Abstract
Purpose
To analyze tear protein profile variations in patients with keratoconus (KC) and to compare them with those of control subjects.
Subjects and methods
Tears from 12 normal subjects and 12 patients with KC were analyzed by two-dimensional gel electrophoresis (2-DE) and liquid chromatography–mass spectrometry (LC–MS). Analysis of the 2-DE gels was performed using Progenesis SameSpots software (Nonlinear Dynamics). Proteins exhibiting high variation in expression levels (P-value <0.05) were identified using matrix-assisted laser desorption/ionization–TOF spectrometry. For LC–MS analysis, a label-free quantification approach was used. Tears were digested with trypsin, subjected to data-independent acquisition (MSE) analysis, and identified proteins were relatively quantified using ProteinLynx Global Server software (Waters).
Results
The 2-DE and LC–MS analyses revealed a significant decrease in the levels of members of the cystatin family and an increase in lipocalin-1 in KC patients. A 1.43-fold decrease was observed for cystatin-S by 2-DE, and 1.69- and 1.56-fold for cystatin-SN and cystatin-SA by LC–MS, respectively. The increase in lipocalin-1 was observed by both methods with fold changes of 1.26 in the 2-DE approach and 1.31 according to LC–MS. Significant protein upregulation was also observed for Ig-κchain C and Ig J chain proteins by 2-DE. Levels of lipophilin-C, lipophilin-A, and phospholipase A2 were decreased in tears from KC patients according to LC–MS. Serum albumin was found to be increased in KC patients according to LC–MS.
Conclusion
The results show differences in the tear protein profile of KC and control subjects. These changes are indicative of alterations in tear film stability and in interactions with the corneal surface in KC patients.
Keywords: keratoconus, tear, liquid chromatography, mass spectrometry, profile
Introduction
Keratoconus (KC) is a bilateral and progressive degenerative corneal ectasia. In this disorder, the keratoconic cornea assumes a conical shape, inducing corneal thinning, irregular astigmatism, myopia, and central or paracentral conical protrusion.1
The reported incidence of KC is 1 in 2000 in the general population, and it is the main reason for corneal transplantation in the Western world.2 Nevertheless, the etiology and the factors governing disease progression are currently unknown. It has been suggested that thinning of the cornea may be due to defective formation or destruction of the extracellular matrix as a consequence of altered or abnormal levels of fibronectin and type VI collagen.3 In the early stages of KC, the cell membrane is disrupted, the basal cell layer disappears,4 and particulate material is deposited between the surface of basal epithelial cells and Bowman's layer.5 As the basal cells degenerate, they may release proteolytic enzymes, which can destroy underlying tissue.6
Biochemical and immunohistological studies of KC corneas have suggested that the loss of corneal stroma could be caused by increased levels of proteases and other catabolic enzymes, or by decreased levels of protease inhibitors, such as α1-proteinase inhibitor and α2-macroglobulin.7 On the other hand, Lema and Duran8 found that in the tears of patients with KC, proinflammatory cytokines such as IL-6 and TNF-α, together with matrix metalloproteinase (MMP)-9, are overexpressed and that this overexpression is related to the degree of progression of KC, despite the fact that this disease is typically considered to be non-inflammatory in nature.
Proteomic-based techniques can provide quantitative measurements of changes that occur under physiologically altered conditions. Two-dimensional gel electrophoresis (2-DE)-based separation, followed by image analysis and mass spectrometry (MS), is currently the standard method for protein separation, relative quantification, and identification and it is widely used to compare protein expression profiles in healthy with diseased conditions.9, 10 Recently, alternative gel-free and MS-based protein quantification strategies have emerged.11 Gel-free methods can effectively overcome some of the limitations of 2-DE, such as multiple protein identification in a single spot or resolution problems at high and low pH. Therefore, both 2-DE and gel-free methods can be used as complementary approaches to reveal richer information about protein expression profiles.
Thus, the aim of this study was to identify proteins that are specifically altered in tears from patients with KC compared with healthy subjects using proteomics techniques. These newly identified proteins are likely to have a significant role in the disease and may therefore shed fresh light on the events occurring in tears that intervene in the pathophysiology of KC.
Subjects and methods
Subjects
Patients and control subjects were recruited from the Cornea and Ocular Surface Unit, ICQO, Bilbao, Vizcaya, Spain. This research was conducted by medically qualified personnel after approval by the ICQO Ethical Committee and in compliance with the tenets of the Declaration of Helsinki. Informed consent was obtained from all patients after the nature and possible consequences of the study had been explained. Tear samples were obtained from 12 patients with KC and from 12 controls who did not present any ocular disease. The exclusion criteria included the presence or history of any systemic or ocular disorder or condition (including ocular surgery, trauma, and disease), which could possibly interfere with the interpretation of the results. Current or recent use of topical ophthalmic or systemic medications that could affect tear status, and the wearing of contact lenses, was also motive of exclusion from this study.
Ophthalmic examinations consisted of best-corrected visual acuity measurements, slit-lamp examination, and corneal topography. Orbscan II (Bausch and Lomb, Salt Lake City, UT, USA) was used for corneal topography. Topographic data were evaluated by means of Rabinowitz criteria for the diagnosis of KC: central corneal power, inferior–superior dioptric asymmetry, and central corneal power differences between the two eyes. The stage of KC was graded as mild when the steepest keratometric reading (K2) was <45 diopters (D), moderate if K2 was between 45 and 52 D, and severe with K2>52 D. We considered K2 as the quantitative clinical variable to assess the severity of KC.
Tear samples
For all experiments, tears were collected using sterile 10 μl micropipettes (BLAUBRAND, Intramark, Wertheim, Germany). After collection, the tear sample was introduced into a 0.5-ml tube (Eppendorf, Fremont, CA, USA) and the samples were centrifuged at 13 000 g for 15 min and stored frozen at −80°C until use. The total protein concentration in each tear sample was measured using Bradford Reagent (Sigma-Aldrich, St Louis, MO, USA) in a NanoDrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's instructions.
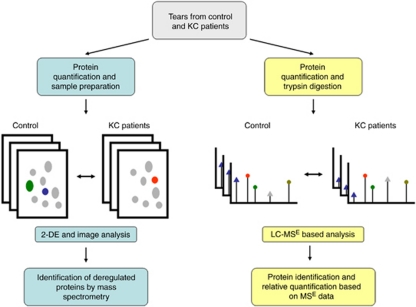
Tear proteins in control and KC patients were semiquantified using two experimental approaches, 2-DE and liquid chromatography–MS (LC–MS). Figure 1 is a work flow diagram of the experiments performed in this study.
Figure 1.
Flowchart of the experimental approach employed in this study. The left branch summarizes the 2-DE-based analysis. Gray spots represent proteins whose expression level is unaltered in KC samples. Green, blue, and red spots represent differentially expressed proteins, which are identified by MS. The right branch illustrates the LC–MS-based approach. Triangles indicate differentially expressed peptides, whereas circles represent peptides whose expression remains unaltered in KC samples. The colour reproduction of this figure is available at the Eye journal online.
Two-dimensional gel electrophoresis
Two-dimensional electrophoresis was carried out on tears from six controls subjects and six patients with KC. In all, 40 μg of protein from each sample was separated in immobilized pH gradient strips with an 11-cm format (3–10 pH range). First-dimension protein separation was performed as follows: 250 V for 20 min, followed by 8000 V for 2 h, and finally 20 000 V/h. The second-dimension SDS-PAGE separation was carried out using a Precast Criterion 8–16% gradient gel in a Dodeca Criterion Cell apparatus (Bio-Rad Laboratories, Hercules, CA, USA). The gels were run at 200 V for 90 min. SDS-polyacrylamide gels were then stained with SYPRO Ruby (Molecular Probes, Carlsbad, CA, USA) and the gel images were captured on a VersaDoc Model 4000 Imaging System (Bio-Rad Laboratories).
Gel images were exported as tiff files with PDQuest 2D gel image analysis software (Bio-Rad Laboratories) and analyzed using Progenesis Work Station Software (Nonlinear Dynamics, Newcastle, UK). The images were aligned and automatically analyzed by comparing the control group with the KC group gels. Subsequently, the spots found to be differently expressed between groups were detected and ordered according to the P-value for the Student's t-distribution (P<0.05). Normalized volume was calculated as the estimated volume of each spot. Spot volume was defined as the optical density of a spot for the area occupied by a spot. Differences in intensity were determined to be significant based on the normalized value. Subsequently, all protein spots showing statistically significant differences in protein expression were manually excised from the gels and subjected to protein identification.
Matrix-assisted laser desorption/ionization–TOF analyses
Spots were cut from 2-DE gels and subjected to in-gel trypsin digestion according to Shevchenko12 with minor modifications. After 30 min, the supernatant was removed and discarded. Then, 20 μl of 50 mM NH4HCO3 was added to the gel piece and the digestion was allowed to proceed at 37°C overnight. Supernatants of each sample were pooled and dried by vacuum centrifugation. Before MS analysis, pellets were resuspended in 10 μl 0.1% trifluoroacetic acid (TFA).
Recovered peptides were purified before MALDI analysis by custom-made nano-columns as described by Gobom,13 with some modifications including a column consisting of 100–300 nl of POROS R2 material (PerSeptive Biosystems, Framingham, MA, USA). The column was equilibrated with 0.1 % TFA and the bound peptides subsequently eluted directly onto the MALDI target with 0.5 μl CHCA solution (20 μg/μl in acetonitrile, 0.1% TFA, 70:30, vol/vol).
Peptide mass fingerprinting was performed on a Bruker Ultraflex TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). Positively charged ions were analyzed in reflector mode using delayed extraction. The spectra were obtained by randomly scanning the sample surface. Typically 600–800 spectra were averaged to improve the signal-to-noise ratio. Spectra were externally calibrated, resulting in a mass accuracy of <50 p.p.m. Protein identification was performed by searching in a non-redundant protein database (UniProtKB/Swiss-Prot, European Bioinformatics Institute, Hinxton, UK) using the Mascot search engine (Matrix Science Ltd, London, UK). MALDI–TOF analysis was performed by the proteomic platform of CIC bioGUNE, which is a member of the ProteoRed network.
Liquid chromatography–mass spectrometry
LC–MS analyses were carried out on tears from six different control subjects and six patients with KC. In all, 10 μg of tear proteins was digested with trypsin (recombinant, proteomics grade, Roche Diagnostics, Penzberg, Germany) overnight at 37°C in the presence of 0.1% RapiGest (Waters, Mildford, MA, USA). After RapiGest inactivation, protein concentration was diluted to a final concentration of 0.1 μg/μl with 0.1% formic acid. LC–MSE analysis was performed in a SYNAPT HDMS mass spectrometer (Waters) interfaced with a NanoAcquity UPLC System (Waters). In all, 5 μl (0.5 μg of protein) of each sample was loaded onto a Symmetry 300 C18, 180 μm × 20 mm precolumn (Waters) and washed with 0.1% formic acid for 3 min at a flow rate of 5 μl/min. The precolumn was connected to a BEH130 C18, 75 μm × 200 mm, 1.7 μm (Waters) and equilibrated in 3% acetonitrile and 0.1% formic acid. Peptides were directly eluted onto a NanoEase emitter (Waters). Mass spectra were acquired using a data-independent acquisition mode (MSE) described by Silva et al.14 Briefly, 1 s alternate MS acquisitions were performed at low (6 eV) and high (12-35 eV ramping) collision energies, and the radio frequency offset was adjusted such that the MS data were acquired from m/z (mass to charge ratio) 300 to 2000. [Glu1]-fibrinopeptide B (Sigma-Aldrich) at a concentration of 50 fmol/μl was sprayed through the NanoLockSpray (Waters) source and sampled every 30 s. Obtained spectra were processed with ProteinLynx Global Server 2.4 Build RC7 (Waters) using the doubly protonated monoisotopic ion of [Glu1]-fibrinopeptide B for mass correction. Protein identification was obtained with the embedded database search algorithm of the program,15 and a human UniProtKB/Swiss-Prot database (version 2010_05) was used. For protein identification, the following parameters were adopted: carbamidomethylation of C as fixed modification; N-terminal acetylation, N and Q deamidation, and M oxidation as variable modifications; and 1 missed cleavage and automatic precursor and fragment error tolerance. A maximum false-positive rate of 4% was allowed.
Relative quantification of protein
Data sets were normalized using the ‘auto-normalization' function of ProteinLynx Global Server software,16 and quantitative analyses were performed by comparing the normalized peak area/intensity of identified peptides in control vs diseased samples. The differentially expressed protein data set was filtered by considering only those identified proteins that replicated in at least three out of six biological samples for each condition. Furthermore a 95% up- or downregulation likelihood ([P>1]<0.05 or [P>1]>0.95) and a fold change higher than 50% (ratio KC/control <0.66 or >1.5) were considered to be indicative of significantly altered levels of expression.
Results
No statistically significant age- or sex-related differences were detected between KC patients and control subjects. Patients with KC presented a mean age of 33.2±5.11 years (8 women and 4 men) and control individuals presented a mean age of 29.33±1.75 years (7 women and 5 men). Four patients were considered to have mild KC (K2<45 D), seven patients had moderate KC (45 D<K2<52 D), and severe KC (K2>52 D) was diagnosed in one patient. In this study, we did not find any correlation between protein expression and KC grade or age.
In contrast, we found that the concentration of total protein in KC tears was reduced (2.8 μg/μl) in comparison with that of the control group (4.25 μg/μl). These differences were statistically significant (P=0.006).
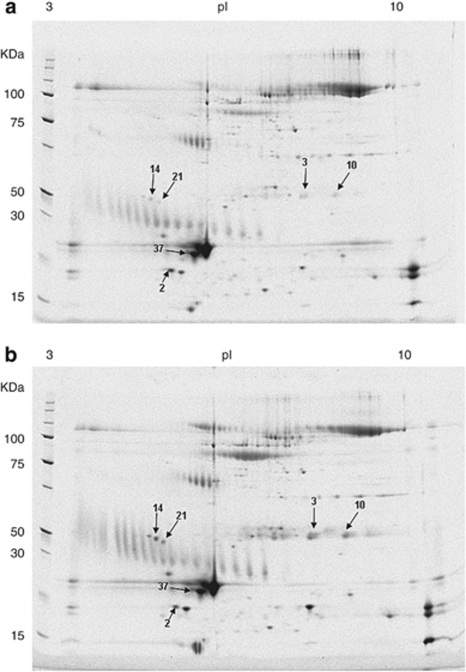
Tear proteins in control and KC patients were semiquantified using two experimental approaches, 2-DE and LC–MS. Tear samples were collected and randomly divided before their use in the 2-DE approach (Figure 2) or in the LC–MS-based quantification experiments to avoid experimental bias. In the gel-based approach, after protein quantification, samples from control and KC patients were run in parallel to 2-DE gels and protein spots, which were determined to be differentially expressed, excised, and identified by MS. In a similar way, in the gel-free approach, protein content in tears was quantified and samples were serially analyzed by LC–MSE. In this case, control and KC samples were alternated in consecutive LC runs. Relative protein quantification is based on signal intensity comparison and is performed after protein identification.
Figure 2.
Representative SYPRO Ruby-stained 2-DE gels of tear proteins. Total protein (40 μg) from tear samples was separated on a pH 3–10 immobilized pH gradient strip in the first dimension and by 8–16% SDS-PAGE in the second dimension. (a) 2-DE gel loaded with healthy individual tear samples. (b) 2-DE gel with KC patient tear samples. Arrows illustrate identified protein spots. Cystatin-S (2), Ig-κ chain C region (3, 10), Ig J chain (14, 21), and lipocalin-1 (37).
Both techniques, 2-DE and LC–MSE, have an optimum performance under certain conditions of total protein amount. Sample under- or overloading can compromise the sensitivity, resolution, and dynamic range of the analysis and therefore the quantification results. In this kind of approaches, the same amount of total protein has to be used in the experimental analysis. If the initial protein concentration is different among samples, the possible effect of this fact has to be taken into account.
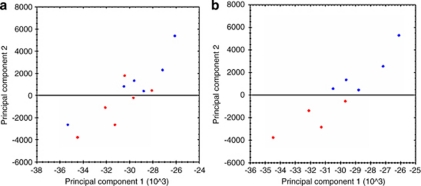
In the 2-DE proteomic analyses, the first step was to carry out the principal component analysis (PCA), which indicates gel distribution and similarities between gels (Figure 3). This analysis indicated that the gels corresponding to the control group (lower quadrant, red symbols) were quite different to those of the group of patients with KC (upper quadrant, blue symbols), with both groups in turn being similar among themselves. On the basis of PCA analysis, two samples from the control group and one sample from the KC group were excluded because they exhibited a high distribution variation within their respective groups (Figures 3a and b).
Figure 3.
Principal component analysis of spot volume data. The figure illustrates the two principal components (PC1 and PC2) that explain the majority of variation in the data set, plotted against each other. Each individual gel is displayed by means of filled circles (score plot) in which control gels (red) and KC gels (blue) are indicated. The closer the circles, the more similar the proteomes. Two samples from the control group and one sample from the KC group were excluded because they exhibited a high distribution variation within their respective groups (a, b).
Comparing the whole proteome of control vs KC groups, a total of six changed protein spots were identified (Figure 2). Spot quantification was assessed by means of computerized densitometry analysis using image analysis software to calculate the relative percentage of each spot of interest within the spots in the gel image taken as the whole.
Statistical analysis indicated that these deregulated spots are statistically valid according to their Student's t-values (P<0.05). The deregulated spots were subjected to MALDI–TOF identification and four different proteins were identified (Table 1). Ig-κ chain C region (P01834) and Ig J chain (P01591) were identified twice in two spots, with different isoelectric points indicating the presence of two distinctly modified forms of these proteins. Expression of both immunoglobulins was significantly increased in KC patients. Lipocalin-1 (P31025) was also significantly increased in KC tears. However, the expression of cystatin-S (P01036), a protein belonging to the cystatin protein family and present in tears, was found to be reduced in KC patients.
Table 1. Proteins whose expression is significantly deregulated in keratoconus tears according to the 2D gel electrophoresis method.
LC–MS analyses were performed using a relatively new data acquisition mode, MSE, or data-independent acquisition.14 Protein identifications were filtered following replication rate, as described in the Subjects and methods section. In the control samples, a total of 33 proteins were identified in three or more samples and 67% of them (22 proteins) were identified in all the six control samples. In the case of KC patients, 39 proteins were identified in three or more samples and 64% of them (25 proteins) in all the six. A total of 27 proteins were identified in both conditions, and therefore were subjected to relative quantification analysis (Table 2). On the basis of the significant regulation level considered in this study, (95% up- or downregulation likelihood and 50% fold change), six proteins were considered to be differentially expressed in the KC samples. Two lipophilins, lipophilin-A and -C, were considered to be downregulated in KC tears, with ratios of 0.45 and 0.25, respectively. Phospholipase A2 was also found to be downregulated in KC tears, with a ratio of 0.52. Protein levels of two isoforms of human cystatin, cystatin-SN and cystatin-SA, were decreased in patient samples, with ratios of 0.59 and 0.64, respectively. Results obtained in the 2-DE analysis indicate that cystatin-S protein was also downregulated in KC patients, which in conjunction with the LC–MS results suggest a general decrease in proteins of the cystatin family. Lipocalin-1, a protein that was found to be upregulated in patient samples by 2-DE analysis, was also found to be upregulated according to the LC–MS analysis, with an upregulation likelihood >95%, although the fold change was slightly <50 % (KC/control ratio=1.31; Table 2, italics). Finally, levels of serum albumin were also found to be increased in KC tears according to the LC–MS approach.
Table 2. Relative quantification of proteins identified in three or more samples in control and KC patients.
| Accession | Entry name | Protein name | Replicate rate: control | Replicate rate: patient | Ratio: patient/ control | Regulation likelihood |
|---|---|---|---|---|---|---|
| O75556a | SG2A1_HUMAN | Mammaglobin-B/lipophilin-C | 6 | 6 | 0.25 | 0 |
| O95968a | SG1D1_HUMAN | Secretoglobin family 1D member 1/lipophilin-A | 6 | 6 | 0.45 | 0 |
| P14555a | PA2GA_HUMAN | Phospholipase A2, membrane associated | 6 | 4 | 0.52 | 0 |
| P01037a | CYTN_HUMAN | Cystatin-SN | 5 | 4 | 0.59 | 0 |
| P09228a | CYTT_HUMAN | Cystatin-SA | 3 | 3 | 0.64 | 0.02 |
| Q5VSP4 | LC1L1_HUMAN | Putative lipocalin-1-like protein 1 | 5 | 5 | 0.7 | 0.14 |
| P61769 | B2MG_HUMAN | β2-Microglobulin | 5 | 6 | 0.73 | 0.04 |
| P01833 | PIGR_HUMAN | Polymeric immunoglobulin receptor | 6 | 6 | 0.75 | 0 |
| P01591 | IGJ_HUMAN | Immunoglobulin J chain | 6 | 6 | 0.76 | 0 |
| P25311 | ZA2G_HUMAN | Zinc-α2-glycoprotein | 6 | 6 | 0.77 | 0 |
| Q96DA0 | ZG16B_HUMAN | Zymogen granule protein 16 homolog B | 4 | 6 | 0.88 | 0.17 |
| Q9GZZ8 | LACRT_HUMAN | Extracellular glycoprotein lacritin | 6 | 6 | 0.89 | 0.16 |
| P01036 | CYTS_HUMAN | Cystatin-S | 6 | 6 | 0.9 | 0.24 |
| P12273 | PIP_HUMAN | Prolactin-inducible protein | 6 | 6 | 0.92 | 0.11 |
| Q16378 | PROL4_HUMAN | Proline-rich protein 4 | 4 | 6 | 0.92 | 0.09 |
| P01877 | IGHA2_HUMAN | Ig-α2 chain C region | 6 | 6 | 0.93 | 0.25 |
| P02788 | TRFL_HUMAN | Lactotransferrin | 6 | 6 | 0.99 | 0.22 |
| P01876 | IGHA1_HUMAN | Ig-α1 chain C region | 6 | 6 | 1 | 0.54 |
| P10909 | CLUS_HUMAN | Clusterin | 4 | 4 | 1.03 | 0.53 |
| P61626 | LYSC_HUMAN | Lysozyme C | 6 | 6 | 1.03 | 0.87 |
| P01834 | IGKC_HUMAN | Ig-κ chain C region | 6 | 6 | 1.08 | 0.76 |
| Q08380 | LG3BP_HUMAN | Galectin-3-binding protein | 3 | 3 | 1.11 | 0.61 |
| Q99935 | PROL1_HUMAN | Proline-rich protein 1 | 6 | 6 | 1.25 | 0.98 |
| P03973 | SLPI_HUMAN | Antileukoproteinase | 6 | 5 | 1.26 | 0.89 |
| P31025 | LCN1_HUMAN | Lipocalin-1 | 6 | 6 | 1.31 | 1 |
| P01842 | LAC_HUMAN | Ig-γ chain C regions | 6 | 6 | 1.35 | 0.93 |
| P02768a | ALBU_HUMAN | Serum albumin | 6 | 6 | 3.29 | 1 |
Indicates proteins whose expression is considered to be deregulated based on a >1.5-fold change and a >95% regulation likelihood ([P>1]<0.05 or [P>1]>0.95) filtering criteria. In italics, data regarding the lipocalin-1 protein.
Discussion
Studies of the tear proteome by electrophoretic separation and MS techniques have permitted an identification of the pattern of tear proteins associated with specific pathological conditions17, 18 and the identification of over 500 proteins whose expression is altered in the pathology.18, 19, 20 In the present study, we have employed 2-DE and LC–MS techniques to identify proteins whose expression is altered in the tears of patients with KC. Our objective was to identify proteins that may have a significant role in this pathology, thereby throwing fresh light on the specific events that occur in tears and may intervene in the pathophysiology of KC.
The proteins that we identified and quantified included cystatin, lipocalin-1, lipophilin-A and -C, and phospholipase A2. We found a decrease in the levels of cystatin family proteins in KC tears. Cystatins are natural inhibitors of cysteine proteinases. These proteinases are one of the most abundant protein-degrading enzymes in mammalian cells.21 They are involved in the initial phases of degradation of intracellular proteins and can provoke tissue degradation after being liberated into the extracellular medium. The activity of cysteine proteinases is controlled by their physiological inhibitors, the cystatins, which are known to be generally present in tears.22 Extracellular cystatins have a protective role against the damaging effects of lysosomal proteinases, which can be secreted under physiological conditions for the degradation and regeneration of tissues, as well as under pathological conditions associated with infection by bacteria or viruses, or with the corneal and conjunctival epithelium.23 The decreased levels of cystatins that we found in KC tears is indicative of an increase in the degradation of tear proteins, which would explain the decrease that we found in the concentration of total protein in KC tears.
Tear lipocalin is a protein that is synthesized by the lacrimal gland and constitutes 15–30% of the total protein content of the tear film. Its principal action is associated with its capacity to bind tear lipids, such as cholesterol, fatty acids, phospholipids, and glycolipids.24, 25 Lipocalin under normal conditions favors the binding of these lipids, thus avoiding the contamination and desiccation of the corneal epithelium26, 27 and provides optimal levels of surface tension.
To date, few researchers have studied the implication of lipocalin in corneal pathology. Glasson28 found a higher concentration of this protein in patients who presented intolerance to contact lenses compared with control subjects. This would seem to contradict the protective role of this protein in the maintenance of the stability and integrity of the epithelium. Nevertheless, the authors speculated that this increase in lipocalin could be the result of an increase in the concentration of lipids derived from processes of peroxidation, which occur in the tears of subjects who wear contact lenses.28 On the basis of our results, we suggest that the increase in lipocalin-1 in the tears of patients with KC is not due to contact lens wear because our patients were not contact lens wearers, as this was a study exclusion criterion; however, this increase in lipocalin-1 may be due to an increase in the concentration of lipids in the tears derived from the peroxidation processes occurring in the keratoconic corneas,29 and it could affect the stability of the corneal epithelium.
Secretory lipophilins are ‘lipid-loving' proteins that are major constituents of several mammalian secretions including the tears of humans. These proteins are small molecules with anti-inflammatory properties that bind steroids and are subjected to their regulation. Although the functions of secretory lipophilins are imperfectly understood, their abundance in glandular secretions and hormone-responsive tissues suggests that they deserve considerably more attention than they have received to date.30 Our results showed a decrease in the levels of lipophilin-A and -C in tears from KC patients vs controls as reported by Versura et al in dry eye patients.31 Thus, decreased levels of lipophilin-A and -C in tears from KC patients may lead to altered tear stability due to higher levels of free lipid in tears. When the lipid layer is abnormal, tear film evaporates faster, which in turn leads to rapid tear dissociation times and finally to epithelial cell damage.
Moreover, lipophilins are proteins with anti-inflammatory properties. When their concentration in the tear decreases, inflammatory processes may be more active in the ocular surface.8 Understanding these processes are essential to establish more specific treatments for these patients.
We also found a similar decrease in the concentration of phospholipase A2 in tear samples from KC patients. Phospholipase A2 is known to catalyze the hydrolysis of phospholipid in tears.32 A decrease in the concentration of phospholipase A2 could promote tear film instability by increasing the abundance of phospholipid in tears.
Determining the route of hydrolysis of phospholipids in the tears could be an appealing strategy for future research in KC.
The presence of serum albumin in tears is indicative of blood–ocular barrier failure in conjunctival vessels, and its value rapidly increases under various pathophysiological conditions.33 Our data showed a significant increase (greater than threefold) in albumin in tears from KC tears vs controls, indicating that exudation of albumin from blood vessels had occurred.
Finally, we also found overexpression of κ-C chain and J chain immunoglobulins in pathological tears. Organisms may regulate the synthesis of these proteins in response to aggression induced by pathology, thus leading to elevated levels associated with the pathological stage. Nevertheless, these postulates should be verified in posterior studies because of the limited number of samples.
The globally reduced level of proteins in KC tears found in our study is also of interest. Previously, Rabinowitz34 reported a reduction in the total amount of protein in KC corneas, which appears to be due to higher levels of lysosomal degradative enzymes and MMPs, in addition to a reduction in protease inhibitors in corneal and conjunctival tissue. Recent studies have demonstrated the involvement of proteases in KC.35 The decrease in the amount of tear proteins that we found may well be due to increased levels of proteases in tears.
The present study illustrates the utility and importance of analyzing components of tears as a source of knowledge about the pathogenic mechanisms implicated in ocular surface disorders. Studies of KC have demonstrated the presence of alterations in the corneal epithelium36 and stroma.37 But complementary and useful information can also be obtained by biochemical analysis of tears, which are relatively easy to obtain. The stable tear film is prerequisite for the proper functioning of optical and metabolic eye. It is important to know and understand the terms related to tear stability, phenomena involved in different proposed theories, and techniques to evaluate it to make more accurate diagnosis, enabling effective treatments.
We have shown that the protein composition of tears is altered in KC either by increased levels of proteases in tears, as there is a decrease in total protein concentration in the tear, or a decrease in protein with anti-inflammatory properties. Our analyses shows that tear proteomic techniques can assist in etiological studies of KC. The differential expression of tear film proteins, such as cystatin, lipocalin-1, lipophilin, and phospholipase A2, can be found in KC subjects. These findings suggest that further testing could help determine whether these molecules have a role in the etiology of KC.
Our study demonstrates that the protein composition of tears is altered in KC; what remains to be elucidated is whether this alteration is a consequence of events in the corneal epithelium or a direct contributor to the development of the disease.
Acknowledgments
We are grateful for financial support from the Spanish Ministerio de Ciencia y Tecnología (SAF2007-62060), Basque Government Grupos Consolidados (IT437-10), Ayudas Investigación Enfermedades Raras (BIOEF), Red Patología Ocular (RETICS-RD07/0062), and Basque Country Government (INTEK program W6088). Proteomics Core Facility-SGiker is a member of ProteoRed and is supported by UPV/EHU, MICINN, GV/EJ, and ESF.
The authors declare no conflict of interest.
References
- Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- Kennedy RH, Bourne WM, Dyer JA. A 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol. 1986;101:267–273. doi: 10.1016/0002-9394(86)90817-2. [DOI] [PubMed] [Google Scholar]
- Kenney MC, Nesburn AB, Burgeson RE, Butkowski RJ, Ljubimov AV. Abnormalities of the extracellular matrix in keratoconus corneas. Cornea. 1997;16:345–351. [PubMed] [Google Scholar]
- Teng CC. Electron microscope study of the pathology of keratoconus: I. Am J Ophthalmol. 1963;55:18–47. doi: 10.1016/0002-9394(63)91645-3. [DOI] [PubMed] [Google Scholar]
- Iwamoto T, Devoe AG. Particulate structures in keratoconus. Arch Ophthalmol Rev Gen Ophthalmol. 1975;35:65–76. [PubMed] [Google Scholar]
- Kao WW, Vergnes JP, Ebert J, Sundar-Raj CV, Brown SI. Increased collagenase and gelatinase activities in keratoconus. Biochem Biophys Res Commun. 1982;107:929–936. doi: 10.1016/0006-291x(82)90612-x. [DOI] [PubMed] [Google Scholar]
- Sawaguchi S, Twining SS, Yue BY, Wilson PM, Sugar J, Chan SK. Alpha-1 proteinase inhibitor levels in keratoconus. Exp Eye Res. 1990;50:549–554. doi: 10.1016/0014-4835(90)90044-u. [DOI] [PubMed] [Google Scholar]
- Lema I, Durán J. Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology. 2005;112:654–659. doi: 10.1016/j.ophtha.2004.11.050. [DOI] [PubMed] [Google Scholar]
- O'Farrel PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Steel LF, Haab BB, Hanash SM. Methods of comparative proteomic profiling for disease diagnostics. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:275–284. doi: 10.1016/j.jchromb.2004.10.072. [DOI] [PubMed] [Google Scholar]
- Bachi A, Bonaldi T. Quantitative proteomics as a new piece of the systems biology puzzle. J Proteomics. 2008;71:357–367. doi: 10.1016/j.jprot.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Gobom J, Nardhoff E, Mirgorodskaya E, Ekman R, Roepstorff P. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 1999;34:105–116. doi: 10.1002/(SICI)1096-9888(199902)34:2<105::AID-JMS768>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- Chambery A, Colucci-D'Amato L, Vissers JP, Scarpella S, Langridge JI, Parente A. Proteomic profiling of proliferating and differentiated neural mes-c-myc A1 cell line from mouse embryonic mesencephalon by LC-MS. J Proteome Res. 2009;8:227–238. doi: 10.1021/pr800454n. [DOI] [PubMed] [Google Scholar]
- Tomosugi N, Kitagawa K, Takahashi N, Sugai S, Ishikawa I. Diagnostic potential of tear proteomic patterns in Sjögren's syndrome. J Proteome Res. 2005;4:820–825. doi: 10.1021/pr0497576. [DOI] [PubMed] [Google Scholar]
- De Souza GA, Godoy LM, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biol. 2006;7:R72. doi: 10.1186/gb-2006-7-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grus FH, Podust VN, Bruns K, Lackner K, Fu S, Dalmasso EA. SELDI-TOF_MS ProteinChip array profiling of tears from patients with dry eye. Invest Ophthalmol Vis Sci. 2005;46:863–876. doi: 10.1167/iovs.04-0448. [DOI] [PubMed] [Google Scholar]
- Zhou L, Beuerman RW, Chan CM, Zhao SZ, Li XR, Yang H, et al. Identification of tear fluid biomarkers in dry eye syndrome using iTRAQ quantitative proteomics. J Proteome Res. 2009;8 (11:4889–4905. doi: 10.1021/pr900686s. [DOI] [PubMed] [Google Scholar]
- Bond JS, Butler PE. Intracellular proteases. Annu Rev Biochem. 1987;56:333–364. doi: 10.1146/annurev.bi.56.070187.002001. [DOI] [PubMed] [Google Scholar]
- Barka T, Asbell PA, Van der Noen H, Prasad A. Cystatins in human tear fluids. Current Eye Res. 1991;10:25–34. doi: 10.3109/02713689109007608. [DOI] [PubMed] [Google Scholar]
- Takahasi M, Honda K, Ogawa K, Barka T. Localization of cystatins in the exorbital lacrimal gland of the rat. Acta Histochem Cytochem. 1990;22:738. doi: 10.1111/j.1755-3768.1992.tb02143.x. [DOI] [PubMed] [Google Scholar]
- Gasymov OK, Abduragimov AR, Prasher P, Yusifov TN, Glasgow BJ. Tear lipocalin: Evidence for a scavenging function to remove lipids from the human corneal surface. Invest Ophthalmol Vis Sci. 2005;46:3589–3596. doi: 10.1167/iovs.05-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow OK, Abduragimov AR, Farahbakhsh Z, Faull KF, Hubbell WL. Tear lipocalins bind a broad array of lipid ligands. Curr eye Res. 1995;14:363–372. doi: 10.3109/02713689508999934. [DOI] [PubMed] [Google Scholar]
- Holly FJ, Lemp MA. Wettability and wetting of corneal epithelium. Exp Eye Res. 1971;11:239–250. doi: 10.1016/s0014-4835(71)80028-3. [DOI] [PubMed] [Google Scholar]
- Sharma A. Energetics of corneal epithelial cell-ocular mucus-tear film interactions: some surface-chemical pathways of corneal defense. Biophys Chem. 1993;47:87–99. doi: 10.1016/0301-4622(93)80036-i. [DOI] [PubMed] [Google Scholar]
- Glasson MJ, Stapleton F, Wilcox MDP. Lipid, lipase and lipocalin differences between tolerant and intolerant contact lens wearers. Curr Eye Res. 2002;25:227–235. doi: 10.1076/ceyr.25.4.227.13482. [DOI] [PubMed] [Google Scholar]
- Kenney MC, Brown DJ. The cascade hypothesis of keratoconus. Cont Lens Anterior Eye. 2003;26:139–146. doi: 10.1016/S1367-0484(03)00022-5. [DOI] [PubMed] [Google Scholar]
- Lehrer RI, Nguyen T, Zhao C, Ha CX, Glasgow BJ. Secretory lipophilins: a tale of two species. Ann NY Acad Sci. 2000;923:59–67. doi: 10.1111/j.1749-6632.2000.tb05519.x. [DOI] [PubMed] [Google Scholar]
- Versura P, Nanni P, Bavelloni A, Blalock WL, Piazzi M, Roda A, et al. Tear proteomics in evaporative dry eye disease. Eye. 2010;24:1396–1402. doi: 10.1038/eye.2010.7. [DOI] [PubMed] [Google Scholar]
- Van den Bosch H. Intracellular phospholipases A. Biochim Biophys Acta. 1980;604:191–246. doi: 10.1016/0005-2736(80)90574-x. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Fullard RJ, Willcox MD, Baleriola-Lucas C, Bestawros F, Sweeney D. Fibronectin in the tear film. Invest Ophthalmol Vis Sci. 1996;37:459–467. [PubMed] [Google Scholar]
- Rabinowitz YS, Dong L, Wistow G. Gene expression profile studies of human keratoconus cornea for NEIBank: a novel cornea-expressed gene and the absence of transcripts for aquaporin 5. Invest Ophthalmol Vis Sci. 2005;46:1239–1246. doi: 10.1167/iovs.04-1148. [DOI] [PubMed] [Google Scholar]
- Balasubramanian SA, Pye DC, Willcox MDP. Are proteinases the reason for keratoconus. Current Eye Research. 2010;35:185–191. doi: 10.3109/02713680903477824. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Vorum H, Fagerholm P, Birkenkamp-Demtröder K, Honoré B, Ehlers N, et al. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp Eye Res. 2006;82:201–209. doi: 10.1016/j.exer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Wilson SE, Liu JJ, Mohan RR. Stromal-epithelial interactions in the cornea. Prog Retin Eye Res. 1999;18:293–309. doi: 10.1016/s1350-9462(98)00017-2. [DOI] [PubMed] [Google Scholar]