization, FRET and TEM studies indicate that the rapamycin-induced linkage and concentration of the OMM and ER-targeted linker halves is confined to the ER-mitochondrial contact zones, specifically labeling the native interface (Fig3E middle) but subsequently expanding by a “zippering” mechanism (Fig3E lower).
Efficient and heterogeneous ER-mitochondrial Ca2+ transfer among individual mitochondria
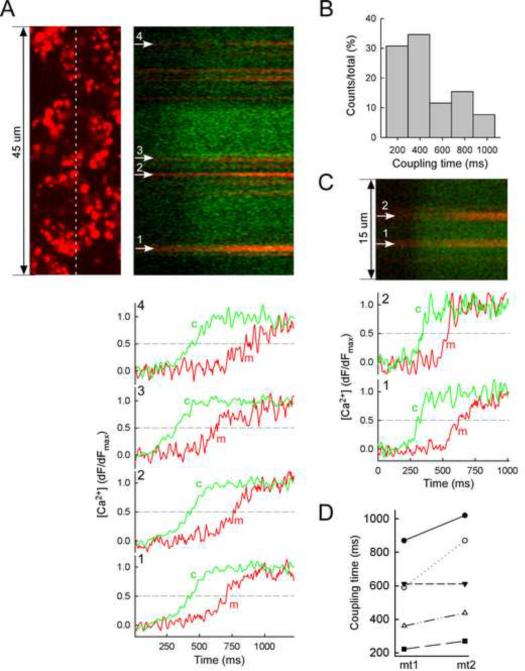
During IP3 receptor (IP3R)-mediated Ca2+ mobilization from the ER, Ca2+ is transferred to the mitochondrial matrix to stimulate mitochondrial energy metabolism (Hajnoczky et al., 1995; Rizzuto et al., 1993). To monitor the IP3R-mitochondrial Ca2+ transfer at millisecond resolution and down to the level of single mitochondria, confocal linescanning was performed, with fluo3 in the cytoplasmic buffer and rhod2 compartmentalized in the mitochondrial matrix in IP3-stimulated (7.5μM) permeabilized RBL-2H3 cells (Csordas et al., 1999; Pacher et al., 2002). Each cell showed an IP3-induced increase in [Ca2+]c closely followed by a rise in [Ca2+]m (Fig4A). The delay of the [Ca2+]m elevations, a measure of the coupling's efficacy, was <500ms in most cases (Fig4B). Intriguingly, when adjacent mitochondria were studied in a single cell, similar local [Ca2+]c elevations supported [Ca2+]m increases that showed distinct kinetics (p<0.05, n=5, Fig4CD). We reasoned that these functionally coupled [Ca2+]c and [Ca2+]m responses are reflections of the ER-mitochondrial structural connections and their heterogeneity between single mitochondria indicates that heterogeneity exist both at the structural and functional level.
Fig 4. Millisecond resolution of the coupling between IP3-induced [Ca2+]c and [Ca2+]m signals in individual mitochondria.
[Ca2+]m and [Ca2+]c were recorded in permeabilized RBL-2H3 cells as the fluorescence of the rhod2 accumulated to the mitochondria and fluo4 (10 μM) dissolved into the cytosolic buffer, respectively. Fluorescence was recorded using a confocal microscope in line-scan mode (6ms/line).
(A) An xy-image of the rhod2 distribution in three neighboring cells is shown in (upper left) with the corresponding linescan recording of the rhod2 (red) and fluo4 (green) fluorescence on the right. The time courses of the IP3-induced [Ca2+]c and [Ca2+]m increases at the positions of the numbered mitochondrial spots are shown in the bottom. The traces are normalized to the maximum fluorescence increase evoked by IP3 (7.5 μM, added at 0 ms). The time courses and the line scan image share their time (x) axis.
(B) The histogram shows the distribution of coupling times (difference in the halftime) recorded from 26 different mitochondrial areas from 4 independent experiments.
(C) Line-scan image and time courses from two distinct mitochondrial spots in the same cell are shown.
(D) Comparison of coupling times in pairs of mitochondria (<5 μm distance) from 5 different cells is shown.
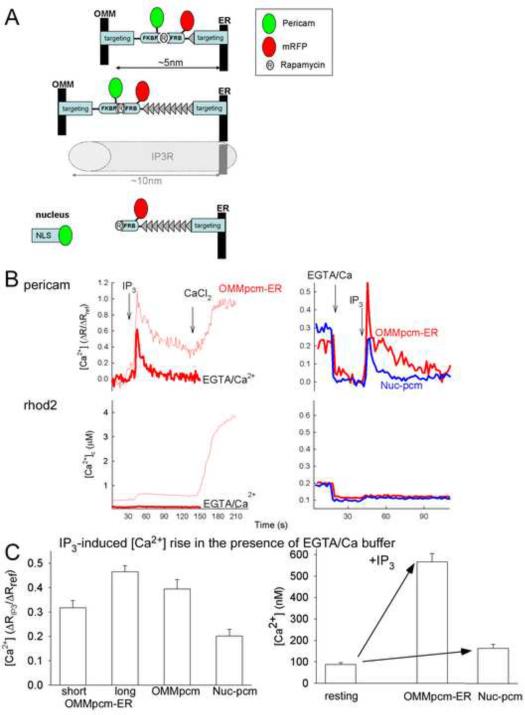
Measurement of ER-mitochondrial interface [Ca2+] ([Ca2+]ER-mt) with pericam inserted into interorganellar linkers
Activation of the mitochondrial Ca2+ uptake mechanism referred as uniporter requires supramicromolar [Ca2+]c (Gunter and Pfeiffer, 1990; Kirichok et al., 2004). This has been confirmed in both RBL-2H3 and H9c2 cells (Csordas et al., 1999; Szalai et al., 2000). The high [Ca2+]c requirement for uniporter activation during IP3R-mediated Ca2+ release can be met at the ER-mitochondrial contacts, via the high [Ca2+]c microdomain occurring in the vicinity of open IP3Rs (Rizzuto et al., 1993). Although this concept is supported by indirect evidence (Csordas et al., 1999; Rizzuto et al., 1998), direct evaluation of the [Ca2+]ER-mt has not been possible. To make this measurement feasible, a Ca2+ sensitive fluorescent protein, ratiometric pericam (Nagai et al., 2001) was placed in the OMM-targeted FKBP12 instead of mRFP (Fig5A) and mRFP was used instead of CFP in the ER-targeted FRB domain to avoid spectral overlap (Fig5A). Upon co-transfection of H9c2 and RBL-2H3 cells with these constructs, the pericam fluorescence showed its expected mitochondrial distribution, whereas the mRFP decorated the ER (FigS2A). As observed before, rapamycin elicited rapid colocalization of the majority of the mRFP with pericam (FigS2A) to form an OMM-(pericam)FKBP-FRB-ER (OMM-pcm-ER) bridge at the ER-mitochondrial interface. First, the response of these mito-ER localized reporters was tested in permeabilized cells. Here, the pericam excitation ratio (F485nm/F420nM) showed a transient increase in response to IP3, and to the subsequent addition of CaCl2, confirming the Ca2+ sensitivity of the OMM-pcm-ER complex (Fig5B left).
Fig 5. Measurement of [Ca2+]ER-mt with pericam-tagged inducible linkers in permeabilized RBL-2H3 cells.
(A) Constructs used to monitor [Ca2+] at the ER-OMM interface and in the nucleus: OMM-targeted linker module tagged with ratiometric pericam, while the ER-targeted module is labeled with mRFP (upper), OMM-targeted pericam with a longer version of the ER-targeted module (middle) and pericam targeted to the nuclear matrix (Nuc-pcm) combined with the longer ER-targeted module (lower).
(B) Wide-field recordings of IP3-induced [Ca2+] responses detected by the differently targeted pericams. In addition, to the cell-bound pericams, [Ca2+] in the bulk cytosolic buffer was also recorded using rhod2 (bottom panels). Rapamycin (100nM) was added during cell permeabilization (7–10min) in each run. The IP3-induced rhod2 response was well below 1 μM, whereas the OMM-targeted linker pericam response peaked as high as the effect of a 20μM CaCl2 pulse that raised the [Ca2+]c to ~4μM (left, thin red lines). The bulk [Ca2+]c increase calculated from the rhod2 signal (obtained in the intercellular regions to avoid the overlap with the cellular mRFP fluorescence) is similar to the [Ca2+] rise predicted by the Winmax program (Csordas et al., 1999). When the Ca2+ buffering strength was enhanced by adding EGTA and CaCl2 (EGTA/Ca2+, 100 μM and 40 μM, respectively) the IP3-induced rhod2 response was abolished but the OMM-pcm-ER response was present (thick red lines). The traces represent the means of 20 (standard) and 8 (EGTA/Ca2+) individual cells. The panels on the right compare the IP3-induced [Ca2+] increase detected by the OMM-pcm-ER (red trace) and Nuc-pcm (blue trace) in the presence of EGTA/Ca2+-buffer. The traces represent the means of 17 and 18 individual cells. The effect of IP3 [Ca2+]pcm is normalized to the change evoked by the CaCl2 pulse (Rref, no EGTA: +20μM, EGTA/Ca2+: +60μM). The axis title shown on the left side also applies to the graphs on the right side
(C) Left: The cumulated magnitudes of IP3-induced [Ca2+] increases detected by OMM-pcm-ER with the short and long ER-targeted modules, by an OMM-targeted pericam lacking rapamycin-binding domain (OMMpcm) and by Nuc-pcm and normalized to the reference Ca2+ pulse ([CaCl2] 100μM, increasing [Ca2+]c to approx. 4 μM). Right: Translation of the IP3-induced [Ca2+] rises to nM values (right). The data represent the means of 8–12 separate recordings.
Effect of rapamycin on OMM-FKBP-pcm localization and ER and mitochondrial Ca2+ handling is shown in FigS2.
Based on the crystal structure, the rapamycin-inducible linker was estimated to span 5–6 nm distance (Varnai and Balla, 2007). In addition, transmission EM studies of constitutive OMM-ER linker expressing cells have shown that this linker shortened the ER-mitochondrial gap width to <7nm (Csordas et al., 2006). Thus, the contacts stabilized and labeled by the drug-inducible version of this linker could make this space too narrow to accommodate the IP3R that was estimated to protrude 10 nm from the ER membrane. Therefore, we also created a longer version of the ER-targeted linker to allow for the presence of IP3Rs at the interface labeled by the OMM-pcm-ER bridge (Fig5A). TEM analysis, has confirmed that the ER-mitochondrial gap is approx. 15 nm when this longer linker is used with rapamycin (16.8±0.7 nm, n=70). To compare the responses of the linker-bound pericam to a refernce signal, Ca2+ measurements with a nucleus-targeted ratiometric pericam (Nuc-pcm) were also performed in the permeabilized cells ((Nagai et al., 2001), Fig5A). In these experiments rhod2, a fluorescent Ca2+ probe with a wide dynamic range was included in the cytoplasmic buffer and was recorded simultaneously with pericam, to provide information on the global [Ca2+]c changes. Importantly, rapamycin treatment (added during permeabilization) did not evoke a significant change in the IP3-induced [Ca2+]c and [Ca2+]m elevations (FigS2B), indicating that neither the ER nor the mitochondrial Ca2+ handling was affected by this drug during the time frame of the experiments.
Addition of IP3 caused elevation of global [Ca2+]c from 250 nM to 700 nM, while CaCl2 addition (20 μM) resulted in a further increase to 4 μM as measured by the rhod2 signal (Fig5B, lower left, thin line). In contrast, the IP3-induced [Ca2+]ER-mt rise measured by OMM-pcm-ERlong was comparable to the CaCl2-induced [Ca2+]ER-mt increase (Fig5B upper left, thin line, n=33). Thus, the OMM-pcm-ER reported on a relatively higher [Ca2+] after IP3, consistent with a high localized [Ca2+] close to the IP3Rs. Furthermore, this result also indicates that the IP3-induced [Ca2+]ER-mt signal can reach levels as high as the overall [Ca2+]c reached after CaCl2 addition that is >4 μM based on the rhod2 calibration. However, because of the high affinity of pericam for Ca2+ (see Fig6C), the calibration of the OMM-pcm-ER signal in the >4 μM [Ca2+] range may not be accurate, partly because of its signal can be contaminated by the general [Ca2+]c increase. To address this problem, the IP3-induced increase in bulk [Ca2+]c, was suppressed by inclusion of an EGTA/Ca2+ buffer in the medium (Fig5B left, thick lines). This buffer clamped [Ca2+]c at 100 nM and prevented the IP3-induced increase in bulk [Ca2+]c as shown by the lack of increase in the rhod2 signal (Fig5B lower, thicker trace). Importantly, IP3 could still evoke a large [Ca2+]ER-mt transient under these conditions (Fig5B upper left, thick line). When the signal from the OMM-pcm-ER was compared to that of Nuc-pcam, the former showed a significantly larger transient, indicating that the [Ca2+]ER-mt signal is greater than [Ca2+]Nuclear one, a measure of the general [Ca2+]c signal (Fig5B right). Because of the size of the IP3-induced Ca2+ signal relative to that evoked by Ca2+ addition was a good measure of how the pericam reported on localized [Ca2+]ER-mt we calculated this ratio for all reporter constructs. In total, the IP3-induced [Ca2+] increases measured with the short and long version of the OMM-pcam-ER and with Nuc-pcm were 32±3%, 47±2%, and 20±3% of the increase evoked by elevation of the bulk [Ca2+]c by addition of CaCl2 (n=4, 12, 8, respectively; Fig5C). The response detected by OMM-pcm-ERlong was significantly larger than the response detected by the OMM-pcm-ERshort (p<0.01) and both were larger than that of Nuc-pcm. Notably, an OMM targeted pericam devoid of the FKBP domain also showed a larger response than the Nuc-pcm (39±4%, p<0.05, n=3). Calibration of the responses in terms of nM [Ca2+] further increased the difference between OMM-pcm-ERlong and Nuc-pcm (Fig5C), because of the slightly higher Ca2+ affinity of the Nuc-pcm construct (Kd= 259±27 nM vs. 460±80 nM, see pericam calibration below). Collectively, these data suggest that: (1) an IP3-induced high [Ca2+] microdomain can detected on the mitochondrial surface by OMM-pcm (2) the [Ca2+]ER-mt can be monitored using OMM-pcm-ER, (3) the [Ca2+]ER-mt rise exceeds 4 μM during IP3
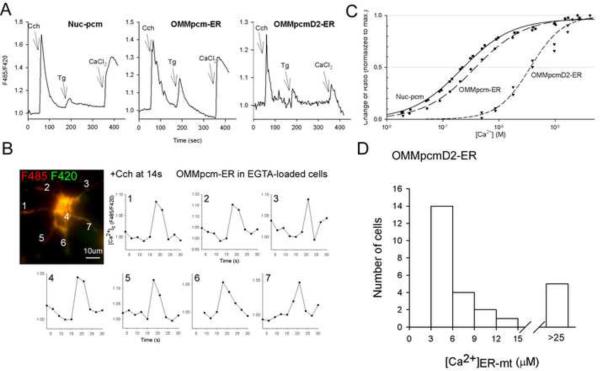
Fig 6. Measurement of IP3R-linked [Ca2+]ER-mt signal in intact cells.
RBL-2H3 cells transiently overexpressing type1 muscarinic receptor and either the long version of the OMM-pcm-ER linker pair or Nuc-pcm were stimulated with saturating dose of carbachol (Cch 100 μM) in Ca2+-free extracellular buffer. In turn, SERCA inhibitor, Tg 2 μM) was added to deplete the ER Ca2+ store and thus open the store-operated Ca2+ entry channels. Three minutes later as a reference, maximum Ca2+ entry was provoked by addition of 5mM CaCl2. To avoid saturation of the Ca2+ probes, the low-affinity OMM-pcmD2-ER was also tested. Rapamycin was added for 5–6 min before the start of the run.
(A) The time courses recorded with the three different probes are shown. The traces are the pericam ratios normalized to the baseline and represent the means of 3–4 parallel recordings (each collected and averaged from 3–5 cells).
(B) To prevent the Cch-induced [Ca2+]c signal, EGTA/AM-loaded cells were used and the spatiotemporal pattern of the [Ca2+]ER-mt signal was visualized. The image shows the distribution of the OMM-pcm-ER fluorescence (merge of excitation at 485nm in red and 420nm in green). Time courses of the OMM-pcm-ER ratio change (normalized to the baseline) during Cch stimulation recorded at the numbered areas are shown on the graphs.
(C) [Ca2+] calibration plots of Nuc-pcm, OMM-pcm-ER and OMMl-pcmD2-ER ratio as determined from experiments in permeabilized cells using stepwise Ca2+ additions.
(D) The Cch-induced [Ca2+]ER-mt rise detected by OMM-pcmD2-ER translated to μMs. The histogram shows the distribution of the calculated magnitudes of [Ca2+]ER-mt responses to Cch from a total of 26 individual cells.
Effect of EGTA/AM loading on the global [Ca2+] signal and the calibration of the OMM-pcm(D2)-ER ratio in terms of nM(μM) concentration is shown in FigS3.
IP3-induced Ca2+ release and (4) the [Ca2+]ER-mt increase is compromised if the gap width is <7 nm in the contact area. Therefore, only the long version of linker construct was used in subsequent experiments.
[Ca2+]ER-mt signaling in intact cells
Carbachol stimulation of RBL-2H3 cells expressing the M1 muscarinic receptor and either Nuc-pcm + mRFP-FRB-ER or OMM-FKBP-pcm + mRFP-FRB-ER resulted in similar transients in the ratio (Fig6A, left and middle). However, when the cells were preloaded with EGTA/AM (5μM), the Nuc-pcm response was practically abolished, and only a slow and small increase remained (FigS3A). By contrast, the OMM-pcm-ER still showed rapid transients (Fig6B). The subcellular spatiotemporal pattern of the transients is shown in Fig6B. The [Ca2+]ER-mt signal could be detected in the cell body and in every protrusions. The onset was slower in some regions (3,7) than in others (1,2,5,6) and it was associated with particularly rapid decay (Fig6B). Because the high affinity of OMM-pcm-ER only allowed measurements of [Ca2+]ER-mt in EGTA-loaded cells, we created a low affinity version of the OMM-FKBP-pcm. For this purpose we introduced a set of mutations into the calmodulin and calmodulin binding peptide sequences of the pericam construct, which have been found to result in lowering the Ca2+ affinity of the D2 cameleon (Palmer 2006). The in situ Ca2+ titration curves in Fig6C show that the new pericam construct (pcmD2) displays a significantly lower affinity for Ca2+ than the Nuc-pcm and the OMM-pcm-ER. Half-maximal ratio changes were attained at 3,140±536 nM, 259±27 nM and 460±80 nM [Ca2+] for OMM-pcmD2-ER, Nuc-pcm and OMM-pcm-ER, respectively (n=7, 4 and 10).
Carbachol stimulation of RBL-2H3 cells expressing the M1 muscarinic receptor and OMM-FKBP-pcmD2 + mRFP-FRB-ER resulted in [Ca2+] transients (Fig6A right), which were narrow as compared to the transients recorded by Nuc-pcm or OMM-pcm-ER (durations at half heights; 9±1 s, 22±1 s and 25±3 s for OMM-pcmD2-ER, Nuc-pcm and OMM-pcm-ER, respectively; n=26, 30 and 8). Furthermore, store-operated Ca2+ entry induced by Tg resulted in a relatively small change in the OMM-pcmD2-ER ratio compared to the signal obtained with the Nuc-pcm or OMM-pcm-ER reporters (Fig6A right). These results further supported the notion that the OMM-pcmD2-ER sensed a high [Ca2+] microdomain at the ER-mitochondrial interface and, unlike the OMM-pcm-ER, its signal did not get saturated by Ca2+. The average [Ca2+]ER-mt peak evoked by carbachol was calculated to be ~9 μM. None of the cells showed a signal ≤3 μM, and most cells showed 3–9μM and several cells gave close to maximal increases in the OMM-pcmD2-ER ratio (≥95%) indicating that the [Ca2+]ER-mt could have risen to 25 μM (Fig6D). Importantly, these numbers represent only the lower limit of the [Ca2+]ER-mt because it is unlikely that the entire amount of OMM-FKBP-pcmD2 on the mitochondrial surface was moved to the ER-mitochondrial interface after rapamycin addition. These results suggest that the [Ca2+]ER-mt can rise to several tens of micromolar during IP3R activation. To determine whether the interface area is better exposed to ER Ca2+ release than the whole OMM, the [Ca2+]ER-mt was compared to [Ca2+]OMMI recorded by the OMMFKBP-pcmD2 in the absence of rapamycin (1) or in the absence of the ER partner (2). To minimize the linkage-induced expansion of the interface area, rapamycin was added only for 3min. The mean [Ca2+]ER-mt was 8.4 μM (n=111), whereas [Ca2+]OMM (1) was 5.6 μM (n=33) and [Ca2+]OMM (2) was 5.2 μM (n=28). Although, [Ca2+]ER-mt appeared to be larger than [Ca2+]OMM the difference was not significant because of the broad range of the data.
Discussion
While previous studies implied that ER-mitochondrial contacts are the generic sites of the local Ca2+ transfer, the present work provides direct demonstration of a high [Ca2+]c microdomain at the ER-mitochondrial interface and correlates [Ca2+]ER-mt with some physical properties of the interface. The biphasic dependence of the efficiency of Ca2+ transfer on the space that separates the two organelles as evidenced by our variable length linkers indicate that certain structural arrangements can support optimal Ca2+ transfer owing to the space requirements for the Ca2+ transport channels. It is reasonable to assume that distinct processes may have their own optimal spatial requirements; while some separation is needed for the Ca2+ delivery, tighter associations are likely to better support lipid and protein transfer between these organelles. Therefore, distinct functions of the ER-mitochondrial interface in calcium coupling, lipid transfer and protein trafficking could be supported by different subsets of contact sites. A deeper exploration of the ER-mitochondrial coupling in live cells were made possible by the invention of drug-inducible interorganellar linkers that can accommodate different fluorescent proteins for colocalization, FRET and chemical signal measurements. This genetic approach offers a powerful new toolkit for the study of various inter-organellar contact sites in live cells.
To study the ER-mitochondrial contacts in living cells, previous studies have used colocalization of ER-and mitochondrion-specific fluorophores/fluorescent proteins (Rizzuto et al., 1998). The present application of the inducible ER-mitochondrial linkers is a specific approach to the visualization of the interface. Based on the results obtained with this strategy, essentially all mitochondria seem to have a contact with the ER, at least in some cell types. Interestingly, a considerable fraction of the peri-PM mitochondria also appear to be coupled to the PM through an ER stack. Of note, this indicates the association of mitochondria with the ER subdomains that control store-operated Ca2+ entry (Korzeniowski et al., 2009; Wu et al., 2006) and provides the structural basis for the observation that the entering Ca2+ crosses the ER before it is taken up by the mitochondria (Demaurex et al., 2009). At the level of individual mitochondria, the contacts are heterogeneous and commonly appear as discrete spots of various size and shape. Paralleling the spatial organization of the interface, the IP3R-mediated Ca2+ release is effectively delivered to each mitochondrion. However, individual mitochondria show heterogeneous kinetics for the [Ca2+]m rise, which is unlikely to result entirely from heterogeneity in local Ca2+ release (see Fig4CD) or from heterogeneity in the driving force of mitochondrial Ca2+ uptake (Pacher and Hajnoczky, 2001). Thus, the observed variability in the area of the contact zones with the ER might be relevant for the heterogeneity in the [Ca2+]m signal.
Based on measurements of the mitochondrial intermembrane space [Ca2+], Rizzuto, Pozzan et al have suggested that the [Ca2+]ER-mt signal attains several micromoles/L (Rizzuto et al., 1998). Furthermore, titration of the IP3-induced [Ca2+]m signal by added Ca2+ has indicated that mitochondria would be exposed to 10–30 μM [Ca2+] (Csordas et al., 1999). In the present work, targeting of pericam to the ER-mitochondrial interface resulted in an opportunity for direct measurement of the [Ca2+]ER-mt. Resistance of the OMM-pcm-ER responses to buffering of the [Ca2+]c made clear that the pericam fluorescence reflects a local [Ca2+] signal. Regarding the magnitude of the [Ca2+]ER-mt signal, saturation of the pericam during IP3-induced Ca2+ release has indicated that the [Ca2+]ER-mt rise attains at least several micromolar concentrations. Better quantitation of the [Ca2+]ER-mt signal was achieved by introduction of a set of mutations to the pericam's Ca2+ binding domain, which resulted in lower affinity for Ca2+. The IP3-linked [Ca2+]ER-mt spikes exceeded 3 μM in every cell, and caused several-fold higher signal in many cells. Since the mean [Ca2+]ER-mt increase was 9 μM and this value is likely to involve the contribution of a fraction of pericam that remains outside the interface, it is a safe estimation that the [Ca2+]ER-mt signal peaks in the ten micromolar concentration range. Since the dynamic range of the low affinity pericam is significantly narrower than that of the pericam, recording of [Ca2+]ER-mt for subsets of mitochondria with this probe remains difficult. However, the spatial distribution of the OMM-pericam-ER response in EGTA-loaded cells indicates the presence of a high [Ca2+]ER-mt elevation throughout the cell. Finally, the dependence of the measured [Ca2+]ER-mt rise on the linker length indicates that the closest contacts are not the most effective in the local Ca2+ signal delivery, presumably because these site can not provide space for the Ca2+ channels. These sites may have another role in the ER-mitochondrial interface function, which does not involve bulky proteins like the IP3R or RyR. Thus, it is likely that the different functions of the ER-mitochondrial associations are assigned to contacts with distinct physical properties. Collectively, our results suggest that both the area and gap width of the ER-mitochondrial interface are important determinants of the [Ca2+]ER-mt signaling and are likely to affect differently the other functions of the junctions.
Methods
Chemicals
Standard chemicals were purchased from Fisher Scientific, Sigma-Aldrich or EMD Biosciences. IP3 was from LC Laboratories (Woburn, MA). Fluorescent Ca2+-indicator dyes, BODIPY FL Thapsigargin and Pluronic F-127 were from Molecular Probes or Teflabs. Chelex 100 Sodium form was purchased from BioRad.
DNA constructs
Nuclear ratiometric pericam was a gift from Dr. Atsushi Miyawaki (RIKEN). For targeting the FKBP12 protein to the cytoplasmic surface of the mitochondria (OMM), the N-terminal sequence of the mAKAP1 (34–63) was used and it was fused to the N-terminus of the human FKBP12 protein through a linker (DPTRSANSGAGAGAGAILSR). The fusion proteins were tagged with mRFP, YFP or pericam using the pEGFP-N1 plasmid backbone. To create mAKAP1-FKBP-pericamD2 three mutations (F68L, M72L es Q104E) were introduced to the calmodulin and calmodulin binding peptide sequences of mAKAP1-FKBP-pericam which have been shown to result in decreased affinity for Ca2+ (Design 2; (Palmer et al., 2006)). The constructs used for targeting the CFP or mRFP-tagged FRB protein to the cytoplasmic surface of the ER were described elsewhere (Varnai and Balla, 2007). For PM targeting of the FRB protein, the N-terminal palmitoylation/myristoylation signal of the Lyn protein (MGCIKSKGKDSAGA) was used and it was fused to the N-terminus of the FRB fragment with a linker of DPTRSANSGAGAGAGAILSR between the two proteins. This fusion protein was tagged with CFP or mRFP using the pEGFP-N1 plasmid backbone.
Cells
RBL-2H3 and H9c2 cells were cultured and transiently transfected with plasmid DNA as described earlier (Csordas et al., 2006; Saotome et al., 2008).
For imaging experiments, the cells were pre-incubated in a serum-free extracellular medium (ECM, 121 mM NaCl, 5 mM NaHCO3, 10 mM Na-HEPES, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 2 mM CaCl2, 10 mM glucose, pH7.4) containing 2% BSA and were loaded with rhod2/AM or fura2/AM for measurements of [Ca2+]m and [Ca2+]c, respectively as described earlier (Csordas and Hajnoczky, 2003; Pacher et al., 2002; Szalai et al., 2000). To increase cytoplasmic Ca2+ buffering, cells were loaded with 50 μM EGTA/AM together with 0.003% pluronic F127 for 30 min at room temperature. For intact cell recordings, at the end of the preincubation or dye-loading period the cells were washed into a fresh ECM containing 0.25% BSA and transferred to the temperature-regulated stage of the microscope (Csordas and Hajnoczky, 2003). For the recordings where store-operated Ca2+ entry was used as a reference signal, just before the recording, a Ca2+ free ECM was added ([Ca2+]≤1 μM) while the standard ECM was supplemented with 2mM CaCl2. For permeabilized cell experiments, the transfected or rhod2/AM-loaded cells were washed with a Ca2+-free extracellular salt solution containing 100 μM EGTA/TRIS and transferred to the imaging chamber in 1ml intracellular medium (ICM, composed of 120 mM KCl, 10 mM NaCl, 1 mM KH2PO4, 20 mM Tris-HEPES at pH 7.2 (Csordas and Hajnoczky, 2003)). Plasma membrane permeabilization was carried out using 25μg/ml digitonin or saponin 40μg/ml. After 7–10 min permeabilization period (35°C), the cells were washed into fresh ICM supplemented with MgATP 2mM and succinate/TRIS.
Live Cell Imaging
Confocal imaging was carried out using a BioRad Radiance 2100 system with Kr/Ar (488 nm and 568 nm lines for green/yellow and red fluorescent probes, respectively) and He/Cd (442 nm line, for CFP) ion laser sources fitted to an Olympus IX70 inverted microscope. To avoid bleed-through from the 488 nm excitation to the cyan emission window, when the rapamycin-inducible linkage formation was imaged, only the mRFP- and CFP-tagged (568nm and 442 nm excitation, respectively) linker modules were scanned simultaneously, and the ER was labeled with BODIPY FL-Tg (1 μM for 3–5 min) and imaged afterwards in a separate recording of the same field (488nm excitation). High-resolution images were taken using a 60× oil objective (NA 1.45). The scanned area was 512×512 pixels at 5–10× zooming factor (41×41–20.5×20.5 μm box size). The scanning speed was 166 lines per second. Image stacks were created from the Z-series (250nm step size) recordings and filtered using a 3×3 median filter to reduce speckle noise using the NIH's Image J software. Contrast and brightness were optimized in the brightest slice of the stack and the same settings were applied to the rest of the sections. 3D reconstruction was performed using either Voxx from Indiana University (http://www.nephrology.iupui.edu/imaging/voxx/, FigS1B) or a plugin of ImageJ (Volume Viewer for Fig2A). Two or 3 color slice stacks were loaded into the software. Maximum intensity rendering was used with the same isometric viewpoint for both datasets. Lookup tables were modified to reduce background noise.
Line scanning of mitochondrial rhod2 and cytosolic fluo3 in permeabilized RBL-2H3 cells was carried out as described (Pacher et al., 2002).
Fluorescence wide field imaging of [Ca2+]c, [Ca2+]Nuc and [Ca2+]ER-mt was carried out using back-illuminated CCD cameras (Roy et al., 2009; Saotome et al., 2008). When only pericam was imaged, 485/20 nm and 420/30 nm excitation filters and a 500 nm long-pass beam splitter were used and an image pair was obtained in every 3s. When pericam was recorded simultaneously with rhod2, 490/20nm, 415/30nm and 580/20nm excitation filters were combined with a dual dichroic and a dual emission filter (535/30nm and 630/60nm) and an image triplet was obtained in every 1.5s.
The excitation ratio of F485nm and F420nm or F490nm and F415nm was used to describe [Ca2+] changes measured by pericam. Notably, the pericam targeted to the OMM, in particular pericamD2 displayed very narrow dynamic range at 485nm compared to the 420nm excitation. Direct Ca2+ calibration of the pericam signals was not feasible due to the pH-sensitivity of the 485nm signal and the substantial pH shift associated with the sequential addition of high CaCl2 (1–2mM) and EGTA/TRIS 10mM (pH 8.7) usually used for this purpose with other Ca2+ indicators. The present calibration approach (see Supplementary Fig2) is based on the postulations that (1) the resting [Ca2+] is in equilibrium between the cytoplasm/cytosolic buffer and the ER-mt interface or the nuclear matrix and (2) for the plateau phase of [Ca2+] increases caused by Ca2+ entry or Ca2+ addition [Ca2+]c=[Ca2+]ER-mt=[Ca2+]nuc. [Ca2+]c measured with fura2 in intact cells and with rhod2 in permeabilized cells was calibrated as described (Csordas et al., 2006; Roy et al., 2009). Therefore, the increase in the OMM-pericam-ER or nuclear pericam ratio in response to CaCl2 addition can be back-calibrated from simultaneous (permeabilized cells) or parallel (intact cells) recording of [Ca2+]c. This value was then used as a reference (ΔRRef) to the transient ratio increases caused by IP3R-mediated Ca2+ release (ΔRCch or ΔRIP3) in the Ca2+ saturation curve of each pericam (see the scheme in FigS3B).
Ca2+ saturation curves of various pericam constructs (Fig5C) were determined in permeabilized cells where the initial very low [Ca2+] in the buffer could be chelated by 100 μM EGTA without a significant shift in the pH. To simultaneously record [Ca2+]c with the pericam fluorescence, rhod2/FA 300 nM was added to the buffer. To avoid interference by mitochondrial or ER Ca2+ accumulation with the equilibrium of [Ca2+]ER-mt with [Ca2+]c, the Ca2+ ionophore, ionomycin (10 μM) was included in the buffer. Titration was carried out by stepwise addition of CaCl2 (10–20 μM pulses, a cumulative 150–300 μM in 12–14 steps). At the end of the recording rhod2 was calibrated using sequential addition of CaCl2 1mM and EGTA/TRIS 10 mM (pH 8.7) and [Ca2+]c was calculated in nM using a Kd value of 1μM as described earlier (Csordas and Hajnoczky, 2003; Csordas et al., 2006). The ratio levels for each step after normalization to the total range were plotted against the corresponding molar [Ca2+] levels calculated from the rhod2 fluorescence. Curve fitting to the data points was done using Sigmaplot (sigmoid curve, 3-parameter Hill plot) and the curves were used as the Ca2+ saturation curves. To avoid interference from the ER-targeted RFP, rhod2 fluorescence for the calculation of [Ca2+]c was obtained from cell-free areas.
Transmission electron microscopy
RBL-2H3 cells co-transfected with AKAP1-FKBP-YFP and mRFP-FRB-9x-Sac1 (24h) were enriched via fluorescence-associated cell sorting (FACS). Sorting was based on YFP fluorescence. Transfection and sorting efficacy was assessed via fluorescence imaging of the cells before and after sorting, respectively. Transfection efficacy was 30–50% with >80% co-transfection rate, while after sorting >90% of the cells expressed both YFP and mRFP and displayed rapamycin-induced redistribution.
Sorted cells were plated to plastic Petri dishes and were kept in culture for 3 hours. Then, after a preincubation in 2%BSA-ECM (30 min, 37°C) the cells were treated with rapamycin (100 nM in 0.25%BSA-containing ECM) for 0, 5 or 30 min. The treatment was ended by the application of 2% gluteraldehyde fixative (2% glutaraldehyde, 1% tannic acid in 0.1M Na-cacodylate buffer; all from Electron Microscopy Sciences). After 10 min fixation period the cells were detached by scraping and processed for TEM as described earlier (Pacher et al., 2000). Ultrathin sections of the embedded cells were imaged using a FEI Tecnai 12 Twin G2 TEM fitted with a high-resolution bottom-mounted CCD camera (Orca-HR from Hamamatsu Photonics). Image analysis and morphometric evaluation was done as described earlier (Csordas et al., 2006).
Statistics
Experiments were carried out with ≥3 different cell preparations and with each preparation the measurements were done at least in triplicates. Data are presented as means ± S.E.M. Significance of differences from the relevant controls was calculated by Student's t test.
Supplementary Material
Acknowledgements
The authors thank Sudipto Das for his expert technical help. This work was supported by a Thomas Jefferson Pilot Research Award to G.C. and by NIH grants (DK051526 and RC2AA019416) to G.H. The research of T.B. was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health. P.V. was supported by grants from the Hungarian Scientific Research Fund (OTKA NF-68563) and the Medical Research Council (ETT 494/2009) of Hungary.
Footnotes
GC and PV contributed equally to this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Breckenridge DG, Stojanovic M, Marcellus RC, Shore GC. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Hajnoczky G. Plasticity of mitochondrial calcium signaling. J Biol Chem. 2003;278:42273–42282. doi: 10.1074/jbc.M305248200. [DOI] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Thomas AP, Hajnoczky G. Quasi-synaptic calcium signal transmission between endoplasmic reticulum and mitochondria. Embo J. 1999;18:96–108. doi: 10.1093/emboj/18.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Demaurex N, Poburko D, Frieden M. Regulation of plasma membrane calcium fluxes by mitochondria. Biochim Biophys Acta. 2009;1787:1383–1394. doi: 10.1016/j.bbabio.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Goetz JG, Nabi IR. Interaction of the smooth endoplasmic reticulum and mitochondria. Biochem Soc Trans. 2006;34:370–373. doi: 10.1042/BST0340370. [DOI] [PubMed] [Google Scholar]
- Gunter TE, Pfeiffer DR. Mechanisms by which mitochondria transport calcium. Am J Physiol. 1990;258:C755–786. doi: 10.1152/ajpcell.1990.258.5.C755. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Hager R, Thomas AP. Mitochondria suppress local feedback activation of inositol 1,4, 5-trisphosphate receptors by Ca2+ J Biol Chem. 1999;274:14157–14162. doi: 10.1074/jbc.274.20.14157. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouaville LS, Ichas F, Holmuhamedov EL, Camacho P, Lechleiter JD. Synchronization of calcium waves by mitochondrial substrates in Xenopus laevis oocytes. Nature. 1995;377:438–441. doi: 10.1038/377438a0. [DOI] [PubMed] [Google Scholar]
- Jouaville LS, Pinton P, Bastianutto C, Rutter GA, Rizzuto R. Regulation of mitochondrial ATP synthesis by calcium: evidence for a long-term metabolic priming. Proc Natl Acad Sci U S A. 1999;96:13807–13812. doi: 10.1073/pnas.96.24.13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski MK, Szanda G, Balla T, Spat A. Store-operated Ca2+ influx and subplasmalemmal mitochondria. Cell Calcium. 2009;46:49–55. doi: 10.1016/j.ceca.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella CA, Buttle K, Rath BK, Marko M. Electron microscopic tomography of rat-liver mitochondria and their interaction with the endoplasmic reticulum. Biofactors. 1998;8:225–228. doi: 10.1002/biof.5520080309. [DOI] [PubMed] [Google Scholar]
- Merkwirth C, Langer T. Mitofusin 2 builds a bridge between ER and mitochondria. Cell. 2008;135:1165–1167. doi: 10.1016/j.cell.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Csordas P, Schneider T, Hajnoczky G. Quantification of calcium signal transmission from sarco-endoplasmic reticulum to the mitochondria. J Physiol. 2000;529(Pt 3):553–564. doi: 10.1111/j.1469-7793.2000.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Hajnoczky G. Propagation of the apoptotic signal by mitochondrial waves. Embo J. 2001;20:4107–4121. doi: 10.1093/emboj/20.15.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Thomas AP, Hajnoczky G. Ca2+ marks: miniature calcium signals in single mitochondria driven by ryanodine receptors. Proc Natl Acad Sci U S A. 2002;99:2380–2385. doi: 10.1073/pnas.032423699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, Baker D, Tsien RY. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Pinton P, Ferrari D, Rapizzi E, Di Virgilio F, Pozzan T, Rizzuto R. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. Embo J. 2001;20:2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993;262:744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pozzan T. Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol Rev. 2006;86:369–408. doi: 10.1152/physrev.00004.2005. [DOI] [PubMed] [Google Scholar]
- Roy SS, Madesh M, Davies E, Antonsson B, Danial N, Hajnoczky G. Bad targets the permeability transition pore independent of Bax or Bak to switch between Ca2+-dependent cell survival and death. Mol Cell. 2009;33:377–388. doi: 10.1016/j.molcel.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnoczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci U S A. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore GC, Tata JR. Two fractions of rough endoplasmic reticulum from rat liver. I. Recovery of rapidly sedimenting endoplasmic reticulum in association with mitochondria. J Cell Biol. 1977;72:714–725. doi: 10.1083/jcb.72.3.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai G, Csordas G, Hantash BM, Thomas AP, Hajnoczky G. Calcium signal transmission between ryanodine receptors and mitochondria. J Biol Chem. 2000;275:15305–15313. doi: 10.1074/jbc.275.20.15305. [DOI] [PubMed] [Google Scholar]
- Szalai G, Krishnamurthy R, Hajnoczky G. Apoptosis driven by IP(3)-linked mitochondrial calcium signals. Embo J. 1999;18:6349–6361. doi: 10.1093/emboj/18.22.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE. Molecular and cell biology of phosphatidylserine and phosphatidylethanolamine metabolism. Prog Nucleic Acid Res Mol Biol. 2003;75:69–111. doi: 10.1016/s0079-6603(03)75003-x. [DOI] [PubMed] [Google Scholar]
- Varnai P, Balla T. Visualization and manipulation of phosphoinositide dynamics in live cells using engineered protein domains. Pflugers Arch. 2007;455:69–82. doi: 10.1007/s00424-007-0270-y. [DOI] [PubMed] [Google Scholar]
- Voelker DR. Genetic and biochemical analysis of non-vesicular lipid traffic. Annu Rev Biochem. 2009;78:827–856. doi: 10.1146/annurev.biochem.78.081307.112144. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Meisinger C, Pfanner N. Cell biology. Connecting organelles. Science. 2009;325:403–404. doi: 10.1126/science.1178016. [DOI] [PubMed] [Google Scholar]
- Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.