Abstract
The lantibiotic nisin inhibits growth of vegetative Gram-positive bacteria by binding to lipid II, which disrupts cell wall biosynthesis and facilitates pore formation. Nisin also inhibits the outgrowth of bacterial spores, including spores of Bacillus anthracis, whose structural and biochemical properties are fundamentally different from those of vegetative bacteria. The molecular basis of nisin inhibition of spore outgrowth had not been identified, as previous studies suggested that inhibition of spore outgrowth involved either covalent binding to a spore target or loss of membrane integrity; disruption of cell wall biosynthesis via binding to lipid II had not been investigated. To provide insights into the latter possibility, the effects of nisin were compared with those of vancomycin, another lipid II binding antibiotic that inhibits cell wall biosynthesis but does not form pores. Nisin and vancomycin both inhibited the replication of vegetative cells, but only nisin inhibited the transition from a germinated spore to a vegetative cell. Moreover, vancomycin prevented nisin’s activity in competition studies, suggesting that the nisin-lipid II interaction is important for inhibition of spore outgrowth. In experiments with fluorescently labeled nisin, no evidence was found for a covalent mechanism for inhibition of spore outgrowth. Interestingly, mutants in the hinge region (N20P/M21P and M21P/K22P) that still bind lipid II but cannot form pores had potent antimicrobial activity against vegetative B. anthracis cells but did not inhibit spore outgrowth. Therefore, pore formation is essential for the latter activity but not the former. Collectively, these studies suggest that nisin utilizes lipid II as the germinated spore target during outgrowth inhibition and that nisin-mediated membrane disruption is essential to inhibit spore development into vegetative cells.
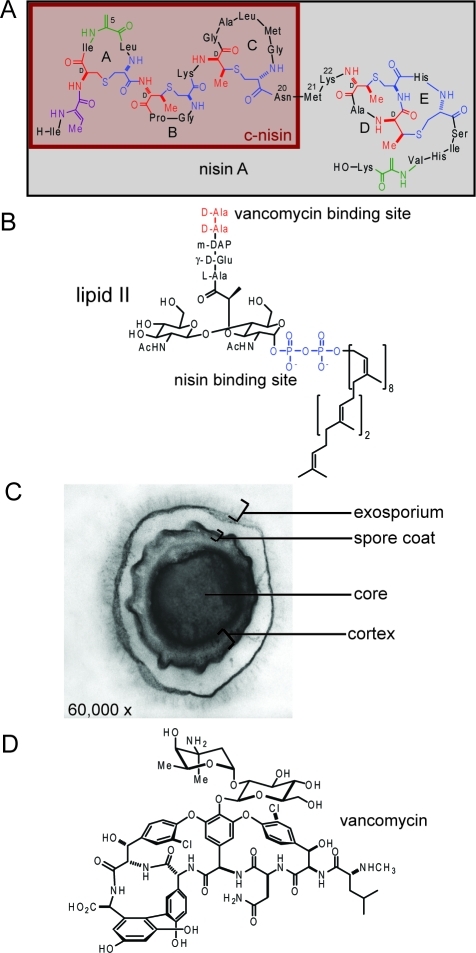
Lantibiotics are (methyl)lanthionine-containing antimicrobial peptides produced by a range of bacteria.(1) Nisin A is a 34-amino-acid polycyclic peptide produced by Lactococcus lactis sbsp. lactis ATCC 11454 (Figure 1A). It has emerged as the prototype for studying the antibacterial properties and structure–activity relationships of the lantibiotics.2,3 Nisin is ribosomally synthesized as a linear precursor peptide and then post-translationally modified, generating two dehydroalanines, one dehydrobutyrine, and five (methyl)lanthionine cross-links.1,3
Figure 1.
Structures of nisin, vancomycin, and lipid II. (A) The structure of nisin A. Dehydroalanine (Dha) and dehydrobutyrine (Dhb) are indicated in green and magenta, respectively. Lanthionine and methyllanthionine cross-links are indicated in red and blue, respectively. The numbers indicate the locations of the amino acids that were mutated. The N-terminal fragment resulting from chymotrypsin cleavage (c-nisin) is highlighted in maroon. (B) The structure of lipid II in B. anthracis. The vancomycin binding site is indicated in red, and the nisin binding site is indicated in blue. m-DAP: diaminopimelic acid. (C) Transmission electron microscopy image of a B. anthracis Sterne 7702 endospore. (D) The structure of vancomycin.
Nisin acts upon Gram-positive bacteria by two distinct mechanisms.(2) The compound forms pores in cell membranes(4) and inhibits cell wall biosynthesis by disruption of transglycosylation via binding to and mislocalization of lipid II.5,6 Lipid II (Figure 1B) is an essential intermediate for cell wall biosynthesis. With two mechanisms of action, nisin has been relatively unaffected by the emergence of microbial resistance, despite widespread and persistent use as a food preservative.3,7
Nisin also inhibits the outgrowth of germinated bacterial spores.8−12 Organisms from several families of bacteria, including the Bacillacaea, form small endospores in nutrient-deprived conditions, allowing survival over extended periods of time, which would not be possible as vegetative cells.13−15 The endospore structure provides protection in a wide range of environments including protection against oxidation, radiation, desiccation, heat, and extremes in pH.(16) The endospore consists of a dual membrane separated by a thick peptidoglycan cortex and two proteinaceous layers, the inner and outer spore coats, all surrounding an inner core containing the DNA (Figure 1C).13−15 Upon encountering more favorable conditions, dormant spores germinate and undergo sequential developmental changes resulting in outgrowth to vegetative cells.13−15 For pathogenic spore-forming bacteria, inhibition of spore outgrowth immediately after germination initiation is an ideal time for antibiotic intervention, because the spore has lost its dormancy and protective properties but has not yet started to produce the virulence factors that contribute to subversion of the host’s immune response and the progression of disease.14−19
A recent study with Bacillus anthracis suggested that membrane disruption by nisin prevents the establishment of a membrane potential and oxidative metabolism to inhibit outgrowth of germinated spores.(9) Whether these processes are mediated by lipid II was not established, nor could inhibition of cell wall biosynthesis be ruled out as a mode of action. At present, little is known regarding lipid II content and accessibility on spores. Moreover, several studies have suggested that covalent binding to a protein target mediates nisin’s inhibitory action on spore outgrowth.8,10,20 To better understand the mechanism of inhibition of spore outgrowth, the effects of nisin, vancomycin, and nisin analogues (Figure 1A,D) on germination and outgrowth of B. anthracis Sterne 7702 spores were compared; vancomycin and the nisin analogues bind to lipid II but do not form pores. These studies show that nisin does bind lipid II on spores but that lipid II binding is not sufficient for outgrowth inhibition, whereas membrane disruption is essential. In contrast, lipid II binding is sufficient for antimicrobial activity against vegetative cells.(6) Using fluorescently labeled nisin analogues, no evidence was found for a covalent mechanism of inhibition.
Methods
Spore Preparations
Spores were prepared from B. anthracis Sterne 7702 or B. anthracis 34F2, as described previously.(21) Both strains do not contain the plasmid pX02 that encodes the genes for capsule production. Enumeration of spores was performed using a Petroff-Hauser hemocytometer under a light microscope at 400x magnification (Nikon Alphaphot YS). A typical spore preparation yielded 10 mL of spores at a concentration of 2.0 × 109 spores/mL.
Labeling of Nisin and Vancomycin
Nisin was purified and characterized as described previously.(9) Nisin was reacted with NHS-fluorescein (Pierce) or with NHS-BODIPY-633 (Invitrogen) to generate analogues with a single fluorescein group (f-nisin) or a single BODIPY group (b-nisin) after purification. The reactions were carried out according to the manufacturer’s protocols except that a 0.75:1 molar ratio of NHS-fluorophore and nisin was used. Labeling reactions were stopped with the addition of 100-fold molar excess of Tris after a 3 h reaction time at 25 °C. Reactions were analyzed via matrix assisted laser desorption ionization-time-of-flight (MALDI-TOF) and electrospray ionization (ESI) mass spectrometry (MS) (Applied Biosystems), which indicated that all nisin was consumed. Proteolytic digest of the fluorescently labeled compounds with trypsin and chymotrypsin and subsequent analysis by LC–MS showed that the label was located predominantly (>85%) at the N-terminus of b-nisin and at the C-terminal Lys of f-nisin. Vancomycin was labeled with NHS-rhodamine (Pierce) to obtain r-vancomycin, with NHS-fluorescein to generate f-vancomycin, or with NHS-BODIPY-633 to produce b-vancomycin as previously described.(22) Vancomycin labeling occurs on the amino group of the vancosamine sugar (Figure 1D).(22) All labeled compounds were purified by reverse phase-high performance liquid chromatography (RP-HPLC) utilizing a C4 semipreparative column (Waters) with a linear gradient of 0–100% acetonitrile with 0.1% trifluoroacetic acid over 40 min. Acetonitrile, TFA, and water were removed from fractions containing nisin or vancomycin by rotary evaporation followed by lyophilization. Prior to use, lyophilized nisin and vancomycin were dissolved in 0.1 M MOPS pH 6.8 to yield the desired concentration.
Truncation of Nisin with Chymotrypsin
Proteolysis of nisin with chymotrypsin was performed as previously described.(23) Reactions were analyzed by MALDI-TOF MS. The N-terminal chymotryptic segment (c-nisin, Figure 1A) was purified from full length nisin and the C-terminal fragment by RP-HPLC utilizing a C4 semipreparative column (Waters) with a linear gradient of 0–100% acetonitrile over 40 min. Acetonitrile, TFA, and water were removed from fractions containing c-nisin by rotary evaporation followed by lyophilization.
Culturing B. anthracis Spores
B. anthracis 34F2 mutants were graciously provided by Dr. David Popham (Virginia Tech). B. anthracis Sterne 7702, B. anthracis 34F2, B. anthracis 34F2 ΔsleB,(24)B. anthracis 34F2 ΔcwlJ1,(24) or B. anthracis 34F2 ΔsleBΔcwlJ1(25) spores at a concentration of 4.0 × 106 spores/mL were incubated in brain heart infusion medium (BHI; BD Bioscience) supplemented with nisin (at 1 or 10 μM), c-nisin (10 μM), nisin variants (S5A, N20P/M21P, or M21P/K22P at 10 μM), vancomycin (at 0.1, 1, 10, or 100 μM), or with 0.1 M 3-(N-morpholino)propanesulfonic acid (MOPS; Sigma) at pH 6.8 as a control. For nongerminating conditions, 0.1 M MOPS was substituted for BHI medium. All incubations were performed at 37 °C under aeration at 180 rpm on a rotary shaker and under ambient CO2.
Differential Interference Contrast (DIC) and Epi-fluorescence Microscopy
Live epi-fluorescence microscopy was performed by mounting samples on glass slides in 0.5% agarose under coverslips. Images were collected using an Applied Precision assembled DeltaVision EpiFluorescence microscope containing an Olympus Plan Apo 100x oil objective with NA 1.42 and a working distance of 0.15 mm. Images were processed and Pearson’s coefficient colocalization analysis of 50 spores per condition was performed using SoftWoRX Explorer Suite. Acquisition of epi-fluorescence images utilized FITC (Ex: 490/20; Em: 528/28), rhodamine (Ex: 555/28; Em: 617/73), and Cy5 (Ex: 640/20; Em: 685/40) to visualize fluorescein, rhodamine, and BODIPY-633 fluorescence, respectively.
Functional Competition Assay
Competitive binding of nisin and vancomycin was evaluated utilizing a functional assay in which vancomycin prevented nisin-mediated loss of membrane potential via lipid II binding.(26) Spores were incubated in BHI medium (germinating conditions), in the presence of 10 mM l-Ala and inosine (germinating conditions), or in 0.1 M MOPS pH 6.8 (nongerminating conditions) with 3,3′-diethyloxacarbocyanine iodide (DIOC2) for 60 min at 37 °C followed by incubation with 0 or 100 μM vancomycin for 2 min. Nisin was added to cultures and the effect on membrane potential disruption was immediately assayed with flow cytometry through 10,000 counts to observe the population of spores that exhibited reduced membrane potential in the presence of nisin. The MIC of vancomycin against B. anthracis is 0.5–3.5 μM,27−29 but the antimicrobial effects of vancomycin under the conditions used here are not manifested until spores have been germinated and incubated in the presence of vancomycin for 90 min, which is well after the time frame of interest in this investigation. Experiments investigating spore hydration, oxidative metabolism, membrane potential, and membrane integrity were performed as previously described.(9) Procedures used for competition binding experiments are described in the Supporting Information.
Site-Directed Mutagenesis
Mutagenesis of nisA was performed using the QuikChange mutagenesis kit from Stratagene. Reactions with complementary mutagenic primers (Supplementary Table S1) were performed as described in the QuikChange protocol with pRSFDuet-1nisAB as the plasmid template (30) for the generation of NisA S5A and M21P mutants. After cycling of the reaction mixture 18 times in a thermal cycler (MJ Research), the resulting mixture was digested with DpnI (New England Biolabs), and the resulting DNA was used to transform supercompetent E. coli DH5α cells. The resulting mutant plasmids were isolated, and the entire gene was sequenced to ensure that only the appropriate mutations were introduced. For double mutations, a second round of site-directed mutagenesis was performed utilizing pRSFDuet-1nisAB-M21P as the plasmid template for the generation of NisA N20P/M21P and M21P/K22P mutants.
Heterologous Production of Nisin and Nisin Mutants
Electro-competent E. coli BL21(DE3) cells were cotransformed with pRSFDuet-1 containing nisA variants and the nisB gene and pACYCDuet-1 containing the nisC gene(30) (Supplementary Table S2). Overnight culture grown from a single colony transformant was used as inoculum to grow 2 L of terrific broth medium (0.12% Pancreatic Digest of Casein, 0.24% Yeast Extract, BD Biosciences; 0.094% K2HPO4, 0.022% KH2PO4, Fisher Chemical) containing 50 mg/L kanamycin and 25 mg/L chloramphenicol at 37 °C until the OD600 nm reached about 0.6. The incubation temperature was then changed to 18 °C, and the culture was induced with 0.5 mM IPTG. The induced cells were shaken continually at 18 °C for an additional 18 h. The cells were harvested by centrifugation (10,000 × g for 10 min, Beckman JA-10 rotor). Purification of the modified precursor peptides is described in the Supporting Information.
Cleavage of Modified Prenisin with Trypsin and Purification of Nisin Variants
Modified prenisin (500 μM) and trypsin (30 μM, Worthington Biochemicals) were incubated at RT for 3 h with 150 rpm mixing on a platform shaker. The resulting mixture was checked by MALDI-TOF MS, and the desired proteolytic fragment corresponding to mature nisin or its variants were observed-calculated for heterologously expressed nisin (h-nisin): 3352.5152 m/z (M + H), observed 3352.5005 m/z; calculated for h-nisin S5A: 3354.5608 m/z (M + H), observed 3354.4946 m/z; calculated for h-nisin N20P/M21P: 3301.5673 m/z (M + H), observed 3301.7642 m/z; and calculated for h-nisin M21P/K22P 3287.9826 m/z (M + H), observed: 3287.8738 m/z. Nisin and variants were purified from the cleavage reaction via RP-HPLC utilizing a C4 semipreparative column with a linear gradient of 0–100% acetonitrile with 0.1% TFA over 40 min. The fractions containing nisin variants were lyophilized and analyzed by MALDI-TOF and ESI MS.
Results and Discussion
Fluorescently Labeled Nisin Analogues Behave Similarly to Wild-type Nisin
To further evaluate the mechanism by which nisin inhibits the outgrowth of germinated B. anthracis spores, bodipy-labeled nisin (b-nisin) and fluorescein-labeled nisin (f-nisin) were synthesized to probe nisin binding to spores using fluorescence microscopy. The fluorescein label, which was located predominantly at the C-terminal lysine residue of nisin, or the bodipy label, which was located predominantly at the N-terminus, did not have major deleterious effects on nisin activity, as fluorescently labeled nisin inhibited B. anthracis growth with IC90 values of 5.4 and 6.5 μM for b-nisin and f-nisin, respectively, which correspond to a 6- and 7-fold loss in activity, respectively, compared to unmodified nisin.(9) These findings are consistent with observations in previous reports using nisin labeled with 5-(aminoacetamido)fluorescein at the C-terminal carboxylate, which also resulted in an active analogue.(5) Additional experiments that confirm very similar activities of fluorescently labeled nisin analogues and wild-type nisin are presented in later sections. Importantly, control studies revealed that these labeled nisin analogues neither induced nor inhibited germination initiation, as monitored by the loss of optical density associated with spore hydration during germination (not shown), analogous to unmodified nisin.9,10
The binding of b-nisin was compared with a known lipid II binding probe, fluorescein-vancomycin, (f-vancomycin).(22) Vancomycin interacts with the d-Ala-d-Ala part of lipid II,2,31,32 which is distinct from the binding site of nisin that features the pyrophosphate group (Figure 1B).(33) Fluorescently labeled vancomycin has been used previously to investigate lipid II localization on vegetative bacilli.22,34 Like nisin, vancomycin (at concentrations up to 100 μM) neither induced nor inhibited spore germination (Supplementary Figure S1). To verify that fluorescently labeled nisin can be used to detect the localization of lipid II, vegetative B. anthracis cells were incubated with f-nisin or b-nisin. Previous studies have demonstrated that nisin relocalizes lipid II to patches in bacilli.(5) In our experiments, f-nisin and b-nisin also localized in patches on B. anthracis (Supplementary Figure S2). Collectively, these experiments provide support that the fluorescently labeled nisin analogues bind to lipid II.
Nisin Binds to Lipid II of Germinated Spores
In the presence of both b-nisin and f-vancomycin (both at 1 μM), the outgrowth of germinated spores was inhibited, and b-nisin and f-vancomycin were associated with spores (Figure 2A), although with distinct binding patterns. Vancomycin was localized diffusely over the entire surface of germinated spores, whereas nisin was localized in a more punctate fashion and predominantly near the poles of the germinated spores, where colocalization with vancomycin was observed (Figure 2A). Co-localization analysis utilizing Pearson’s coefficient confirmed that vancomycin was localized at sites where nisin was found at all time points (n = 50; Supplementary Figure S3). It is possible that the clustering of b-nisin near the poles reflects the localization of lipid II that is required for the biosynthesis of new cell wall during the outgrowth process. However, we cannot rule out that the observed strong localization at the poles may be the result of nisin-induced relocalization of lipid II.
Figure 2.
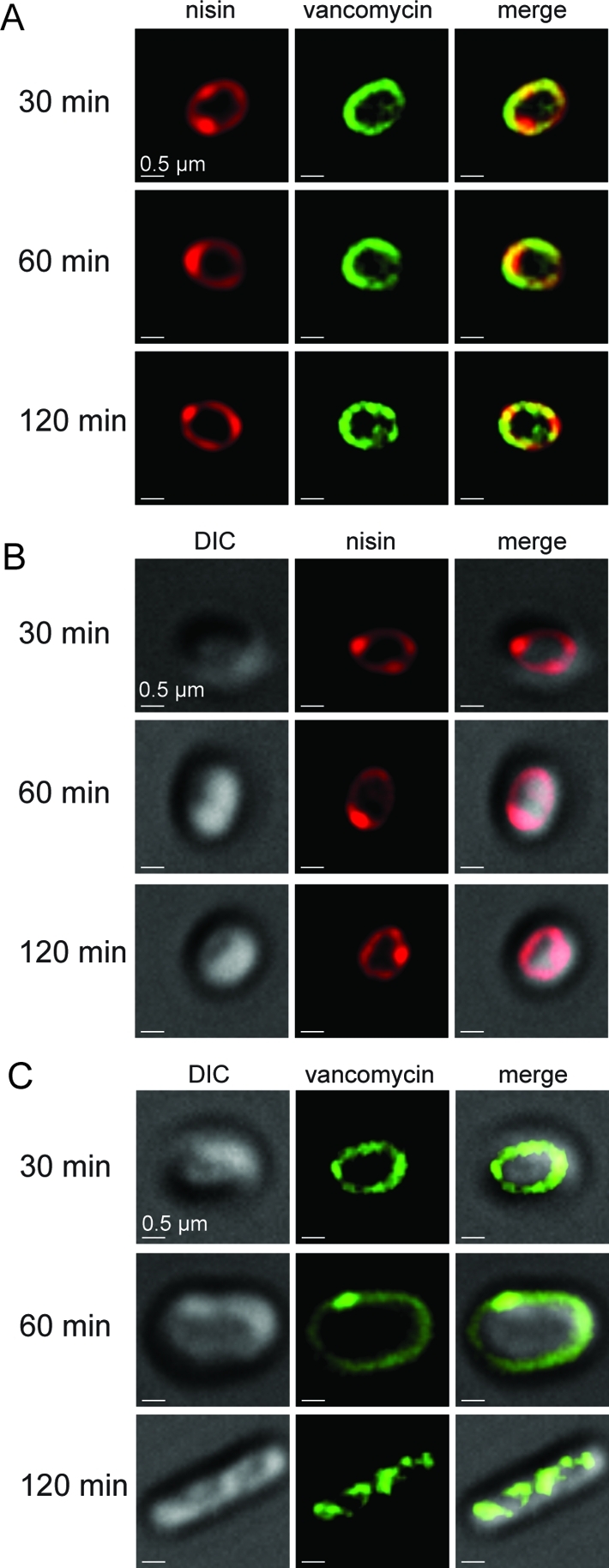
Nisin and vancomycin binding to spores. At time 30, 60, and 120 min, samples were removed and visualized by epi-fluorescence microscopy. (A) Incubation of germinated spores with 1 μM bodipy-nisin (b-nisin, red) and fluorescein-vancomycin (f-vancomycin, green). Co-localization is indicated by yellow in merged images. (B) Incubation of germinated spores with 1 μM b-nisin (red). (C) Incubation of germinated spores with 1 μM f-vancomycin (green). For each panel, a single spore is shown for clarity, but the image is representative of all B. anthracis spores within that sample.
The distinct binding patterns of b-nisin and f-vancomycin were not caused by intrinsic differences between bodipy and fluorescein, because switching the labels (e.g., using fluorescein-labeled nisin and bodipy-labeled vancomycin) resulted in similar binding patterns (Supplementary Figure S4A–C). Furthermore, the patterns of nisin and vancomycin binding were independent of the order in which the compounds were introduced (Supplementary Figure S5A). Finally, the incubation of spores with b-nisin under germinating conditions in the absence of vancomycin also resulted in punctate localization at the pole of the spore with some diffuse fluorescence associated with the spore membrane (Figure 2B).
Complete colocalization of nisin and vancomycin was not expected since vancomycin binds the d-Ala-d-Ala structure present in both the pentapeptide of the non-cross-linked cell wall and cortex in addition to the pentapeptide of lipid II,(26) whereas nisin will bind to the pyrophosphate that is only present in lipid II (Figure 1B).(33) Reflective of these differences in binding specificities, nisin did not detectably associate with dormant spores (Supplementary Figure S5B), indicating that lipid II was not accessible prior to germination initiation. In contrast, vancomycin was associated with dormant spores (Supplementary Figure S5B), which may be due to the accessibility of non-cross-linked peptidoglycan within the spore cortex.
Incubation of spores with f-vancomycin (1 μM) in the absence of nisin showed relatively uniform labeling at 30 min, with more punctuate labeling at 60 min (Figure 2C). Outgrowth of spores into bacilli altered the localization of vancomycin to bands that cross the long axis of the bacilli at 120 min (Figure 2C). This localization of vancomycin in outgrown spores is similar to lipid II localization observed previously in vegetative bacilli of B. subtilis and reflects the helical localization of lipid II along the long axis of the rod-shaped cell.22,34
To provide support that b-nisin binds to lipid II associated with germinated spores, competition binding experiments were performed. These studies revealed that a 100-fold molar excess of unlabeled vancomycin significantly reduced binding of b-nisin to the surface of germinated spores (Figure 3A), presumably by preventing nisin interaction with lipid II as a consequence of steric hindrance. Likewise, a 100-fold molar excess of unmodified nisin significantly inhibited the binding of f-vancomycin to the surface of spores under germinating conditions (Figure 3B). As mentioned above, complete competition is not expected since these two compounds, in addition to sharing lipid II as target, also have nonoverlapping binding sites (non-cross-linked cell wall for vancomycin and the membrane for nisin). Importantly, 100-fold molar excess of unmodified nisin also significantly reduced the binding of b-nisin to germinated spores (Figure 3C), providing additional evidence that b-nisin and wild-type nisin bind to the same target.
Figure 3.
Competition assays with nisin and vancomycin. (A) Competition assay of spore binding by unlabeled vancomycin and b-nisin. * indicates a p-value <0.05 between b-nisin-only treated spores (1 μM) and spores pretreated with unlabeled vancomycin (100 μM) prior to b-nisin addition. ** indicates a p-value <0.05 between b-nisin-treated (1 μM) spores and spores treated with unlabeled vancomycin (100 μM). (B) Binding competition assay of unlabeled nisin in competition with b-vancomycin. * indicates a p-value <0.05 between b-vancomycin (1 μM) treated spores and spores pretreated with unlabeled nisin (100 μM) prior to b-vancomycin addition. ** indicates a p-value <0.05 between b-vancomycin (1 μM) treated spores and spores treated with unlabeled nisin (100 μM). (C) Binding assay of unlabeled c-nisin in competition with b-nisin. * indicates a p-value <0.05 between b-nisin (1 μM) treated spores and spores pretreated with unlabeled c-nisin (100 μM) prior to b-nisin addition. * indicates a p-value <0.05 between control (0 μM nisin) spores and spores pretreated with unlabeled nisin or unlabeled c-nisin (100 μM) prior to b-vancomycin addition. In A–C, the data are plotted as the mean fluorescent intensity (MFI) associated with the binding of the labeled antibiotic. (D) At time 0, 5, and 10 h, samples were removed and visualized by DIC microscopy. For each panel, a single spore is shown for clarity, but the image is representative of all other B. anthracis spores within that sample. The data are representative of those from three independent experiments with 10 μM nisin or c-nisin.
Alternative Approaches with Fluorescently Labeled Nisin Analogues
In an effort to determine whether the time-dependent labeling patterns observed with fluorescently labeled nisin and vancomycin reflect degradation and processing of the cortex and cell wall, a series of cortex hydrolase mutants were evaluated for their ability to alter nisin and vancomycin localization. Previous studies demonstrated that single knockouts of sleB or cwlJ1 slowed the rate of cortex hydrolysis while the double mutant, ΔsleBΔcwlJ1, eliminated cortex hydrolysis in a B. anthracis 34F2 background.24,25 In the presence of both b-nisin and f-vancomycin (both at 1 μM), the outgrowth of germinated spores of these mutants were inhibited, and b-nisin and f-vancomycin were associated with spores (Supplementary Figure S6A), with a similar binding pattern observed for the spore variants as shown above for B. anthracis 7702 (Figure 2). Thus, although the structure of the cell wall is changing during cortex hydrolysis, the accessible binding sites for vancomycin and nisin do not appear to change much. We note that a complementary approach to change the lipid II content of outgrowing spores by using the lipid II biosynthesis inhibitors bacitracin and fosfomycin was not possible because B. anthracis is highly resistant to these compounds.35−37
Although the collective results indicate that nisin binds to lipid II on the spore surface, the microscopy data do not reveal whether lipid II is the only nisin target in germinated spores. Previous studies suggested that nisin may bind covalently to a protein target during outgrowth inhibition through a Michael-type addition of a Cys of a spore protein to Dha5 of nisin.8,10,20 However, extensive efforts to confirm and identify a cognate protein receptor using fluorescently or biotin-labeled nisin as probes were unsuccessful (data not shown), consistent with earlier results emerging from mutagenesis of Dha5.(38) Our results also argue against a covalent mechanism involving the other electrophilic sites in nisin.
Lipid II Binding Is Not Sufficient to Inhibit Outgrowth
To evaluate whether lipid II binding alone is sufficient for nisin-dependent outgrowth inhibition, we studied the action of a purified amino-terminal chymotryptic fragment of nisin (c-nisin, Figure 1A) that retained the A, B, and C rings of the full-length parent compound. c-Nisin contains the necessary N-terminal portion of nisin required for lipid II binding(33) but is unable to form pores.(38) Competition assays between c-nisin and b-nisin revealed that the binding of b-nisin to germinated spores was significantly reduced in the presence of 100-fold molar excess of c-nisin (Figure 3C), once more providing evidence that b-nisin recognizes and binds lipid II. When incubated with germinated spores, c-nisin did not demonstrate significant inhibition of spore outgrowth (Figure 3D, Supplementary Table S4), although a slight delay in the elongation of vegetative bacilli was observed. These data indicate that while the chymotrypsin-derived amino terminal nisin fragment binds lipid II, this fragment alone does not inhibit the outgrowth of germinated spores. This result is different from a previous study, which demonstrated that nisin fragments missing the D and E rings still inhibited B. subtilis spore outgrowth.(38) Possible explanations for the different observations include differences in spore structure between B. subtilis and B. anthracis or differences in the methods to determine spore outgrowth. In the study on B. subtilis, outgrowth was measured by monitoring optical density at 600 nm, whereas the current work utilized microscopy to provide direct visual observation of spore outgrowth. Consistent with the observations that nonpore forming, lipid II-binding nisin analogues do not inhibit spore outgrowth, vancomycin was also incapable of inhibiting the outgrowth of germinated spores (Figure 2). Together, these results support the model that lipid II binding alone is not sufficient to inhibit the outgrowth of germinated spores.
Lipid II Binding Is Associated with Loss of Membrane Potential
Nisin prevents the establishment of a membrane potential in germinated spores.(9) Although the results presented here support binding of nisin to lipid II, they do not establish whether lipid II is important for nisin-dependent membrane potential dissipation. To evaluate this possibility, the membrane potential was first measured with germinated spores in the presence of vancomycin, monitoring the uptake of the cationic membrane potential dye DiOC2 upon the establishment of a membrane potential utilizing flow cytometry.(9) In these experiments, vancomycin did not cause much of a decrease in membrane potential (Figure 4A,B). In contrast, nisin prevented the establishment of a membrane potential in a large fraction of the population (Figure 4A–C). Furthermore, a significant decrease of nisin-dependent reduction of the trans-membrane potential of spores was observed in the presence of a 100-fold molar excess of vancomycin (Figure 4A). Moreover, germination of spores in the presence of c-nisin resulted in significantly less reduction of spore trans-membrane potential (Figure 4C). Because these studies were carried out under conditions where vancomycin or c-nisin reduced the binding of b-nisin to spores (Figures 3A–C), these results suggest that nisin-dependent reduction of the trans-membrane potential requires binding to lipid II in newly germinated spores.
Figure 4.
Lipid II binding is not sufficient to inhibit membrane potential establishment via membrane disruption. (A) Functional competition assay using nisin-induced dissipation of the membrane potential as read-out. The data is plotted as the population of spores exhibiting a native membrane potential per 10,000 spores as observed by flow cytometry using the DiOC2 dye. Preincubation with vancomycin blocks nisin-mediated membrane potential dissipation. * indicates a p-value <0.05 between nisin (1 μM) treated spores and spores pretreated with vancomycin (100 μM) prior to nisin addition. ** indicates a p-value <0.05 between nontreated spores and spores treated with vancomycin (100 μM) only. (B, C) At time 0 (prior to the addition of 10 μM nisin, c-nisin, or vancomycin) and 30 min aliquots were removed from the cultures and evaluated for membrane potential by measuring the DiOC2-associated B. anthracis fluorescence by flow cytometry. Data are rendered as the fold change in membrane potential relative to spores in the presence of 0.1 M MOPS pH 6.8 at the indicated time point.
Lipid II Binding Is Associated with Nisin-Dependent Alterations in Membrane Integrity
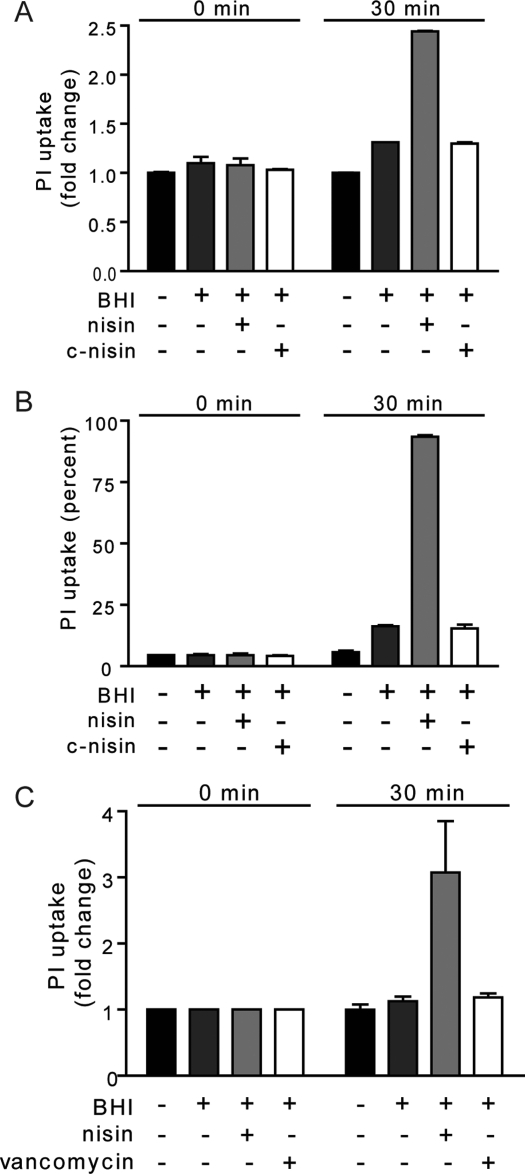
Nisin disrupts the membrane integrity of germinated spores,(9) but the data thus far do not determine whether nisin-lipid II interactions are required for this activity. Therefore, experiments were conducted to examine the effect on membrane integrity monitored by the uptake of the membrane-impermeable dye propidium iodide (PI) upon nisin-induced membrane disruption utilizing flow cytometry.(9) Nisin induced significantly more PI uptake than c-nisin in over 92% of the population of spores (Figure 5A,B). Vancomycin was not able to induce an increase in PI uptake (Figure 5C). In addition, only nisin was able to render the germinated spore metabolically inactive prior to outgrowth (Supplementary Figure S8). Collectively, these results suggest that an association between nisin and lipid II is important for the disruption of membrane integrity.
Figure 5.
C-Terminal region of nisin is essential for membrane disruption. At time 0 (prior to the addition of BHI as germinant and 10 μM nisin, c-nisin, or vancomycin) and 30 min, aliquots were removed from the cultures and evaluated for the following: (A, C) Membrane disruption by measuring PI uptake by the entire population of B. anthracis spores using flow cytometry. (B) Population of spores that exhibited an increase in fluorescence associated with PI uptake. The data are plotted as the percent of spores that took up PI. Data in panels A and C are rendered as the fold change in PI uptake relative to spores in the presence of 0.1 M MOPS pH 6.8 at the indicated time point.
Modification of the Nisin Hinge Region Results in Loss of Outgrowth Inhibition
Previous studies have reported conflicting results as to whether Dha5 is essential for outgrowth inhibition.8,10,20,38 Therefore, experiments were conducted to determine the necessity of Dha5 for membrane disruption resulting in outgrowth inhibition. Nisin and nisin-Dha5Ala were heterologously produced in E. coli(30) (h-nisin and h-nisinS5A, respectively). Both compounds inhibited membrane potential establishment and spore outgrowth and increased PI uptake (Figure 6, Supplementary Table S3) to the same extent. These results show that Dha5 is not essential for spore outgrowth inhibition, as reported previously by Moll and co-workers,(38) and that the mutation does not change the mechanism of inhibition.
Figure 6.
A flexible hinge region, not Dha5, is essential for preventing establishment of a membrane potential, inhibition of spore outgrowth, and membrane disruption. At time 0 and 30 min, aliquots were removed from the cultures and evaluated for the following: (A) Membrane potential by measuring the fluorescence of DiOC2-associated B. anthracis using flow cytometry. (B) Membrane disruption by measuring PI uptake by B. anthracis using flow cytometry. (C) Population of spores exhibiting an increase in fluorescence associated with PI uptake. The data are plotted as the percent of spores that took up PI. (A, B) Data are rendered as the fold change in membrane potential (A) or PI uptake (B) relative to spores in the presence of 0.1 M MOPS pH 6.8 at the indicated time point. (D) At time 0, 5, and 10 h, samples were removed and visualized by DIC microscopy. For each panel, a single spore is shown for clarity, but the image is representative of all other B. anthracis spores within that sample. The data are representative of those from three independent experiments. All experiments were performed with 10 μM nisin or analogue.
A second region of interest for mutation is the hinge region between the three N-terminal rings and the two C-terminal rings. The double mutants N20P/M21P and M21P/K22P retain lipid II binding affinity but not pore-forming activity.5,6,39 These mutants were prepared in this study by heterologous expression in E. coli (h-nisin N20P/M21P and h-nisin M21P/K22P) and analyzed for their ability to disrupt the spore membrane and inhibit membrane potential establishment and spore outgrowth. As depicted in Figure 6, both mutants lost these activities (see also Supplementary Table S5). Importantly, IC99 experiments against M. flavus indicated that the heterologously expressed nisin variants have the same biological activities as those purified from L. lactis(6) and that these variants have high activity against vegetative cells of B. anthracis Sterne 7702 (Supplementary Table S6) . These data corroborate the findings with the truncated analogue c-nisin and demonstrate that the ability to form pores in the spore membrane is essential for outgrowth inhibition. In addition, they show that membrane disruption is not essential for inhibiting growth of vegetative cells but is essential for inhibiting spore outgrowth.
Conclusions
Previous work implicated nisin-mediated alterations in the membrane integrity and prevention of the establishment of a membrane potential as the mechanism by which nisin inhibits spore outgrowth.8−10,26 Whether or not interactions with lipid II were important for these activities was not determined. Utilizing fluorescently labeled nisin and vancomycin, competition experiments, and nisin analogues, lipid II was identified in this study as the target for nisin to induce inhibition of spore outgrowth. Lipid II binding alone is not sufficient for outgrowth inhibition, however, which requires membrane disruption. Modification of Dha5 to Ala does not result in the loss of outgrowth inhibition and membrane disruption. We anticipate that lantibiotics such as subtilin,40,41 epidermin,(42) and haloduracin(43) also inhibit spore outgrowth via pore formation and that lipid II binding may also be involved in nisin inhibition of other spore-forming bacteria from the genera Bacillus and Clostridium, including those of medical relevance such as C. botulinum and C. difficile.
Germinated spores no longer have antibiotic resistance associated with spore dormancy, yet they have not yet begun virulence factor expression. Therefore, germination is an ideal point in the spore-forming bacterial life cycle for antibiotic intervention to eliminate or prevent infection.(9) Current antibiotic treatments require that the endospore be fully converted into a vegetative cell with a functional metabolism as current antibiotics target protein synthesis, cell wall biosynthesis, specific metabolic pathways, or DNA replication. Cell wall biosynthesis and DNA replication inhibition will prevent bacteremia by preventing growth, but drugs targeting these processes will not eliminate the effects of bacterial toxins on the host. Therefore, ribosomal inhibitors are required in conjunction with growth-inhibiting antimicrobials to prevent toxin synthesis to eliminate toxemia.44,45 The unique ability of nisin to inhibit spore outgrowth prior to toxin production(9) suggests that nisin and other membrane-targeting antimicrobials may have potential for the treatment of infections by spore-forming pathogens.
Acknowledgments
The authors thank Dr. B. Pilas and B. Montez (UIUC) for assistance with flow cytometry, and Dr. D. Popham (Virginia Tech) for supplying the B. anthracis 34F2 mutant strains. This work was supported by an NIH-NIAID Award to the Western Regional Center for Excellence for Biodefense and Emerging Infectious Diseases Research U54-AI057156 (S.R.B.; P.I. D. Walker), by a Chemical Biology Interface Training Grant from the National Institutes of Health (5 T32GM070421 to I.M.G.), and by the Howard Hughes Medical Institute (to W.A.V.).
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Chatterjee C.; Paul M.; Xie L; van der Donk W. A. (2005) Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–683. [DOI] [PubMed] [Google Scholar]
- Breukink E.; de Kruijff B. (2006) Lipid II as a target for antibiotics. Nat. Rev. Drug Discovery 5, 321–332. [DOI] [PubMed] [Google Scholar]
- Lubelski J.; Rink R.; Khusainov R.; Moll G. N.; Kuipers O. P. (2008) Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 65, 455–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhr E.; Sahl H. G. (1985) Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob. Agents Chemother. 27, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasper H. E.; Kramer N. E.; Smith J. L.; Hillman J. D.; Zachariah C.; Kuipers O. P.; de Kruijff B.; Breukink E. (2006) An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313, 1636–1637. [DOI] [PubMed] [Google Scholar]
- Wiedemann I.; Breukink E.; van Kraaij C.; Kuipers O. P.; Bierbaum G.; de Kruijff B.; Sahl H. G. (2001) Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779. [DOI] [PubMed] [Google Scholar]
- Delves-Broughton J.; Blackburn P.; Evans R. J.; Hugenholtz J. (1996) Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69, 193–202. [DOI] [PubMed] [Google Scholar]
- Morris S. L.; Walsh R. C.; Hansen J. N. (1984) Identification and characterization of some bacterial membrane sulfhydryl groups which are targets of bacteriostatic and antibiotic action. J. Biol. Chem. 259, 13590–13594. [PubMed] [Google Scholar]
- Gut I. M.; Prouty A. M.; Ballard J. D.; van der Donk W. A.; Blanke S. R. (2008) Inhibition of Bacillus anthracis spore outgrowth by nisin. Antimicrob. Agents Chemother. 52, 4281–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. C.; Dodd H. M.; Horn N.; Maclean K.; Lian L. Y.; Bycroft B. W.; Gasson M. J.; Roberts G. C. (1996) Structure-activity relationships in the peptide antibiotic nisin: role of dehydroalanine 5. Appl. Environ. Microbiol. 62, 2966–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott V. N.; Taylor S. L. (1981) Effect of nisin on the outgrowth of Clostridium botulinum spores. J. Food Sci. 46, 117–120. [Google Scholar]
- Montville T. J.; De Siano T.; Nock A.; Padhi S.; Wade D. (2006) Inhibition of Bacillus anthracis and potential surrogate bacilli growth from spore inocula by nisin and other antimicrobial peptides. J. Food Prot. 69, 2529–2533. [DOI] [PubMed] [Google Scholar]
- Moir A.; Corfe B. M.; Behravan J. (2002) Spore germination. Cell. Mol. Life Sci. 59, 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. (2003) Spore germination. Curr. Opin. Microbiol. 6, 550–556. [DOI] [PubMed] [Google Scholar]
- Paidhungat M., Setlow P. (2002) Spore Germination and Outgrowth, in Bacillus subtilis and Its Closest Relatives from Genes to Cells (Sonenshein A. L., Hoch J. A., Losick R., Eds.), pp 537–548, American Society for Microbiology, Washington, D.C. [Google Scholar]
- Setlow P. (2006) Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101, 514–525. [DOI] [PubMed] [Google Scholar]
- Guidi-Rontani C.; Levy M.; Ohayon H.; Mock M. (2001) Fate of germinated Bacillus anthracis spores in primary murine macrophages. Mol. Microbiol. 42, 931–938. [DOI] [PubMed] [Google Scholar]
- Guidi-Rontani C.; Pereira Y.; Ruffie S.; Sirard J. C.; Weber-Levy M.; Mock M. (1999) Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol. Microbiol. 33, 407–414. [DOI] [PubMed] [Google Scholar]
- Guidi-Rontani C.; Weber-Levy M.; Labruyere E.; Mock M. (1999) Germination of Bacillus anthracis spores within alveolar macrophages. Mol. Microbiol. 31, 9–17. [DOI] [PubMed] [Google Scholar]
- Liu W.; Hansen J. N. (1993) The antimicrobial effect of a structural variant of subtilin against outgrowing Bacillus cereus T spores and vegetative cells occurs by different mechanisms. Appl. Environ. Microbiol. 59, 648–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojkovic B.; Torres E. M.; Prouty A. M.; Patel H. K.; Zhuang L.; Koehler T. M.; Ballard J. D.; Blanke S. R. (2008) High-throughput, single-cell analysis of macrophage interactions with fluorescently labeled Bacillus anthracis spores. Appl. Environ. Microbiol. 74, 5201–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyanont K.; Doan T.; Lazarus M. B.; Fang X.; Rudner D. Z.; Walker S. (2006) Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. U.S.A. 103, 11033–11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. C.; Leyland M.; Clark J.; Dodd H. M.; Lian L. Y.; Gasson M. J.; Bycroft B. W.; Roberts G. C. (1996) Structure-activity relationships in the peptide antibiotic nisin: antibacterial activity of fragments of nisin. FEBS Lett. 390, 129–132. [DOI] [PubMed] [Google Scholar]
- Heffron J. D.; Orsburn B.; Popham D. L. (2009) Roles of germination-specific lytic enzymes CwlJ and SleB in Bacillus anthracis. J. Bacteriol. 191, 2237–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron J. D.; Lambert E. A.; Sherry N.; Popham D. L. (2010) Contributions of four cortex lytic enzymes to germination of Bacillus anthracis spores. J. Bacteriol. 192, 763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukink E.; Wiedemann I.; van Kraaij C.; Kuipers O. P.; Sahl H.; de Kruijff B. (1999) Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286, 2361–2364. [DOI] [PubMed] [Google Scholar]
- Athamna A.; Athamna M.; Abu-Rashed N.; Medlej B.; Bast D. J.; Rubinstein E. (2004) Selection of Bacillus anthracis isolates resistant to antibiotics. J. Antimicrob. Chemother. 54, 424–428. [DOI] [PubMed] [Google Scholar]
- Athamna A.; Massalha M.; Athamna M.; Nura A.; Medlej B.; Ofek I.; Bast D.; Rubinstein E. (2004) In vitro susceptibility of Bacillus anthracis to various antibacterial agents and their time-kill activity. J. Antimicrob. Chemother. 53, 247–251. [DOI] [PubMed] [Google Scholar]
- Turnbull P. C.; Sirianni N. M.; LeBron C. I.; Samaan M. N.; Sutton F. N.; Reyes A. E.; Peruski L. F. Jr. (2004) MICs of selected antibiotics for Bacillus anthracis, Bacillus cereus, Bacillus thuringiensis, and Bacillus mycoides from a range of clinical and environmental sources as determined by the Etest. J Clin Microbiol 42, 3626–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Yang X.; Garg N.; van der Donk W. A. (2010) Production of lantipeptides in Escherichia coli. J. Am. Chem. Soc. 133, 2338–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M.; Jones P. G.; Kennard O.; Williams D. H.; Smith G. A. (1978) Structure of vancomycin and its complex with acetyl-d-alanyl-d-alanine. Nature 271, 223–225. [DOI] [PubMed] [Google Scholar]
- Molinari H.; Pastore A.; Lian L. Y.; Hawkes G. E.; Sales K. (1990) Structure of vancomycin and a vancomycin/d-Ala-d-Ala complex in solution. Biochemistry 29, 2271–2277. [DOI] [PubMed] [Google Scholar]
- Hsu S. T.; Breukink E.; Tischenko E.; Lutters M. A.; de Kruijff B.; Kaptein R.; Bonvin A. M.; van Nuland N. A. (2004) The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11, 963–967. [DOI] [PubMed] [Google Scholar]
- Daniel R. A.; Errington J. (2003) Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113, 767–776. [DOI] [PubMed] [Google Scholar]
- Parsonage D.; Newton G. L.; Holder R. C.; Wallace B. D.; Paige C.; Hamilton C. J.; Dos Santos P. C.; Redinbo M. R.; Reid S. D.; Claiborne A. (2010) Characterization of the N-acetyl-α-d-glucosaminyl l-malate synthase and deacetylase functions for bacillithiol biosynthesis in Bacillus anthracis. Biochemistry 49, 8398–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedar G. C.; Brown-Driver V.; Reyes D. R.; Hilgers M. T.; Stidham M. A.; Shaw K. J.; Finn J.; Haselbeck R. J. (2008) Comparison of the essential cellular functions of the two murA genes of Bacillus anthracis. Antimicrob. Agents Chemother. 52, 2009–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard R.; El Ghachi M.; Mengin-Lecreulx D.; Chippaux M.; Denizot F. (2005) BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance. J. Biol. Chem. 280, 28852–28857. [DOI] [PubMed] [Google Scholar]
- Rink R.; Wierenga J.; Kuipers A.; Kluskens L. D.; Driessen A. J.; Kuipers O. P.; Moll G. N. (2007) Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C-terminal truncation. Appl. Environ. Microbiol. 73, 5809–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D.; Connor P. M.; Cotter P. D.; Hill C.; Ross R. P. (2008) The generation of nisin variants with enhanced activity against specific Gram-positive pathogens. Mol. Microbiol. 69, 218–230. [DOI] [PubMed] [Google Scholar]
- Liu W.; Hansen J. N. (1992) Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J. Biol. Chem. 267, 25078–25085. [PubMed] [Google Scholar]
- Parisot J.; Carey S.; Breukink E.; Chan W. C.; Narbad A.; Bonev B. (2008) Molecular mechanism of target recognition by subtilin, a class I lanthionine antibiotic. Antimicrob. Agents Chemother. 52, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotz H.; Josten M.; Wiedemann I.; Schneider U.; Gotz F.; Bierbaum G.; Sahl H. G. (1998) Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30, 317–327. [DOI] [PubMed] [Google Scholar]
- Oman T. J.; van der Donk W. A. (2009) Insights into the mode of action of the two-peptide lantibiotic haloduracin. ACS Chem Biol 4, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. W.; Tierney B. C.; Aranas A.; Rosenstein N. E.; Franzke L. H.; Apicella L.; Marano N.; McNeil M. M. (2005) An overview of adverse events reported by participants in CDC’s anthrax vaccine and antimicrobial availability program. Pharmacoepidemiol. Drug Saf. 14, 393–401. [DOI] [PubMed] [Google Scholar]
- Shepard C. W.; Soriano-Gabarro M.; Zell E. R.; Hayslett J.; Lukacs S.; Goldstein S.; Factor S.; Jones J.; Ridzon R.; Williams I.; Rosenstein N. (2002) Antimicrobial postexposure prophylaxis for anthrax: adverse events and adherence. Emerging Infect. Dis. 8, 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.