Abstract
OBJECTIVE
Obesity, insulin resistance, and type 2 diabetes form a tightly correlated cluster of metabolic disorders in which adipose is one of the first affected tissues. The role of hypoxia and hypoxia-inducible factor 1 (HIF1) in the development of high-fat diet (HFD)–induced obesity and insulin resistance was investigated using animal models.
RESEARCH DESIGN AND METHODS
Mice with adipocyte-specific targeted disruption of the genes encoding the HIF1 obligatory subunits Hif1α or Arnt (Hif1β) were generated using an aP2-Cre transgene with the Cre/LoxP system. The mice were fed an HFD for 12 weeks and their metabolic phenotypes were determined. Gene expression patterns in adipose tissues were also determined by microarray and quantitative PCR.
RESULTS
On an HFD, adipocyte-specific ARNT knockout mice and adipocyte-specific HIF1α knockout mice exhibit similar metabolic phenotypes, including reduced fat formation, protection from HFD-induced obesity, and insulin resistance compared with similarly fed wild-type controls. The cumulative food intake remained similar; however, the metabolic efficiency was lower in adipocyte-specific HIF1α knockout mice. Moreover, indirect calorimetry revealed respiratory exchange ratios were reduced in adipocyte-specific HIF1α knockout mice. Hyperinsulinemic-euglycemic clamp studies demonstrated that targeted disruption of HIF1α in adipocytes enhanced whole-body insulin sensitivity. The improvement of insulin resistance is associated with decreased expression of Socs3 and induction of adiponectin.
CONCLUSIONS
Inhibition of HIF1 in adipose tissue ameliorates obesity and insulin resistance. This study reveals that HIF1 could provide a novel potential therapeutic target for obesity and type 2 diabetes.
In obesity, oxygen supply cannot meet the demand of expanding adipose, resulting in relative hypoxia within adipose tissue, increased lactate production, and hypoperfusion in both obese human and animal models (1,2). Hypoxia was found to cause insulin resistance in 3T3-L1 adipocytes and human subcutaneous abdominal adipocytes (3). However, the role of hypoxia in adipose tissue during obesity and insulin resistance remains unclear. Regulation of hypoxia-mediated responses is mainly dependent on the hypoxia-inducible factor (HIF) family. HIFs are nuclear transcription factors and function as oxygen-sensitive α subunit and β heterodimers (ARNT). All isoforms of HIFα, HIF1α, HIF2α, and HIF3α require the ubiquitously expressed subunit aryl hydrocarbon nuclear translocator (ARNT or HIF1β) as an obligate heterodimerization partner for activation of target genes. HIF1α, HIF2α, HIF3α, and ARNT are all expressed in adipose tissue (4–6). HIF function is primarily regulated by HIF1α protein stability. Under normoxia, HIF1α is hydroxylated by prolylhydroxylase (PHD). Following hydroxylation, HIF1α is ubiquitinated by the E3 ubiquitin ligase, von Hippel-Lindau tumor suppressor (VHL) and degraded via the proteasome pathway. Conversely, under hypoxia, hydroxylation is inhibited, leading to stabilization of the α subunit, heterodimerization with ARNT, and activation of HIF target genes (7).
To investigate the role of hypoxia in obesity and insulin resistance, adipocyte-specific ARNT knockout (Arnt∆Adipo) mice and adipocyte-specific HIF1α knockout (Hif1α∆Adipo) mice were generated with the Cre/LoxP system using the adipose-specific aP2-Cre transgene (8). Both mouse lines exhibit similar metabolic phenotypes, including reduced fat formation, protection from high-fat diet (HFD)-induced obesity, and insulin resistance, suggesting a role for HIF1 in the pathogenesis of obesity and insulin resistance. Taken together, the findings of this study reveal an essential role of HIF1α in controlling adipose mass and function and provide a potential therapeutic target of obesity and insulin resistance.
RESEARCH DESIGN AND METHODS
Arnt-floxed (ArntF/F) (9) and Hif1α-floxed (Hif1αF/F) (10) mice containing loxP sites flanking exons 6 and 13–15, of the Arnt and Hif1α genes, respectively, were crossed with mice harboring the Cre recombinase under control of the aP2 promoter (aP2-Cre mice). All mice were on the C57BL/6 background, and only male mice were used for experiments. Primers used to assess recombination and routine genotyping for the Arnt and Hif1α allele are listed in Supplementary Table 2. Male mice were housed in temperature- and light-controlled rooms and supplied with water and pelleted NIH-31 chow diet (10% kcal consisting of fat) ad libitum. In the HFD study, 6-week-old male mice were given an HFD (60% kcal consisting of fat; BioServ, Frenchtown, NJ) for 12 weeks. All animal studies were performed in accordance with Institute of Laboratory Animal Resources guidelines and approved by the National Cancer Institute Animal Care and Use Committee.
Metabolic assays.
For glucose tolerance test (GTT), mice were fasted for 16 h, blood was drawn, and mice were injected intraperitoneally with 1 g/kg glucose. For insulin tolerance test (ITT), mice were fasted for 4 h, blood was drawn, and then mice were injected intraperitoneally with 1 unit/kg body wt insulin (Humulin R; Eli Lilly, Indianapolis, IN).
Hyperinsulinemic-euglycemic clamps were performed in awake mice fasted for 12 h as previously described (11) with modifications. Primed-continuous infusion of [3-3H]glucose was used: 2.5 µCi bolus, 0.05 µCi/min during the basal state and 0.1 µCi/min during the clamp period. Insulin (Humulin R) was infused as a bolus of 18 mU/kg over a period of 3 min, followed by continuous insulin infusion at the rate of 3.5 mU/kg lean mass/min (in Hif1αF/F mice) and 9.4 m/kg lean mass/min (in Hif1α∆Adipo mice) to raise plasma insulin concentration to 4 ng/mL.
In vivo insulin stimulation and analysis of insulin signaling.
Mice were fasted overnight, and then injected via the vena cava with 5 units of Humulin R under anesthesia. Liver, quadriceps, and white adipose tissue were collected after 5 min and stored at –80°C until use. Total Akt and phospho-Akt (Ser473) antibodies were from Cell Signaling Technologies (Danvers, MA).
Body composition, food intake, and metabolic rate.
Body composition was measured in nonanesthetized mice using an Echo3-in-1 nuclear magnetic resonance (NMR) analyzer (Echo Medical Systems, Houston, TX). Cumulative food intake was measured in 6- to 8-week-old male mice maintained on regular chow for 2 weeks and 10- to 12-week-old mice fed an HFD for 4–6 weeks. Mice were housed individually in their home cages a week prior to recording food intake. Metabolic efficiency was calculated as the ratio of weight gain to energy consumed during a 2-week period. Total and resting metabolic rates were measured by indirect calorimetry using the Oxymax system (Columbus Instruments, Columbus, OH) (12). Mice had free access to food and water during the measurements and were allowed to adapt to metabolic cages for 24 h prior to data collection. Following an adaptation period, data were recorded for 24 h at 24 and 30°C for an additional 24 h. Four mutant and four control mice were tested at the same time, and each mouse was tested every 20 min. Motor activities were measured by an infrared beam interruption (Opto-Varimex mini, Columbus Instruments), and resting was defined as time points with ambulation equal to zero. Diet-induced thermogenesis and β3 adrenergic thermogenesis were measured as previously described (12,13).
Biochemical assays.
Fasted serum insulin was measured by use of an ELISA kit (Crystal Chem). Fasted serum cholesterol, free fatty acids (FFAs), and triglycerides were measured using reagents from Wako. Adiponectin serum levels were measured with a mouse adiponectin ELISA kit (ALPCO). Serum resistin was measured with an ELISA kit (R&D).
RNA and protein analysis.
Quantitative PCR (qPCR) reactions were carried out on an ABI Prism 7900HT sequence detection system (Applied Biosystems). Primer sequences are listed in Supplementary Table 2. Tissues were lysed by use of radioimmunoprecipitation assay for whole cell extract. The membranes were incubated with antibodies against total Akt, phospho-Akt (Ser473), SOCS3, total STAT3, phospho-STAT3 (Cell Signaling Technologies, Danvers, MA), and Arnt and Histone 1H (Santa Cruz Biotechnology, Santa Cruz, CA). The signals obtained were normalized to β-actin (Millipore Corp, Temecula, CA) for whole cell extracts.
Isolation of adipocytes and macrophage of stromal vascular fraction and microarray analysis.
Adipocytes and macrophage of stromal vascular fraction (SVF-Mϕ) were isolated from epididymal white adipose tissue (WAT) as previously described (14). Dye-coupled cDNAs were purified with a MiniElute PCR purification kit (Qiagen) and hybridized to an Agilent 44 K mouse 60-mer oligo microarray (Agilent Technologies). The procedures were repeated for replicate experiments with independent hybridization and processing and the data processed and analyzed by Genespring GX software (Agilent Technologies).
Histology.
Paraffin-embedded tissue sections were stained with hematoxylin-eosin (H-E) using a standard protocol. Quantification of adipocyte area was done on H-E–stained sections using ImageTool software.
Data analysis.
Results were expressed as means ± SD. Differences between groups were examined for statistical significance with Student t test. A P value of < 0.05 was considered statistically significant.
RESULTS
Generation and characterization of Arnt∆Adipo and Hif1α∆Adipo mice.
For examination of the role of HIF transcription factors in obesity and insulin resistance, Arnt and Hif1α genes were disrupted in adipocytes. To estimate the extent of cell-specific disruption of the Arnt and Hif1α loci, PCR analysis was used. A PCR amplicon for the Arnt-null allele amplified as a 340 base pair product and was detected in genomic DNA isolated from adipocytes or adipose tissue of Arnt∆Adipo mice and not in adipose DNA isolated from ArntF/F mice. In contrast, the floxed allele was the only band detected in adipose tissue from ArntF/F mice and from all nonadipose tissues in Arnt∆Adipo mice (Fig. 1A). The Hif1α-null amplicon amplified as a 355 base pair product was detected in genomic DNA isolated from adipose tissue of Hif1α∆Adipo mice, and was not detected in adipose tissue DNA isolated from Hif1αF/F mice. The null allele was only detected in adipose tissue and not in liver, skeletal muscle, spleen, or kidney from Hif1α∆Adipo mice (Fig. 1B). The expression of Arnt mRNA was specifically decreased in WAT and brown adipose tissue (BAT) by 50% in the Arnt∆Adipo mice compared with ArntF/F mice; no decrease was evident from liver or skeletal muscle. In addition, qPCR showed nearly absent expression of ARNT mRNA in the adipocytes of Arnt∆Adipo mice (Fig. 1C). Similar results were obtained from tissues of Hif1α∆Adipo mice, where an ~88% decrease in HIF1α mRNA from adipocytes was observed (Fig. 1D). Nuclear ARNT protein expression in Arnt∆Adipo mice was also markedly decreased (Supplementary Fig. 1A). To confirm that loss of ARNT was of functional significance, the extent of activation of the ARNT-dependent aryl hydrocarbon receptor (AhR) pathway upon 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) challenge was determined. Induction of the AHR target gene Cyp1a1 was markedly attenuated in WAT and BAT of Arnt∆Adipo mice (Supplementary Fig. 1B and C). However, no significant difference in the extent of induction of CYP1A1 mRNA was noted in liver or skeletal muscle compared with ArntF/F mice (Supplementary Fig. 1D and E). These results demonstrate adipocyte-specific knockout of the Arnt and Hif1α genes in mice.
FIG. 1.
Adipocyte-specific disruption of the Arnt and Hif1a genes via Cre-loxP–mediated recombination. A and B: PCR diagnostic for aP2-Cre–mediated recombination of the Arnt or Hif1α allele in genomic DNA isolated from adipocytes or tissue of ArntF/F and Arnt∆Adipo or Hif1αF/F and Hif1α∆Adipo mice. C and D: qPCR analysis of Arnt mRNA expression in the tissues (left panel) or adipocytes (right panel) from ArntF/F and Arnt∆Adipo or Hif1αF/F and Hif1α∆Adipo mice. For qPCR analysis, the expression was normalized to β-actin. Data are means ± SD. *P < 0.05, **P < 0.01 compared with floxed littermates.
Arnt∆Adipo or Hif1α∆Adipo mice are resistant to diet-induced weight gain.
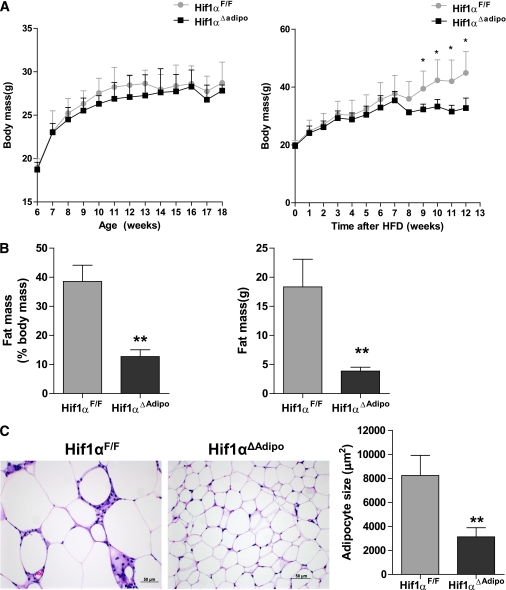
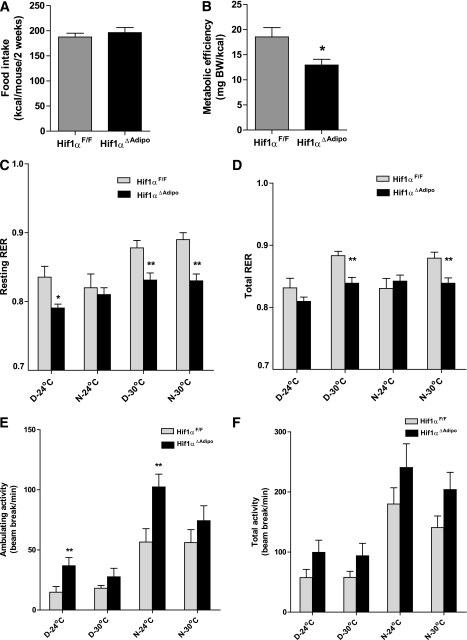
To explore the role of HIF1 in fat metabolism and glucose homeostasis, male mice were fed either a chow diet or HFD. When fed a chow diet, Arnt∆Adipo and Hif1α∆Adipo mice grew at a rate similar to that of ArntF/F and Hif1αF/F mice, respectively. However, 12 weeks of HFD led to weight gain in ArntF/F and Hif1αF/F mice, while Arnt∆Adipo and Hif1α∆Adipo mice were resistant to the HFD-induced weight gain (Supplementary Fig. 2A; Fig. 2A). NMR measurements confirmed that the body fat mass and the ratio of fat and body mass of Arnt∆Adipo and Hif1α∆Adipo mice fed an HFD were decreased compared with ArntF/F and Hif1αF/F mice, respectively (Supplementary Fig. 2B; Fig. 2B). The adipocyte size in Arnt∆Adipo and Hif1α∆Adipo mice was significantly decreased compared with ArntF/F and Hif1αF/F mice, respectively, after 12 weeks of HFD (Supplementary Fig. 2C; Fig. 2C). To explore the mechanism of reduced adiposity in Arnt∆Adipo and Hif1α∆Adipo mice, cumulative food intake, metabolic efficiency, and metabolic rates were measured in young mice maintained on chow diet and in mice fed an HFD for 4–7 weeks, which is before the difference between control and mutant mice became apparent. There were no significant differences in weight, cumulative food intake, or metabolic efficiency between Hif1αF/F and Hif1α∆Adipo mice maintained on chow diet (Supplementary Fig. 3A–C). Similarly, indirect calorimetry performed at 24°C and thermoneutrality (30°C) on the same set of mice did not reveal significant differences in metabolic rate, respiratory exchange ratio (RER), or activity between Hif1αF/F and Hif1α∆Adipo mice fed chow diet (Supplementary Fig. 3D). A short 4-day exposure to HFD caused comparable increases in body weight and resting metabolic rate in Hif1αF/F and Hif1α∆Adipo mice, indicating that adipose-specific inactivation of Hif1α did not alter the acute thermogenic response to HFD (Supplementary Fig. 3E and F). These results indicate that it takes a long time for the adipose tissue to get big enough to be hypoxic and that Hif1α expression was significantly induced, resulting in activation of HIF1 target genes such as Vegf and Glut1 (Supplementary Fig. 7). After 4–6 weeks on an HFD, the cumulative food intake remained similar between Hif1αF/F and Hif1α∆Adipo mice; however, the metabolic efficiency was lower in the Hif1α∆Adipo mice, suggesting an increase in the metabolic rate (Fig. 3A). Indirect calorimetry performed at 24°C on week 7 of HFD did not reveal a significant difference in resting or total oxygen consumption between the genotypes, while at 30°C these parameters tended to be higher in Hif1α∆Adipo mice compared with controls (Supplementary Fig. 3G). Hif1α∆Adipo mice had reduced resting and total RER at 30°C during resting phase (daytime) and active phase (nighttime), suggesting increased fatty acid oxidation (Fig. 3C and D) and a tendency toward increased activity (Fig. 3E and F). All of these changes in Hif1α∆Adipo mice could contribute to the decreased metabolic efficiency and weight gain on an HFD compared with Hif1αF/F mice. However, it is unlikely that resistance to HFD was caused by activated BAT thermogenesis because the response to mild cold measured as the difference in metabolic rate at thermoneutral and room temperature and the response to a maximal dose of the β3-adrenergic agonist CL316243, which specifically stimulates BAT thermogenesis, were similar in both strains (Supplementary Fig. 3H).
FIG. 2.
Disruption of Hif1α protected mice from HFD-induced obesity. A: Typical growth curves of Hif1αF/F and Hif1α∆Adipo mice maintained on chow diet (left panel) or HFD (right panel) (n = 6–8/group). B: Body composition by NMR to show the fat mass and fat mass ratio in Hif1αF/F and Hif1α∆Adipo mice after 12 weeks of HFD (n = 5/group). C: Representative H-E–stained WAT sections and quantification of adipocyte size from Hif1αF/F and Hif1α∆Adipo mice after 12 weeks of HFD (n = 5/group). Data are means ± SD. *P < 0.05, **P < 0.01 compared with floxed littermates. (A high-quality digital representation of this figure is available in the online issue.)
FIG. 3.
Energy balance in Hif1αF/F and Hif1α∆Adipo mice. A and B: Cumulative food intake and metabolic efficiency for 2 weeks in Hif1αF/F and Hif1α∆Adipo mice after 4–6 weeks of HFD. BW, body weight. C and D: Resting and total RER; VCo2/Vo2. E and F: Ambulatory and total activity levels. A–F were measured in the same set of Hif1αF/F and Hif1α∆Adipo mice. Indirect calorimetry (C–F) was performed after 7 weeks of HFD at 24°C and 30°C (thermoneutrality) during resting phase (daytime [D]) and active phase (nighttime [N]) (n = 6–8/group). Data are means ± SEM. *P < 0.05, **P < 0.01 compared with floxed littermates.
HIF1 deficiency in adipocytes improves HFD-induced glucose intolerance and insulin resistance.
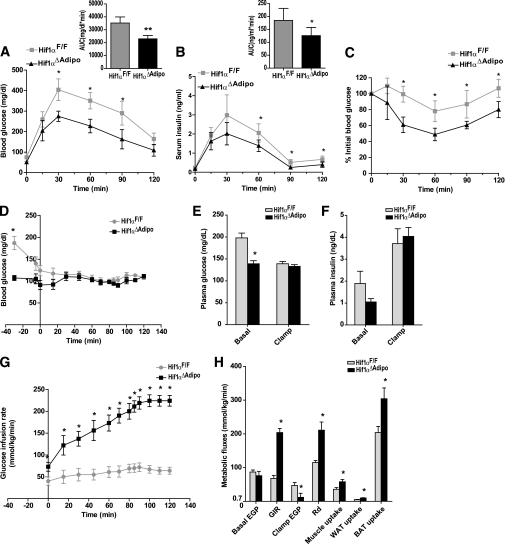
To explore the role of adipocyte HIF1 deficiency in obesity-induced insulin resistance, GTTs and ITTs were performed. When mice were fed a chow diet, there were no significant differences in GTT and ITT between ArntF/F and Arnt∆Adipo mice (Supplementary Fig. 4A and C). However, GTT revealed that after 11 weeks of HFD challenge, Arnt∆Adipo and Hif1α∆Adipo mice displayed significantly reduced blood glucose compared with ArntF/F and Hif1αF/F mice after glucose loading (Supplementary Fig. 4B; Fig. 4A). Moreover, serum insulin was significantly decreased in Hif1α∆Adipo mice during GTT (Fig. 4B). ITT showed a significant improvement in insulin sensitivity by adipose HIF1 disruption (Supplementary Fig. 4D; Fig. 4C). Moreover, fed glucose, fasted glucose, and fasted serum insulin levels were significantly lower in Arnt∆Adipo and Hif1α∆Adipo mice compared with ArntF/F and Hif1αF/F mice, respectively, after 12 weeks of HFD. The calculated homeostasis model assessment (HOMA) measure of insulin resistance was significantly decreased in Arnt∆Adipo and Hif1α∆Adipo mice (Table 1). Arnt∆Adipo and Hif1α∆Adipo mice also had reduced fasted serum triglycerides and FFA levels consistent with improved glucose tolerance and insulin sensitivity in these mice (Table 1). Although body mass was similar between Hif1αF/F and Hif1α∆Adipo mice after 4 weeks, 6 weeks, and 8 weeks of HFD, GTT and ITT revealed that glucose tolerance and insulin sensitivity were improved in Hif1α∆Adipo mice from 4 weeks on an HFD (Supplementary Fig. 5A and B). After 6 weeks of HFD challenge, fasted glucose was similar while fasted insulin levels and HOMA index in Hif1α∆Adipo mice were significantly decreased (Supplementary Fig. 5C). These results indicated that HIF1 disruption in adipocytes improved HFD-induced glucose tolerance and insulin resistance before the onset of the decrease in body mass.
FIG. 4.
Hif1α disruption in adipocytes improved HFD-induced glucose intolerance and insulin resistance. A and B: Blood glucose levels and serum insulin levels in 2-h GTT 12 weeks after HFD (n = 6–8/group). Inset graphs in A and B depict the respective analysis of the area under the curve (AUC). C: ITT 12 weeks after HFD (n = 6–8/group). Data are means ± SD. *P < 0.05 compared with Hif1α∆Adipo littermates. D: Time courses of blood glucose during the hyperinsulinemic-euglycemic clamp. E and F: Plasma glucose and insulin levels in the basal state and during the clamp. G: GIR during the clamp. H: Basal and clamp endogenous glucose production (EGP), GIR, whole-body glucose disposal (Rd), and glucose uptake in skeletal muscle, WAT, and BAT. The hyperinsulinemic-euglycemic clamp (D–H) was performed after 15 weeks of HFD (n = 6/group). Data are means ± SEM. *P < 0.05 compared with floxed littermates.
TABLE 1.
Metabolic parameters after 12 weeks of HFD
| Genotype | ArntF/F | Arnt∆Adipo | Hif1aF/F | Hif1α∆Adipo |
|---|---|---|---|---|
| Fasting glucose (mg/dL) | 150 ± 21.7 | 106 ± 21.5* | 172 ± 32.5 | 131 ± 19.8* |
| Fed glucose (mg/dL) | 269 ± 28.1 | 139 ± 27.9† | 206 ± 23.5 | 149 ± 30.4† |
| Fasting serum insulin (ng/mL) | 3.6 ± 1.4 | 2.1 ± 0.8* | 2.6 ± 0.6 | 1.1 ± 0.2† |
| HOMA index | 31.7 ± 8.6 | 13.8 ± 7.6† | 27.9 ± 10.4 | 8.4 ± 1.8† |
| Fasted serum triglyceride (mg/dL) | 124 ± 42.1 | 78.4 ± 7.5* | 162 ± 38.8 | 99.4 ± 19.8† |
| Fasted serum cholesterol (mg/dL) | 182 ± 37.6 | 169 ± 43.6 | 196 ± 45.8 | 160 ± 24.1 |
| Fasted serum FFA (mmol/L) | 1.88 ± 0.42 | 1.07 ± 0.39* | 1.63 ± 0.35 | 1.09 ± 0.30* |
Data are means ± SD.
*P < 0.05.
†P < 0.01 compared with controls.
A hyperinsulinemic-euglycemic clamp was performed in Hif1αF/F and Hif1α∆Adipo mice after 15 weeks of HFD to further characterize in vivo insulin action. In the basal state, Hif1α∆Adipo mice had significantly reduced plasma glucose and insulin levels; basal endogenous glucose production (EGP) was similar between genotypes (Fig. 4D–F and H). During the clamp, insulin was infused to maintain plasma insulin levels at ∼4 ng/mL (Fig. 4F), and the glucose infusion rate (GIR) was adjusted in order to maintain blood glucose levels in Hif1αF/F and Hif1α∆Adipo mice at similar levels (Fig. 4D, E, and G). GIR was significantly increased in Hif1α∆Adipo mice (Fig. 4G and H), which confirmed improved whole-body insulin sensitivity in Hif1α∆Adipo mice. During the clamp, insulin induced a more marked suppression of EGP in Hif1α∆Adipo mice, suggesting increased insulin sensitivity in the liver. Whole-body glucose disposal (Rd) and glucose uptake into skeletal muscle and adipose tissue were significantly increased in Hif1α∆Adipo mice (Fig. 4H). Taken together, these data suggest that adipose selective inactivation of HIF1 caused increased insulin sensitivity in major insulin target tissues, liver, skeletal muscle, and fat.
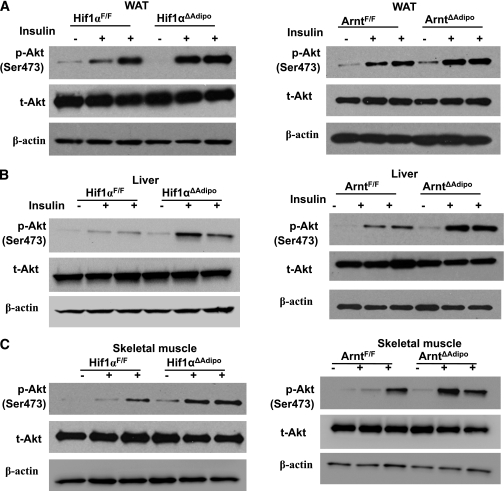
Insulin action was further investigated in WAT, liver, and skeletal muscle (15). Both ARNT and HIF1α deficiency in adipocytes improved insulin signaling pathways in WAT, liver, and skeletal muscle, as revealed by increased phosphorylation of Akt (ser473) (Fig. 5A–C). These findings indicated that HIF1 deficiency in adipocytes improved HFD-induced glucose tolerance and insulin resistance, which is in support of the hyperinsulinemic-euglycemic clamp studies.
FIG. 5.
Arnt and Hif1α disruption in adipocytes enhanced insulin signaling pathways. Insulin-stimulated Akt phosphorylation (Ser473) in WAT (A), liver (B), and skeletal muscle (C) of ArntF/F and Arnt∆Adipo and Hif1αF/F and Hif1α∆Adipo mice.
Expression of genes altered in ARNT- and HIF1-deficient adipose tissue.
After 12 weeks of HFD, HIF1α was significantly elevated, resulting in activation of target genes such as Glut1 and Vegf (Supplementary Fig. 6A). On a chow diet, HIF1 target genes and adiponectin expression were similar between Hif1aF/F and Hif1α∆Adipo mice (Supplementary Fig. 6B). In addition, there was no significant nuclear HIF1α protein expression in WAT from chow-fed wild-type mice, while it was significantly induced in HFD-fed mice and diminished in Hif1α∆Adipo mice (Supplementary Fig. 6C). Thus, the phenotype of Hif1α∆Adipo mice is similar to Hif1aF/F mice on a chow diet but different on an HFD. In contrast to HIF1α protein, HFD treatment did not elevate HIF2α protein expression and there was no increase in Hif1α∆Adipo mice (Supplementary Fig. 6C).
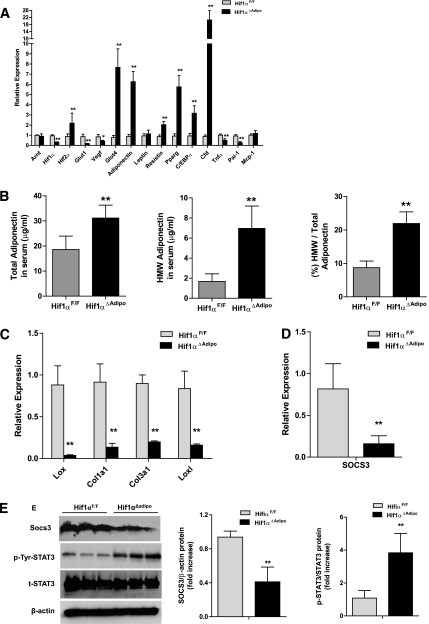
Gene expression profiling of WAT after 12 weeks of HFD in ArntF/F and Arnt∆Adipo mice or Hif1αF/F and Hif1α∆Adipo mice showed that the HIF1 target genes Glut1 and Vegf were decreased in Arnt∆Adipo and Hif1α∆Adipo mice (Supplementary Table 1; Supplementary Fig. 7A; Fig. 6A). Expression of genes involved in adipogenesis and glucose metabolism, Pparg, C/EBPα, Cfd, and Glut4, were upregulated in WAT of Arnt∆Adipo and Hif1α∆Adipo mice compared with ArntF/F and Hif1αF/F mice, respectively, on an HFD. Importantly, total serum adiponectin, the high–molecular weight (HMW) form of adiponectin, and the ratio of HMW to total adiponectin were increased in Arnt∆Adipo and Hif1α∆Adipo mice on a 12-week HFD (Supplementary Fig. 7B; Fig. 6B). HMW adiponectin and ratio of HMW to total adiponectin began to be increased following 6 weeks of HFD before the decrease in body weight (Supplementary Fig. 5D). Previous studies showed that overexpression of a constitutively active form of HIF1α initiates adipose tissue fibrosis and insulin resistance (16). In agreement with this finding, expression of the fibrosis related genes Lox, Col1a1, Col3a1, and Loxl1 were decreased in Arnt∆Adipo or Hif1α∆Adipo mice (Supplementary Table 1; Supplementary Fig. 7C; Fig. 6C). Expression of the inflammation-related genes Tnfα and Pai-1 was downregulated in Arnt∆Adipo and Hif1α∆Adipo mice (Supplementary Table 1; Supplementary Fig. 7A; Fig. 6A). Macrophage marker F4/80 staining of adipose tissue sections revealed that macrophage infiltration to WAT decreased significantly in Arnt∆Adipo and Hif1α∆Adipo mice. Expression of mRNAs encoding the macrophage markers F4/80 and CD68 were also decreased in Arnt∆Adipo mice and Hif1α∆Adipo mice (Supplementary Table 1; Supplementary Fig. 8A and B). Because it was reported that the aP2 promoter is expressed in macrophage (17), the possibility that HIF1 was disrupted in macrophage was investigated by determining Arnt gene disruption in peritoneal macrophage (P-Mϕ) from Arnt∆Adipo mice. Loss of ARNT in peritoneal macrophage of Arnt∆Adipo mice was minimal in contrast to much lower ARNT mRNA levels in Arnt∆Adipo mouse adipocytes (Fig. 7A).
FIG. 6.
Expression of genes related to insulin resistance in Hif1α disrupted WAT. WAT from Hif1αF/F and Hif1α∆Adipo mice after 12 weeks of HFD was analyzed. A: qPCR analysis of various mRNAs. B: Total adiponectin levels, HMW adiponectin, and the ratio of HMW to total adiponectin. C: qPCR analyses of fibrosis-related gene expression. D: qPCR analyses of Socs3 expression. E: Western blot analysis of SOCS3 expression and STAT3 phosphorylation (left panel) and quantitation of SOCS3 expression and STAT3 phosphorylation (right panel). Relative protein levels were normalized to levels for floxed littermates. For qPCR analysis, expression was normalized to β-actin. Data are means ± SD. *P < 0.05, **P < 0.01 compared with floxed littermates.
FIG. 7.
Gene expression in adipocytes and SVF-Mϕ after HFD. A: qPCR analysis of Arnt mRNA expression in the peritoneal macrophages (P-Mϕ) (B) and qPCR analysis of HIF1α, SOCS3, and adiponectin mRNA expression in adipocytes and SVF-Mϕ (C) of Hif1αF/F and Hif1α∆Adipo mice after 12 weeks of HFD. The expression was normalized to β-actin. Data are means ± SD. *P < 0.05, **P < 0.01 compared with floxed littermates.
Previous studies demonstrated that SOCS3 mRNA is elevated in WAT of diet-induced obesity (DIO) mice (18,19). Further, haploinsufficiency of SOCS3 significantly protected mice against the development of DIO and associated metabolic complications (20). Consistent with these findings, SOCS3 mRNA was also found to be downregulated in Arnt∆Adipo and Hif1α∆Adipo mice, as revealed by microarray analysis (Supplementary Table 1). Expression of SOCS3 mRNA in Arnt∆Adipo and Hif1α∆Adipo mice was significantly lower than that in ArntF/F and Hif1αF/F mice, respectively (Supplementary Fig. 7D; Fig. 6D). Expression of SOCS3 protein was also significantly decreased in Hif1α∆Adipo mice (Fig. 6E). Adipocytes and macrophage of stromal vascular fraction (SVF-Mϕ) were prepared from WAT of Hif1α∆Adipo and Hif1αF/F mice after 12 weeks of HFD. In adipocytes, no significant expression of HIF1α mRNA in Hif1α∆Adipo mice was found. After 12 weeks of HFD, expression of SOCS3 in adipocytes of Hif1α∆Adipo mice was ~80% decreased compared with that in Hif1αF/F mice. Adiponectin expression was upregulated in adipocytes of Hif1α∆Adipo mice (Fig. 7B). Consistent with data obtained in P-Mϕ, the expression of HIF1α, SOCS3, and adiponectin (adiponectin expression being nearly undetectable in SVF-Mϕ) in SVF-Mϕ were not significantly different between Hif1α∆Adipo and Hif1αF/F mice after 12 weeks of HFD (Fig. 7C).
Previous studies revealed that SOCS3 can inhibit adiponectin production via JAK2-STAT3–dependent mechanisms in adipocytes (21). Since inactivation of STAT3 may be involved in the inhibition of adiponectin production by SOCS3, tyrosine phosphorylation of STAT3 was assessed in WAT from Hif1αF/F and Hif1α∆Adipo mice. Indeed, tyrosine phosphorylation of STAT3 was significantly induced in parallel with increased adiponectin level in Hif1α∆Adipo mice (Fig. 6E). These results suggest that the SOCS3 and adiponectin pathway is involved, in part, in the improvement of HFD-induced insulin resistance in Hif1α∆Adipo mice.
DISCUSSION
Cellular hypoxia is observed in adipose tissue of obese individuals (22). However, the role of hypoxia in adipose tissue during obesity and insulin resistance remains unclear. In the current study, adipocyte-specific disruption of HIF1α or its heterodimerization partner, ARNT, were shown to protect against HFD-induced obesity and insulin resistance. HIF1 signaling may regulate SOCS3 and adiponectin, thus providing a mechanistic clue by which HIF1 exacerbates whole-body insulin resistance. These findings indicate a central role for adipocyte HIF1 signaling in the pathogenesis of obesity and insulin resistance.
In the obese mouse model, hypoxia occurs specifically in WAT, and the response to adipose hypoxia may lead to insulin resistance (23). HIF1, the main mediator of the hypoxia response in adipose tissue, is almost undetectable in lean mice but significantly increased in obese mice resulting in induction of the HIF1 target genes Glut1 and Pdk1 (2). Moreover, overexpression of a constitutively-active HIF1α in adipose tissue leads to glucose intolerance and insulin resistance (16). These studies are consistent with results of the present work showing that ablation of ARNT or HIF1α in adipose tissue improved HFD-induced obesity and insulin resistance.
In Arnt∆Adipo and Hif1α∆Adipo mice, adipogenesis-related genes such as Pparg, C/ebpα, Cfd, and Glut4 were increased and adipocyte size and mass reduced after HFD challenge, consistent with the notion that enhanced adipogenesis does not correlate with obesity (24,25). These results provide in vivo evidence that hypoxia inhibits adipogenesis in an HIF1-dependent manner (26). The reduced adiposity in Arnt∆Adipo and Hif1α∆Adipo mice was independent of food intake and may be due in part to increased energy expenditure. Arnt∆Adipo and Hif1α∆Adipo mice exhibited higher serum total adiponectin levels. Others found that in obese mice, adiponectin treatment increased energy expenditure and decreased mouse fat mass by upregulating expression of uncoupling protein 2 (UCP2) and increasing fatty acid oxidation (27–30). In addition, decreased vascular endothelial growth factor (VEGF) mRNA in Arnt∆Adipo and Hif1α∆Adipo mice WAT suggests that lower VEGF as a result of loss of HIF1 may also contribute to the decreased adiposity in Arnt∆Adipo and Hif1α∆Adipo mice. Indeed, VEGF inhibitors were shown to decrease adipose mass following HFD (31).
It has previously been claimed that aP2 expression is induced in activated macrophages (17). However, in Hif1α∆Adipo mice the knockout efficiency of HIF1α in SVF-Mϕ was very low—consistent with recent reports showing that the efficiency of Cre recombination in macrophage was much less than that in adipocytes (14,32–34).
Loss of HIF1 in adipose tissue can improve whole-body glucose intolerance after HFD, mainly due to enhanced insulin signaling in WAT, liver, and skeletal muscle. The current study revealed that SOCS3 is regulated by HIF1 in WAT. Inflammatory factors such as tumor necrosis factor-α, interleukin-6, and lipopolysaccharide also inhibit insulin signaling via upregulation of SOCS3 (18,35). SOCS3 can bind to phosphorylated tyrosine 960 of the insulin receptor (36) and inhibit insulin receptor autophosphorylation (37), IRS1 phosphorylation, and downstream insulin signaling (35). SOCS3 deficiency increases insulin-stimulated glucose uptake in adipocytes (19). Therefore, decreased SOCS3 in Arnt∆Adipo and Hif1α∆Adipo mice may account for the improved insulin signaling in WAT. The increased adiponectin expression and secretion was accompanied by a decrease of SOCS3 and activation of STAT3 in Arnt∆Adipo and Hif1α∆Adipo mice. Adiponectin improves insulin signaling in both skeletal muscle and the liver (30). This can explain the present findings that insulin sensitivity in liver and muscle was improved in Arnt∆Adipo and Hif1α∆Adipo mice compared with ArntF/F and Hif1αF/F mice. In addition, HMW adiponectin levels and the ratio of HMW to total adiponectin were increased in Arnt∆Adipo and Hif1α∆Adipo mice. Indeed, HMW adiponectin levels, or the ratio of HMW adiponectin to total adiponectin, are more meaningful markers than total adiponectin levels for predicting insulin resistance and the development of metabolic syndrome (38). Overexpression of SOCS3 in adipose tissue inhibited local insulin action but improved systemic glucose metabolism, and adiponectin production was increased after HFD (39). Whereas these data seem paradoxical given the current results, others reported that SOCS3 is elevated in WAT under the pathological conditions of insulin resistance (18,19) and when SOCS3 is overexpressed SOCS3 levels rise far higher than under normal physiological conditions. In the current study, SOCS3 expression was decreased in Arnt∆Adipo and Hif1α∆Adipo mice after HFD. Differences in the model and experimental conditions may account for these different observations. In addition, consistent with the present results, a recent study showed that pioglitazone exerts its effect to improve whole-body insulin sensitivity through the suppression of SOCS3, which is associated with an increase in STAT3 phosphorylation and adiponectin production in WAT (21). Moreover, Arnt∆Adipo and Hif1α∆Adipo mice also have reduced serum triglycerides and FFAs, which is consistent with studies showing that FFAs and triglycerides impair insulin signaling and induce insulin resistance mainly in liver and muscle (40,41). It is also of interest that the proinflammatory factors tumor necrosis factor-α and plasminogen activator inhibitor-1 were decreased in Arnt∆Adipo and Hif1α∆Adipo mice. These factors are also associated with improved insulin sensitivity of the whole body in Arnt∆Adipo and Hif1α∆Adipo mice after HFD.
In conclusion, the current study clearly shows that HIF1 in adipose tissue plays an important role in the metabolism of lipid and glucose. The HIF1 effects on insulin sensitivity may be due in part to its direct or indirect regulation of SOCS3 in WAT, whereas the mechanism of its effect on adiposity and obesity is still not clear and requires further investigation. Because loss of HIF1 activity improves metabolic function, compounds that inhibit HIF1 function in adipose tissue might have significant therapeutic potential in reducing obesity and insulin resistance.
ACKNOWLEDGMENTS
This study was supported by the National Cancer Institute and National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Programs and National Institutes of Health Grant CA-148828 (to Y.M.S.).
No potential conflicts of interest relevant to this article were reported.
C.J. researched data, contributed to discussion, and wrote the manuscript. A.Q., T.M., T.C., W.J., O.G., and Y.M.S. researched data, contributed to discussion, and reviewed and edited the manuscript. F.J.G. contributed to discussion and reviewed and edited the manuscript.
The authors thank Barbara B. Kahn, Harvard Medical School, for supplying the aP2-Cre mouse line used in the study.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0174/-/DC1.
REFERENCES
- 1.Trayhurn P, Wang B, Wood IS. Hypoxia in adipose tissue: a basis for the dysregulation of tissue function in obesity? Br J Nutr 2008;100:227–235 [DOI] [PubMed] [Google Scholar]
- 2.Ye J, Gao Z, Yin J, He Q. Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of ob/ob and dietary obese mice. Am J Physiol Endocrinol Metab 2007;293:E1118–E1128 [DOI] [PubMed] [Google Scholar]
- 3.Regazzetti C, Peraldi P, Grémeaux T, et al. Hypoxia decreases insulin signaling pathways in adipocytes. Diabetes 2009;58:95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatanaka M, Shimba S, Sakaue M, et al. Hypoxia-inducible factor-3alpha functions as an accelerator of 3T3-L1 adipose differentiation. Biol Pharm Bull 2009;32:1166–1172 [DOI] [PubMed] [Google Scholar]
- 5.Shimba S, Wada T, Hara S, Tezuka M. EPAS1 promotes adipose differentiation in 3T3-L1 cells. J Biol Chem 2004;279:40946–40953 [DOI] [PubMed] [Google Scholar]
- 6.Ye J. Emerging role of adipose tissue hypoxia in obesity and insulin resistance. Int J Obes (Lond) 2009;33:54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivan M, Kondo K, Yang H, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001;292:464–468 [DOI] [PubMed] [Google Scholar]
- 8.Abel ED, Peroni O, Kim JK, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 2001;409:729–733 [DOI] [PubMed] [Google Scholar]
- 9.Tomita S, Sinal CJ, Yim SH, Gonzalez FJ. Conditional disruption of the aryl hydrocarbon receptor nuclear translocator (Arnt) gene leads to loss of target gene induction by the aryl hydrocarbon receptor and hypoxia-inducible factor 1alpha. Mol Endocrinol 2000;14:1674–1681 [DOI] [PubMed] [Google Scholar]
- 10.Tomita S, Ueno M, Sakamoto M, et al. Defective brain development in mice lacking the Hif-1alpha gene in neural cells. Mol Cell Biol 2003;23:6739–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyoshima Y, Gavrilova O, Yakar S, et al. Leptin improves insulin resistance and hyperglycemia in a mouse model of type 2 diabetes. Endocrinology 2005;146:4024–4035 [DOI] [PubMed] [Google Scholar]
- 12.Gavrilova O, Marcus-Samuels B, Reitman ML. Lack of responses to a beta3-adrenergic agonist in lipoatrophic A-ZIP/F-1 mice. Diabetes 2000;49:1910–1916 [DOI] [PubMed] [Google Scholar]
- 13.Chen M, Chen H, Nguyen A, et al. G(s)alpha deficiency in adipose tissue leads to a lean phenotype with divergent effects on cold tolerance and diet-induced thermogenesis. Cell Metab 2010;11:320–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugii S, Olson P, Sears DD, et al. PPARgamma activation in adipocytes is sufficient for systemic insulin sensitization. Proc Natl Acad Sci USA 2009;106:22504–22509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001;414:799–806 [DOI] [PubMed] [Google Scholar]
- 16.Halberg N, Khan T, Trujillo ME, et al. Hypoxia-inducible factor 1alpha induces fibrosis and insulin resistance in white adipose tissue. Mol Cell Biol 2009;29:4467–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makowski L, Boord JB, Maeda K, et al. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med 2001;7:699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emanuelli B, Peraldi P, Filloux C, et al. SOCS-3 inhibits insulin signaling and is up-regulated in response to tumor necrosis factor-alpha in the adipose tissue of obese mice. J Biol Chem 2001;276:47944–47949 [DOI] [PubMed] [Google Scholar]
- 19.Shi H, Tzameli I, Bjørbaek C, Flier JS. Suppressor of cytokine signaling 3 is a physiological regulator of adipocyte insulin signaling. J Biol Chem 2004;279:34733–34740 [DOI] [PubMed] [Google Scholar]
- 20.Howard JK, Cave BJ, Oksanen LJ, Tzameli I, Bjørbaek C, Flier JS. Enhanced leptin sensitivity and attenuation of diet-induced obesity in mice with haploinsufficiency of Socs3. Nat Med 2004;10:734–738 [DOI] [PubMed] [Google Scholar]
- 21.Kanatani Y, Usui I, Ishizuka K, et al. Effects of pioglitazone on suppressor of cytokine signaling 3 expression: potential mechanisms for its effects on insulin sensitivity and adiponectin expression. Diabetes 2007;56:795–803 [DOI] [PubMed] [Google Scholar]
- 22.Hosogai N, Fukuhara A, Oshima K, et al. Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes 2007;56:901–911 [DOI] [PubMed] [Google Scholar]
- 23.Yin J, Gao Z, He Q, Zhou D, Guo Z, Ye J. Role of hypoxia in obesity-induced disorders of glucose and lipid metabolism in adipose tissue. Am J Physiol Endocrinol Metab 2009;296:E333–E342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006;7:885–896 [DOI] [PubMed] [Google Scholar]
- 25.van Eijk M, Aten J, Bijl N, et al. Reducing glycosphingolipid content in adipose tissue of obese mice restores insulin sensitivity, adipogenesis and reduces inflammation. PLoS ONE 2009;4:e4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yun Z, Maecker HL, Johnson RS, Giaccia AJ. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev Cell 2002;2:331–341 [DOI] [PubMed] [Google Scholar]
- 27.Bauche IB, El Mkadem SA, Pottier AM, et al. Overexpression of adiponectin targeted to adipose tissue in transgenic mice: impaired adipocyte differentiation. Endocrinology 2007;148:1539–1549 [DOI] [PubMed] [Google Scholar]
- 28.Fruebis J, Tsao TS, Javorschi S, et al. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc Natl Acad Sci USA 2001;98:2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masaki T, Chiba S, Yasuda T, et al. Peripheral, but not central, administration of adiponectin reduces visceral adiposity and upregulates the expression of uncoupling protein in agouti yellow (Ay/a) obese mice. Diabetes 2003;52:2266–2273 [DOI] [PubMed] [Google Scholar]
- 30.Yamauchi T, Kamon J, Minokoshi Y, et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med 2002;8:1288–1295 [DOI] [PubMed] [Google Scholar]
- 31.Tam J, Duda DG, Perentes JY, Quadri RS, Fukumura D, Jain RK. Blockade of VEGFR2 and not VEGFR1 can limit diet-induced fat tissue expansion: role of local versus bone marrow-derived endothelial cells. PLoS ONE 2009;4:e4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He W, Barak Y, Hevener A, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA 2003;100:15712–15717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wueest S, Rapold RA, Schumann DM, et al. Deletion of Fas in adipocytes relieves adipose tissue inflammation and hepatic manifestations of obesity in mice. J Clin Invest 2010;120:191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, Lawrence JC, Jr, Jung DY, et al. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes 2010;59:1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueki K, Kondo T, Kahn CR. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol Cell Biol 2004;24:5434–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emanuelli B, Peraldi P, Filloux C, Sawka-Verhelle D, Hilton D, Van Obberghen E. SOCS-3 is an insulin-induced negative regulator of insulin signaling. J Biol Chem 2000;275:15985–15991 [DOI] [PubMed] [Google Scholar]
- 37.Senn JJ, Klover PJ, Nowak IA, et al. Suppressor of cytokine signaling-3 (SOCS-3), a potential mediator of interleukin-6-dependent insulin resistance in hepatocytes. J Biol Chem 2003;278:13740–13746 [DOI] [PubMed] [Google Scholar]
- 38.Matsushita K, Yatsuya H, Tamakoshi K, et al. Comparison of circulating adiponectin and proinflammatory markers regarding their association with metabolic syndrome in Japanese men. Arterioscler Thromb Vasc Biol 2006;26:871–876 [DOI] [PubMed] [Google Scholar]
- 39.Shi H, Cave B, Inouye K, Bjørbaek C, Flier JS. Overexpression of suppressor of cytokine signaling 3 in adipose tissue causes local but not systemic insulin resistance. Diabetes 2006;55:699–707 [DOI] [PubMed] [Google Scholar]
- 40.Hulver MW, Dohm GL. The molecular mechanism linking muscle fat accumulation to insulin resistance. Proc Nutr Soc 2004;63:375–380 [DOI] [PubMed] [Google Scholar]
- 41.Leroyer SN, Tordjman J, Chauvet G, et al. Rosiglitazone controls fatty acid cycling in human adipose tissue by means of glyceroneogenesis and glycerol phosphorylation. J Biol Chem 2006;281:13141–13149 [DOI] [PubMed] [Google Scholar]