Abstract
OBJECTIVE
A major feature of type 1 diabetes is the appearance of islet autoantibodies before diagnosis. However, although the genetics of type 1 diabetes is advanced, the genetics of islet autoantibodies needs further investigation. The primary susceptibility loci in type 1 diabetes, the HLA class I and II genes, are believed to determine the specificity and magnitude of the autoimmune response to islet antigens. We investigated the association of glutamic acid decarboxylase autoantibodies (GADA) and insulinoma-associated antigen-2 autoantibodies (IA-2A) with the HLA region.
RESEARCH DESIGN AND METHODS
Associations of GADA and IA-2A with HLA-DRB1, HLA-DQB1, HLA-B, HLA-C, HLA-A, MICA, and 3,779 single nucleotide polymorphisms (SNPs) were analyzed in 2,531 childhood-onset case subjects (median time since diagnosis 5 years). All analyses were adjusted for age-at-diagnosis and duration of diabetes.
RESULTS
GADA and IA-2A were associated with an older age-at-diagnosis (P < 10−19). For GADA, the primary association was with HLA-DQB1 (P = 9.00 × 10−18), with evidence of a second independent effect in the HLA class I region with SNP, rs9266722 (P = 2.84 × 10−6). HLA-DRB1 had the strongest association with IA-2A (P = 1.94 × 10−41), with HLA-A*24 adding to the association, albeit negatively (P = 1.21 × 10−10). There was no evidence of association of either IA-2A or GADA with the highly type 1 diabetes predisposing genotype, HLA-DRB1*03/04.
CONCLUSIONS
Despite genetic association of type 1 diabetes and the islet autoantibodies localizing to the same HLA class II genes, HLA-DRB1 and HLA-DQB1, the effects of the class II alleles and genotypes involved are quite different. Therefore, the presence of autoantibodies is unlikely to be causal, and their role in pathogenesis remains to be established.
Islet autoantibodies, specifically glutamic acid decarboxylase autoantibodies (GADA), insulinoma-associated antigen-2 autoantibodies (IA-2A), and insulin autoantibodies (IAA), are commonly found in type 1 diabetic case subjects at time of diagnosis, with >90% of patients positive for one or more of IA-2A, IAA, or GADA (1–3). Autoantibodies can appear months or even years before diagnosis of disease, with the probability of developing disease increasing with the number of persistent islet autoantibodies present (3–8). Consequently, they can be used to predict the onset of type 1 diabetes in unaffected relatives of type 1 diabetic case subjects (6,9). The major histocompatibility complex (MHC) region, specifically the HLA class II genes, are the primary genetic loci associated with type 1 diabetes (10,11). Recently, a combination of high throughput, dense, single nucleotide polymorphism (SNP) and classical HLA typing has allowed further mapping of the MHC region, revealing independent type 1 diabetes associations at the HLA-B and HLA-A loci, involving multiple alleles, including HLA-B*39, HLA-B*18, HLA-A*24, HLA-A*01, and HLA-A*11 (10,11). Autoantibody expression has also been correlated with HLA class II alleles. However, once unaffected relatives of type 1 diabetic patients were positive for two or more autoantibodies, HLA class II risk genotypes did not add to the prediction of type 1 diabetes diagnosis (6,12).
It seems reasonable to assume that the genetics of islet autoantibodies may be shared in part with the genetics of type 1 diabetes. Others have investigated the association of specific type 1 diabetes–associated HLA class II and I alleles and genotypes with GADA and IA-2A, but these have predominantly been confined to small sample sets. GADA has been shown to be associated with the HLA-DRB1*03-HLA-DQB1*02 haplotype (13,14), whereas HLA-DRB1*04 was shown to be negatively correlated with GADA (14). The largest study to date did not obtain evidence of association between the HLA class II and I genes and GADA (15). The HLA-DRB1*04-HLA-DQB1*0302 haplotype has been reported to be positively associated with IA-2A, whereas there is a reported negative association of HLA-DRB1*03-HLA-DQB1*02 with IA-2A (13–16). Both HLA-DRB1*07-HLA-DQA1*(0201 or 0301) and HLA-DRB1*09-HLA-DQA1*0301 haplotypes have been reported to be associated with increased prevalence of IA-2A in case subjects with type 1 diabetes, which the authors concluded was attributable to the HLA-DQA1 gene driving the association of these haplotypes with IA-2A (17). HLA-A*24 has also been negatively associated with IA-2A (15). However, this was not shown to be independent of the primary HLA class II autoantibody associations nor has it been independently replicated. To date, a comprehensive analysis of the association of these islet autoantibodies with the MHC region, in a large well-powered sample of type 1 diabetes, has not been reported. Here we have fine-mapped, using over 3,700 SNPs and classical HLA genotypes, the association of GADA and IA-2A in the extended MHC region in up to ∼2,500 type 1 diabetic case subjects.
RESEARCH DESIGN AND METHODS
Subjects.
All type 1 diabetic case subjects were recruited for the Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory’s Genetic Resource Investigating Diabetes British, type 1 diabetes collection (www.childhood-diabetes.org.uk/grid.shtml). Case subjects were diagnosed with type 1 diabetes under age 17 years (mean age-at-diagnosis = 7.6 years) and on average had diabetes for 6.8 years (Table 1). The appropriate ethics committees approved the collection of all DNA samples, and written consent was obtained from all individuals or parents of individuals who were too young to consent.
TABLE 1.
Distribution of phenotypes in type 1 diabetic case subjects overall and for each autoantibody subgroup
| Phenotype | All case subjects | GADA |
IA-2A |
||
|---|---|---|---|---|---|
| Positives | Negatives | Positives | Negatives | ||
| N | 2,531 | 1,274 | 1,256 | 1,475 | 1,046 |
| Age-at-diagnosis (years) | |||||
| Mean (SD) | 7.6 (4.0) | 8.5 (3.9) | 6.6 (3.9) | 8.3 (3.7) | 6.6 (4.2) |
| Median | 7 | 9 | 6 | 8 | 6 |
| Age (years) | |||||
| Mean (SD) | 14.4 (8.3) | 14.1 (7.1) | 14.8 (9.3) | 13.7 (6.8) | 15.4 (10.0) |
| Median | 13 | 13 | 13 | 13 | 13 |
| Duration (years) | |||||
| Mean (SD) | 6.8 (7.8) | 5.5 (6.5) | 8.2 (8.7) | 5.4 (6.1) | 8.8 (9.4) |
| Median | 5 | 3.4 | 6 | 4 | 6 |
| Sex | |||||
| Male, N (%) | 1,316 (52) | 579 (45) | 737 (59) | 779 (53) | 530 (51) |
| Female, N (%) | 1,215 (48) | 695 (55) | 519 (41) | 696 (47) | 516 (49) |
Genotyping.
The HLA-DRB1, HLA-DQB1, HLA-A, HLA-B, and HLA-C genes were typed at four-digit resolution using Dynal RELI SSO assays (Invitrogen, Paisley, U.K.). A subset of 2,960 case subjects was genotyped at HLA-DRB1 and HLA-DQB1 using Roche Molecular Systems SSO reverse dot blot technology. As described previously, the MICA-STR was genotyped using PCR followed by capillary electrophoresis of the amplified product (18); the 2,420 SNPs from the Type 1 Diabetes Genetics Consortium (T1DGC) genome-wide association (GWA) study were genotyped using the Illumina 550 K Infinium platform (19), and 1,687 SNPs from the Wellcome Trust Case-Control Consortium GWA study were genotyped using the Affymetrix GeneChip Human Mapping 500 K Array set (20). Three hundred and twenty-eight SNPs that passed quality control were common to both platforms. Follow-up and tag SNPs were genotyped blind to case-control status using the Taqman 5′ nuclease assay (Applied Biosystems, Warrington, U.K.) according to manufacturer’s protocols. Two operators scored genotypes. HLA-DQA1 genotypes were not determined.
Autoantibodies.
Presence of each of the autoantibodies (IA-2A, GADA) in the type 1 diabetic case subjects was tested using plasma stored in aliquots at −80°C. Autoantibodies to GAD and IA-2 were measured in the Department of Clinical Sciences at North Bristol, University of Bristol, between 2006 and 2007, using a radioimmunoassay (21,22). In the 2007 Proficiency Evaluation of the Diabetes Antibody Standardization Program (23), the GADA assay achieved 94% sensitivity with 96% specificity, whereas the IA-2 assay sensitivity was 70% sensitivity with 98% specificity. Presence of GADA and IA-2A was taken as above 14 and 6 World Health Organization units/mL, respectively, which corresponds to the 97.5th percentile of the distribution of these autoantibodies in 2,860 school children from Oxford, U.K. (21,22). We did not measure IAA since most case subjects were recruited after diagnosis and antibodies to exogenous insulin are produced.
Statistical analyses.
All statistical analyses were carried out in STATA (www.stata.com) or R (24). Associations with autoantibodies were tested using logistic regression models with autoantibody status (positive/negative) as the outcome variable and genotype at the test locus and any covariates (sex, duration, age-at-diagnosis; see results) as predictors. Inclusion of the covariates accounted for their effects, such that association of genotype with autoantibody positivity was tested, rather than, for example, association of genotype with decline in autoantibody positivity (as a result of disease duration). A model with and without the genotype variable was compared using a likelihood ratio test to assess significance. Stepwise logistic regression was used to test for independent associations (25). Haplotypes were generated using the R library haplo.stats (available from www.r-project.org) and analyzed by logistic regression using the posterior probabilities as weights and robust variance estimates to account for the dependency between haplotype assignments. Despite being genotyped at four-digit resolution, to maximize statistical power, analyses of the classical loci were confined to two-digit resolution, except for HLA-DQB1*0301 and HLA-DQB1*0302.
RESULTS
Autoantibodies.
GADA were measured in 2,530 type 1 diabetic case subjects, 1,274 (50.4%) of whom were found to have levels of GADA above the 97.5th percentile for the reference population (research design and methods) and were defined as GADA positive. IA-2A were measured in 2,521 type 1 diabetic case subjects, 1,475 (58.5%) of whom had levels above the 97.5th percentile for the reference population (research design and methods) and were defined as IA-2A positive. Of the 2,520 case subjects tested for both IA-2A and GADA, 788 (31.3%) were positive for both autoantibodies and 1,958 (77.7%) were positive for at least one of IA-2A and GADA.
As expected, GADA positivity and IA-2A positivity were both found to be correlated with a shorter duration of diabetes (P = 9.80 × 10−14 and 8.65 × 10−22, respectively, having accounted for age-at-diagnosis effects; Table 1). In addition, GADA positivity and IA-2A positivity were also correlated with a later age-at-diagnosis (P = 3.72 × 10−29 and 7.22 × 10−20, respectively, having accounted for duration of diabetes; Supplementary Data). More females were GADA positive than males (P = 2.91 × 10−12; Table 1). Hence, age-at-diagnosis and duration of disease were included as covariates in the statistical models testing for association at IA-2A and GADA; in addition, sex was included for GADA.
GADA associations.
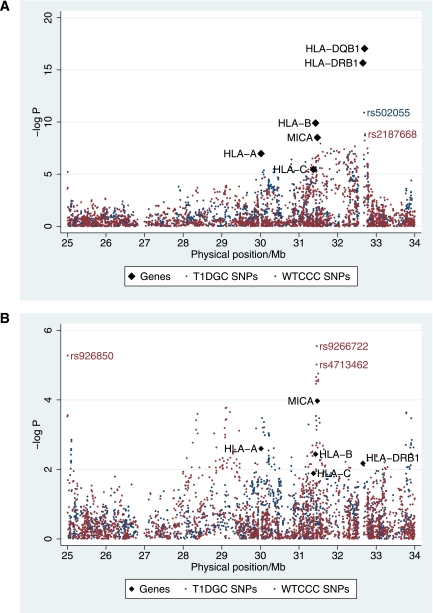
In total 3,779 SNPs between 25 and 33 Mb on chromosome 6p21 containing the extended MHC region, and the HLA-DQB1, HLA-DRB1, HLA-B, HLA-C, HLA-A, and MICA(STR) genes were analyzed for association with GADA. In type 1 diabetes, the strongest association signal was with the HLA class II region (10,11), which was also found to have the strongest signal for association with GADA positivity (Fig. 1A and Table 2). The strongest association with GADA positivity was with HLA-DQB1 (P = 9.00 × 10−18), but HLA-DRB1 was also convincingly associated (P = 3.10 × 10−17; Table 2). Indeed, the two SNPs most associated with GADA positivity (rs502055, located at 32.579 Mb between HLA-DRB1 and HLA-DQA1, P = 6.53 × 10−11; and rs2187668, located at 32.606 Mb in intron 1 of HLA-DQA1, P = 1.28 × 10−9; Fig. 1A) were in linkage disequilibrium (LD) with the type 1 diabetes susceptible allele, HLA-DQB1*02 (r2 > 0.69) and were in strong LD with the HLA-DRB1*03 type 1 diabetes susceptible allele (r2 > 0.85).
FIG. 1.
GADA positivity association map. A: Association of SNPs used in the T1DGC Illumina experiment (using ∼1,218 type 1 diabetic case subjects) and the WTCCC Affymetrix experiment (using ∼1,056 case subjects) and the classical loci (using between 2,408 and 1,275 type 1 diabetic case subjects; Table 2) with GADA positivity. The top SNPs from both the WTCCC and T1DGC experiments are in linkage disequilibrium with HLA-DRB1*03 (r2 = 0.85 and D’ = 1.00 with rs502055 and r2 = 1.00 and D’ =1.00 with rs2187668). B: Association of SNPs and genes with GADA positivity conditional on HLA-DQB1 genotypes.
TABLE 2.
Association of the HLA genes with GADA positivity in type 1 diabetic case subjects
| Gene | Physical position† | Number of samples |
P | P addition of locus to HLA-DQB1 | P addition of locus to HLA-DQB1 and rs9266722 | |
|---|---|---|---|---|---|---|
| GADA positive | GADA negative | |||||
| HLA-DQB1 | 32,273,871–33,498,585 | 1,195 | 1,213 | 9.00 × 10−18* | NA | NA |
| HLA-DRB1 | 32,485,163–32,557,625 | 1,194 | 1,212 | 3.10 × 10−17* | 0.0391 | 0.0050 |
| HLA-B | 31,237,115–31,324,989 | 774 | 776 | 1.26 × 10−10 | 0.0036 | 0.0422 |
| HLA-C | 31,236,526–31,323,369 | 667 | 608 | 3.48 × 10−6 | 0.0130 | 0.0209 |
| MICA (STR) | 31,367,561–31,384,016 | 1,206 | 1,196 | 3.09 × 10−9 | 1.08 × 10−4 | 0.3385 |
| HLA-A | 29,909,037–29,913,661 | 876 | 853 | 1.06 × 10−10* | 0.0025 | 0.0045 |
*The multiplicative allelic effects model was not an appropriate approximation (P = 5.66 × 10−4 for HLA-DQB1, P = 0.0011 for HLA-DRB1, P = 0.0015 for HLA-A); hence the P value for the genotype effects model is reported.
†Physical positions are from GRCh37 (www.t1dbase.org).
There is very little to distinguish the effects of HLA-DRB1 and HLA-DQB1 on GADA positivity (P = 0.002 for addition of HLA-DQB1 to HLA-DRB1 and P = 0.04 for addition of HLA-DRB1 to HLA-DQB1; Table 2), such that either HLA-DRB1 or HLA-DQB1 is sufficient to model the association, but both are not required. This contrasts with type 1 diabetes in which both genes are independently associated with disease and add to the association of each other (P < 3 × 10−240; Supplementary Data and Supplementary Tables 1 and 2). We examined which specific class II alleles and genotypes were involved in the GADA association and whether these were consistent with the type 1 diabetes associations. Despite having full four-digit HLA typing, owing to the rarity of many four-digit alleles, the HLA loci were analyzed at two-digit resolution, except at HLA-DQB1 for the HLA-DQB1*0301 and *0302 alleles, which are known to have opposite effects in type 1 diabetes (odds ratio [OR] [95% CI] = 4.21 [3.46–5.13] and 0.44 [0.36–0.54], respectively; data not shown).
At HLA-DQB1, the 02/02 genotype was most associated with GADA positivity (OR [95% CI] = 2.32 [1.71–3.13]; Table 3), whereas at HLA-DRB1, as expected owing to LD, the 03/03 genotype was most associated with GADA positivity (OR [95% CI] = 2.60 [1.86–3.65]; Table 4). These findings, and that HLA-DRB1*03 was convincingly associated with GADA positivity (P = 4.80 × 10−17; Supplementary Table 4), agree with previous reports in the literature (13,14). Unexpectedly, the highly type 1 diabetes susceptible HLA-DQB1*02/0302 genotype (Supplementary Data and Supplementary Table 1) had a comparable frequency in GADA positives (0.38) and GADA negatives (0.40; Table 3). Similarly, the most type 1 diabetes–associated genotype at HLA-DRB1, HLA-DRB1*03/04 (Supplementary Tables 2 and 3), was not associated with GADA (OR = 1.23 [0.90–1.68] using 04/04 as reference [Table 4]). There was also no suggestion of an HLA-DRB1*03/04 synergic effect with GADA (P = 0.19, compared with P < 7 × 10−20, in type 1 diabetes; Supplementary Data). Both the HLA-DQB1*0302/0302 genotype and the HLA-DRB1*04/04 genotype are highly susceptible for type 1 diabetes (Supplementary Tables 1 and 2), but these genotypes were not associated with GADA positivity (Tables 3 and 4). This is different from type 1 diabetes in which all HLA-DQB1*0302/X and HLA-DRB1*04/X genotypes confer susceptibility (Supplementary Tables 1 and 2). HLA-DQB1*0302 and HLA-DRB1*04 alleles were only (negatively) associated with GADA positivity when they were with specific type 1 diabetes low risk alleles (*06 or *04 at HLA-DQB1 and *01 or *13 at HLA-DRB1; Tables 3 and 4), which contrasts other reports (13,14).
TABLE 3.
Association of HLA-DQB1 genotypes with GADA positivity in type 1 diabetic case subjects
| Genotype |
N (frequency) |
OR [95% CI] | ||
|---|---|---|---|---|
| GADA positives | GADA negatives | Unconditional | ||
| 02/02 | 197 (0.16) | 83 (0.07) | 2.32 [1.71–3.13] | 2.76 [1.77–4.32] |
| 02/0303 | 28 (0.02) | 13 (0.01) | 2.31 [1.15–4.63] | 2.76 [1.28–5.95] |
| 02/04 | 24 (0.02) | 16 (0.01) | 1.70 [0.87–3.34] | 2.03 [0.96–4.30] |
| 0302/0301 | 60 (0.05) | 29 (0.02) | 1.55 [0.96–2.52] | 1.85 [1.03–3.32] |
| 02/05 | 74 (0.06) | 69 (0.06) | 1.09 [0.74–1.59] | 1.30 [0.78–2.14] |
| 02/0302 | 451 (0.38) | 482 (0.40) | 1.00 (reference) | 1.19 [0.81–1.75] |
| Rares | 56 (0.05) | 52 (0.04) | 0.88 [0.57–1.33] | 1.04 [0.61–1.78] |
| 02/0301 | 40 (0.03) | 41 (0.03) | 0.86 [0.53–1.39] | 1.02 [0.57–1.83] |
| 0302/0302 | 67 (0.06) | 74 (0.06) | 0.84 [0.57–1.23] | 1.00 (reference) |
| 02/06 | 28 (0.02) | 37 (0.03) | 0.61 [0.35–1.03] | 0.72 [0.39–1.35] |
| 0301/05 | 25 (0.02) | 39 (0.03) | 0.55 [0.32–0.95] | 0.66 [0.35–1.24] |
| 0302/05 | 76 (0.06) | 130 (0.11) | 0.54 [0.39–0.75] | 0.65 [0.41–1.03] |
| 0302/04 | 19 (0.02) | 46 (0.04) | 0.44 [0.25–0.78] | 0.52 [0.27–1.02] |
| 0302/0303 | 10 (0.01) | 23 (0.02) | 0.43 [0.19–0.95] | 0.51 [0.22–1.21] |
| 0302/06 | 40 (0.03) | 79 (0.07) | 0.39 [0.26–0.60] | 0.47 [0.27–0.80] |
Effect estimates were adjusted for the known covariates: sex, duration of disease, and age-at-diagnosis, whereas genotype frequencies were not. OR are reported with 95% CI using the most common, neutrally associated, genotype HLA-DQB1*02/0302 as reference and also the less common neutral genotype HLA-DQB1*0302/0302 as reference. Rare genotypes (13 in total) were grouped at 0.01 frequency.
TABLE 4.
Association of HLA-DRB1 genotypes with GADA positivity in type 1 diabetic case subjects
| Genotype |
N (frequency) |
OR [95% CI] |
||||
|---|---|---|---|---|---|---|
| GADA positives | GADA negatives | Unconditional | Conditional on HLA-DQB1 | |||
| 03/03 | 163 (0.14) | 63 (0.05) | 2.60 [1.86–3.65] | 3.20 [2.11–4.85] | 3.89 [2.10–7.18] | 12.24 [2.68–55.86] |
| 03/07 | 37 (0.03) | 16 (0.01) | 1.81 [0.96–3.40] | 2.22 [1.13–4.36] | 2.60 [1.17–5.78] | 7.80 [1.62–37.47] |
| 03/04 | 420 (0.35) | 441 (0.36) | 1.00 (reference) | 1.23 [0.90–1.68] | 1.00 (reference) | 3.14 [1.29–7.66] |
| 03/13 | 31 (0.03) | 31 (0.03) | 0.81 [0.47–1.41] | 1.00 [0.55–1.82] | 1.20 [0.51–2.84] | 3.42 [0.84–14.03] |
| 04/07 | 64 (0.05) | 63 (0.05) | 0.95 [0.64–1.41] | 1.17 [0.74–1.85] | 0.92 [0.62–1.37] | 2.92 [1.15–7.39] |
| 04/11 | 22 (0.02) | 8 (0.01) | 2.20 [0.94–5.15] | 2.70 [1.12–6.55] | 1.33 [0.50–3.58] | 2.10 [0.79–5.63] |
| 01/03 | 57 (0.05) | 53 (0.04) | 1.08 [0.70–1.65] | 1.33 [0.81–2.17] | 1.27 [0.66–2.43] | 1.39 [0.49–3.97] |
| 03/09 | 20 (0.02) | 11 (0.01) | 1.85 [0.85–4.00] | 2.27 [1.01–5.10] | 1.68 [0.64–4.40] | 1.66 [0.32–8.60] |
| 04/08 | 22 (0.02) | 48 (0.04) | 0.46 [0.26–0.80] | 0.56 [0.31–1.03] | 0.25 [0.09–0.75] | 1.02 [0.14–7.52] |
| Rares | 96 (0.08) | 114 (0.09) | 0.77 [0.56–1.06] | 0.94 [0.63–1.41] | 0.63 [0.36–1.12] | 1.04 [0.53–2.02] |
| 04/04 | 108 (0.09) | 115 (0.09) | 0.81 [0.59–1.12] | 1.00 (reference) | 0.52 [0.29–0.93] | 1.00 (reference) |
| 03/08 | 16 (0.01) | 11 (0.01) | 1.47 [0.65–3.34] | 1.81 [0.77–4.25] | 1.23 [0.39–3.90] | 0.87 [0.20–3.71] |
| 01/04 | 93 (0.08) | 155 (0.13) | 0.53 [0.39–0.72] | 0.66 [0.45–0.97] | 0.39 [0.20–0.76] | 0.86 [0.32–2.31] |
| 04/13 | 45 (0.04) | 83 (0.07) | 0.40 [0.26–0.60] | 0.49 [0.30–0.79] | 0.42 [0.19–0.94] | 0.74 [0.31–1.75] |
Effect estimates were calculated accounting for the known covariates: sex, duration of disease, and age-at-diagnosis, whereas genotype frequencies were not. Rare genotypes (39 in total) were grouped at 0.01 frequency. HLA-DRB1*0403 and *0407 alleles have opposite effects in type 1 diabetes to the remaining HLA-DBR1*04 alleles; however, they are of such low frequency in the case subjects that an extra subtype was not warranted and so are included with the HLA-DRB1*04 allele. The apparent association of certain HLA-DRB1 genotypes following conditioning on HLA-DQB1 are likely to be attributable to effects outside of the class II region (see main text) and all have very large 95% CIs reflecting uncertainty in the model.
Haplotypes of HLA-DQB1.HLA-DRB1 were also tested for association with GADA (Supplementary Table 5) and reflected the unconditional associations observed (Tables 3 and 4). The strongest association with GADA positivity was with the HLA-DQB1*02.HLA-DRB1*03/HLA-DQB1*02.HLA-DRB1*03 homozygous genotype (OR [95% CI] = 2.62 [1.87–3.67]; Supplementary Table 5); HLA-DQB1*0302.HLA-DRB1*04/HLA-DQB1*05.HLA-DRB1*01, HLA-DQB1*0302.HLA-DRB1*04/HLA-DQB1*0402.HLA-DRB1*08, and HLA-DQB1*0302.HLA-DRB1*04/HLA-DQB1*06.HLA-DRB1*13 were negatively associated with GADA positivity. In contrast, HLA-DQB1*0302.HLA-DRB1*04/HLA-DQB1*05.HLA-DRB1*01 and HLA-DQB1*0302.HLA-DRB1*04/HLA-DQB1*0402.HLA-DRB1*08 were associated with a significant increase in risk of type 1 diabetes (Supplementary Table 1). HLA-B*18 is associated with increased risk of type 1 diabetes, whereas HLA-B*08 is not (10,11). However, although HLA-B*08.HLA-DRB1*03.HLA-DQB1*02 could well be driving the association of HLA-DRB1*03 and HLA-DQB1*02 with GADA positivity, the effect could not strictly be distinguished from HLA-B*18.HLA-DRB1*03.HLA-DQB1*02 or HLA-B*X.HLA-DRB1*03.HLA-DQB1*02 since the 95% CIs overlapped (Supplementary Table 6). The extended DR3 haplotype, HLA-A*01.HLA-B*08.HLA-DRB1*03, was also associated with GADA positivity (P = 1.24 × 10−6; OR = 1.75 [1.40–2.19]).
The association with GADA positivity extended into the class I region, and included HLA-B, HLA-C, HLA-A, and MICA (Table 2 and Fig. 1A). Given the strong LD in the region, these associations were likely to be attributable to the HLA class II effects. HLA-DQB1 was the most associated locus with GADA and so was used to test whether there were additional associations that were outside and independent of the HLA class II region. Having conditioned on HLA-DQB1, there remained some evidence of association around MICA (P = 1.08 × 10−4) including two SNPs, rs9266722 and rs4713462, both at 31.35 Mb (P = 2.84 × 10−6 and 9.66 × 10−6; Fig. 1B), which were in LD with each other (D’ = 0.98; r2 = 0.36). The genotyping of rs9266722 was checked and extended to the full collection with GADA measures (an additional 1,124 samples) to maximize power. The genotyping was 99.90% consistent, and the association of rs9266722 remained in the 2,320 samples tested for association with GADA conditional on HLA-DQB1 (P = 4.84 × 10−6). The SNP, rs9266722, was in LD with HLA-B*15 (r2 = 0.58; D’ = 0.93) and the MICA-STR*A5 allele (r2 = 0.48; D’ = 0.79). However, the association with MICA did not remain once rs9266722, which lies in the pseudo-gene HLA-S located between HLA-B and MICA, was in the model (P = 0.33). Even the MICA-STR*A5 allele alone did not add to the association of rs9266722 (P = 0.16). Conversely, rs9266722 did not add convincingly to the association of the MICA-STR*A5 (P = 0.017). Likewise, the effects of HLA-B and rs9266722 could not be distinguished; HLA-B did not improve the association of HLA-DQB1 and rs9266722 (P = 0.04; Table 2) and rs9266722 did not improve the association of HLA-DQB1 and HLA-B (P = 0.13). At HLA-A, although no alleles were in LD with rs9266722 (r2 >0.04), the effect of rs9266722 could not be distinguished from HLA-A (P = 0.0045 for the addition of HLA-A to a model including HLA-DQB1 and rs9266722; P = 0.0039 for the addition of rs9266722 to a model including HLA-DQB1 and HLA-A). More samples, therefore, are needed to decipher the association with GADA positivity in the class I region.
Once the effect of HLA-DQB1 was accounted for, as well as the association discussed in the class I region, evidence for an additional association with GADA positivity was found with the intronic SNP, rs926850 in FAM65B at 25.0 Mb (P = 3.15 × 10−5; Fig. 1B). To increase power, this SNP was genotyped in an additional 1,101 case subjects and further tested for association with GADA. The evidence did not improve, becoming relatively unconvincing (P = 0.0067), and, hence, associations with GADA were confined to the HLA class II region and a second independent effect in the class I region.
IA-2A associations.
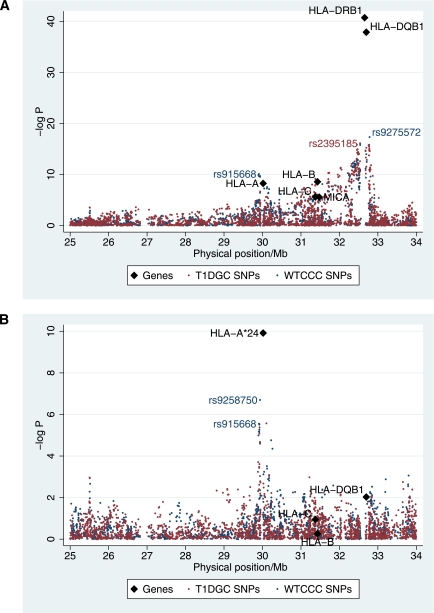
Convincing evidence of the association with IA-2A was obtained in both the HLA class I and II regions (P < 3 × 10−6; Fig. 2A and Table 5). HLA-DRB1 had the strongest evidence for association with IA-2A positivity (P = 1.94 × 10−41; Fig. 2A and Table 5). However, in contrast with the association with type 1 diabetes, there was no evidence of deviation from a multiplicative allelic effects model (P > 0.2 for the class I and II genes). The most associated SNP, rs9275572 (P = 5.07 × 10−17; Fig. 2A), which was genotyped in the WTCCC experiment (and by the T1DGC, P = 5.9 × 10−16), was also in the class II region, between the pseudo-genes MTC03P1 and HLA-DQB3. It was in strong LD with HLA-DRB1*03 (r2 = 0.72; D’ = 1.00).
FIG. 2.
IA-2A positivity association map. A: Association of SNPs used in the T1DGC Illumina experiment (using ∼1,215 type 1 diabetic case subjects) and the WTCCC affymetrix experiment (using ∼1,051 case subjects) with IA-2A and the classical loci (using between 2,399 and 1,268 type 1 diabetic case subjects). The most associated SNP, rs9275572, is in LD with HLA-DRB1*03 (r2 = 0.72, D’ = 1.00). B: Association of SNPs and classical genes with IA-2A, conditional on HLA-DRB1 alleles. The most associated SNP, rs9258750, is in LD with the HLA-A*24 allele (r2 = 0.55, D’ = 0.99).
TABLE 5.
Association of the HLA genes with IA-2A positivity in type 1 diabetic case subjects
| Gene | Physical position* | Number of samples |
P | P addition of locus to HLA-DRB1 | P addition of locus to HLA-DRB1 and HLA-A*24 | |
|---|---|---|---|---|---|---|
| IA-2A positive | IA-2A negative | |||||
| HLA-DQB1 | 32,273,871–33,498,585 | 1,394 | 1,005 | 1.34 × 10−38 | 0.0094 | 0.0320 |
| HLA-DRB1 | 32,485,163–32,557,625 | 1,390 | 1,007 | 1.94 × 10−41 | NA | NA |
| HLA-B | 31,237,115–31,324,989 | 893 | 647 | 2.69 × 10−9 | 0.5518 | 0.8631 |
| HLA-C | 31,236,526–31,323,369 | 752 | 516 | 2.42 × 10−6 | 0.1120 | 0.1290 |
| MICA | 31,367,561–31,384,016 | 1,410 | 983 | 2.67 × 10−6 | 0.2250 | 0.6640 |
| HLA-A | 29,909,037–29,913,661 | 1,005 | 716 | 5.50 × 10−9 | 6.13 × 10−6 | NA |
*Physical positions are from GRCh37 (www.t1dbase.org).
The HLA-DRB1*03 allele (and the HLA-DQB1*02 allele) was negatively associated with IA-2A positivity (OR [95% CI] = 0.44 [0.34–0.58]) for HLA-DRB1*03 using HLA-DRB1*0404 as reference (Table 6). In contrast, HLA-DRB1*0401 (and its associated HLA-DQB1 allele, HLA-DQB1*0302) were strongly associated with IA-2A positivity (OR = 1.72 [1.34–2.20]; Table 6), a major similarity to type 1 diabetes susceptibility (Supplementary Table 2). Interestingly, because the effects of HLA-DRB1*0401 and HLA-DRB1*03 were in opposite directions, the most susceptible type 1 diabetes genotype, HLA-DRB1*03/04, was not associated with IA-2A, having a frequency of 0.331 in the IA-2A positive case subjects and 0.356 in IA-2A negatives (OR = 1.07 [0.74–1.54]) using as a reference the non-HLA-DRB1*03, non-HLA-DRB1*04 homozygous genotype, which had a frequency of 0.064 in IA-2A positives and negatives (Supplementary Table 7). The rare allele HLA-DRB1*09 was associated with IA-2A positivity. However, unlike another report of only 618 samples (17), we found that the type 1 diabetes protective allele, HLA-DRB1*07 (Supplementary Table 2), was negatively correlated with IA-2A (OR = 0.65 [0.45–0.93]; Table 6). The highly type 1 diabetes protective allele, HLA-DQB1*0301, however, was associated with IA-2A positivity (Table 6).
TABLE 6.
HLA-DRB1 and HLA-DQB1 allelic associations with IA-2A positivity in type 1 diabetic case subjects
|
HLA-DRB1 |
HLA-DQB1 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Allele |
N (frequency) |
Unconditional | OR [95% CI] |
Allele |
N (frequency) |
OR [95% CI] |
||
| IA-2A positives | IA-2A negatives | Conditional on HLA-A*24 | IA-2A positives | IA-2A negatives | Unconditional | |||
| 09 | 47 (0.02) | 19 (0.01) | 1.85 (1.02–3.36) | 1.22 [0.60–2.49] | 0301 | 250 (0.09) | 59 (0.03) | 3.09 [2.13–4.49] |
| 0401 | 903 (0.32) | 413 (0.21) | 1.72 (1.34–2.20) | 1.47 [1.08–1.99] | 0303 | 67 (0.02) | 26 (0.01) | 2.55 [1.51–4.29] |
| 11 | 42 (0.02) | 16 (0.01) | 1.35 (0.68–2.64) | 1.40 [0.61–3.24] | 0302 | 1,087 (0.39) | 629 (0.31) | 1.89 [1.49–2.41] |
| 04 | 50 (0.02) | 13 (0.01) | 1.20 (0.83–1.76) | 1.34 [0.86–2.07] | 05 | 284 (0.10) | 198 (0.10) | 1.00 [reference] |
| 0402 | 35 (0.01) | 19 (0.01) | NA | 0402 | 61 (0.02) | 61 (0.03) | 0.85 [0.55–1.31] | |
| 0405 | 58 (0.02) | 46 (0.02) | NA | 06 | 119 (0.04) | 97 (0.05) | 0.74 [0.52–1.04] | |
| 0404 | 229 (0.08) | 166 (0.08) | 1.00 (reference) | 1.00 (reference) | 02 | 920 (0.33) | 940 (0.47) | 0.71 [0.57–0.88] |
| Rare | 57 (0.02) | 37 (0.02) | 0.82 (0.50–1.34) | 0.74 [0.36–1.52] | ||||
| 01 | 271 (0.10) | 174 (0.09) | 0.73 (0.54–0.98) | 0.64 [0.44–0.92] | ||||
| 07 | 141 (0.05) | 96 (0.05) | 0.65 (0.45–0.93) | 0.56 [0.35–0.87] | ||||
| 08 | 64 (0.02) | 64 (0.03) | 0.53 (0.34–0.83) | 0.59 [0.34–1.01] | ||||
| 13 | 113 (0.04) | 94 (0.05) | 0.50 (0.34–0.73) | 0.47 [0.29–0.75] | ||||
| 03 | 770 (0.28) | 857 (0.43) | 0.44 (0.34–0.58) | 0.35 [0.26–0.49] | ||||
Effect estimates were adjusted for the known covariates, duration of disease, and age-at-diagnosis, whereas the listed allele frequencies were not. Rare: alleles 10, 12, 14, 15, 16, and 0403 had a frequency of less than 0.5% and so were combined. NA, not available (the 04 alleles were combined, except 0404, which was neutral, and 0401, which was associated).
In addition to the evidence of association in the class II region, the HLA class I region also showed evidence of association with IA-2A positivity (Fig. 2A and Table 5). The SNP, rs915668 near HLA-A, was the most associated in that region (P = 5.71 × 10−10). However, these class I associations with IA-2A positivity could be attributable to LD with HLA class II alleles, and, hence, we tested the class I variants conditional on class II to ensure they are independent of the primary class II effect. A model including the HLA-DRB1 alleles was only improved by HLA-A (P = 6.13 × 10−6; Table 5) and SNPs in the region adjacent to HLA-A (Fig. 2B). No evidence of additional effects at the other genes and SNPs tested was obtained (Fig. 2B and Table 5). The class II independent effect in the class I region with IA-2A was largely attributable to HLA-A*24 (P = 1.21 × 10−10 for the association of HLA-A*24 alone conditional on HLA-DRB1). Despite being associated with susceptibility to type 1 diabetes (10), HLA-A*24 was negatively associated with IA-2A positivity (OR [95% CI] = 0.44 [0.34–0.57]) using all other non-HLA-A*24 alleles at HLA-A as reference. Strikingly, haplotype analyses showed that HLA-A*24 significantly reduced the positive IA-2A association of HLA-DRB1*04 from (OR [95% CI] = 2.02 [1.49–2.73]) for the HLA-A*02.HLA-DRB1*04 haplotype to a neutral effect for the HLA-A*24.HLA-DRB1*04 haplotype (OR = 0.94 [0.63–1.40]; Supplementary Table 8). Although HLA-DRB1*03 was negatively associated with IA-2A (Table 6), the association was significantly stronger in the presence of HLA-A*24 on the same haplotype (OR = 0.55 [0.39–0.77] HLA-A*02.HLA-DRB1*03 compared with OR = 0.25 [0.16–0.38] for HLA-A*24.HLA-DRB1*03; Supplementary Table 8). Note, however, that HLA-DRB1*03 remained negatively associated with IA-2A positivity even after conditioning on HLA-A*24 (OR = 0.35 [0.26–0.49] using HLA-DRB1*0404 as reference; Table 6).
The HLA-A*01.HLA-B*08.HLA-DRB1*03 extended haplotype was also negatively associated with IA-2A positivity (OR [95% CI] = 0.59 [0.44–0.78]), and again presence of HLA-A*24 increased this association (OR = 0.26 [0.13–0.52]; Supplementary Table 9). The negative association of HLA-DRB1*03 was, as expected given the lack of independent association at HLA-B, unaltered whether it was on the HLA-B*08.HLA-DRB1*03 haplotype or the HLA-B*18.HLA-DRB1*03 haplotype (OR = 0.29 and 0.34, respectively; Supplementary Table 10).
DISCUSSION
We have observed striking associations of islet autoantibodies with HLA alleles and haplotypes. Because levels of autoantibodies reduce after diagnosis of disease (26), there will be an increased false-negative rate for IA-2A and GADA compared with at time of diagnosis. A faster decline in GADA and/or IA-2A positivity in the younger onset case subjects, which we had limited power to detect, may be possible. However, we have adjusted for diabetes duration in our analyses, so the genetic associations we report were not attributable to decline in autoantibody levels (Supplementary Data). Although the false-negative rate reduces the power of our study to detect genetic associations, it does not affect the significant associations obtained. We cannot comment on genetic associations with autoantibody positivity prediagnosis since we only measured autoantibodies at a single time point at or after diagnosis. Some case subjects will have been transiently positive for autoantibodies. However, we were interested in the genetic association of these autoantibodies among case subjects, not in using them for prediction of type 1 diabetes onset; hence this will not affect our study since the samples were positive at the time of measurement. Indeed, our findings are highly consistent with other studies of IA-2A and GADA (13–16), and 30% of our case subjects were sampled within 2 years of diagnosis.
Our data show that although both GADA and IA-2A are very strongly associated with type 1 diabetes, and risk of all three localizes to the MHC region, the alleles and genotypes involved differ between autoantibodies and type 1 diabetes. Our major novel finding is that even though the HLA-DRB1*03/04 (and the HLA-DQB1*02/0302) genotype is highly predisposing for type 1 diabetes, it has very little effect on positivity for GADA or IA-2A, a finding that is also reflected in the 434 case subjects recruited within 1 year of diagnosis (frequency of HLA-DRB1*03/04 was 0.28 on GADA positives, 0.26 in GADA negatives, 0.33 in IA-2A positives, 0.35 in IA-2A negatives). Furthermore, HLA-DRB1*03/04 was not associated with decline in GADA or IA-2A positivity with disease duration (Supplementary Data). HLA-DRB1*03/04 has its strongest effects in children with the youngest age-at-diagnosis. In contrast, in our type 1 diabetic case subjects, both positivity for IA-2A and GADA were associated with an older age-at-diagnosis (Table 1) such that case subjects with the youngest age-at-diagnosis were less likely to be autoantibody positive. This agrees with other studies that also found GADA positivity was associated with an older age-at-diagnosis (13,27).
There are a number of possible explanations for these surprising findings. One possibility is that perhaps HLA-DRB1*03/04 recognizes a yet undiscovered, highly diabetogenic autoantigen, which accelerates disease progression. This leads to a younger age-at-diagnosis, with GADA and IA-2A produced as a result of disease pathogenesis, but not having caused disease. It is believed that HLA-DRB1*03/04 genotype susceptibility is attributable to a HLA DQ transdimer molecule (28), and it presumably binds certain preproinsulin peptides, including from the signal peptide sequence (29,30). The HLA-DRB1*03/04 genotype susceptibility is directed toward a potent, highly pathogenic antipreproinsulin response. Knip et al. (14) have also shown that HLA-DQB1*02/0302 is associated with IAA positivity. IAA has also been shown to be more common in young onset type 1 diabetic case subjects and is often the first autoantibody to appear before age 3 years (8). Functional masking has been observed for HLA class I molecules in a mouse model of multiple sclerosis in which HLA-A3 and HLA-A2 have opposite functional effects on autoreactive CD8+ T cells (31). So, another explanation for the lack of HLA-DRB1*03/04 association with IA-2A is that there is functional antigen competition between the HLA-DRB1*03 and HLA-DRB1*04 associated haplotypes, with one favoring one autoantigen response and the other competing or suppressing that response. Together these alleles maintain an equilibrium that manifests as no association of HLA-DRB1*03/04 with autoantibody positivity. Our results do not support a simple model in which HLA-DRB1*03/04 drives early and rapid β-cell destruction, effectively lowering the amount of GAD and IA-2 antigens available to the autoimmune response, since a negative association of this genotype would be expected.
The positive association of the type 1 diabetes protective allele, HLA-DQB1*0301, with IA-2A positivity was also unexpected. However, in contrast with HLA-DRB1*03/04, which is associated with a younger age-at-diagnosis, both HLA-DQB1*0301 and IA-2A positivity (Table 1) were associated with an older age-at-diagnosis (P = 4.48 × 10−13 and 7.22 × 10−20, respectively). The frequency of HLA-DQB1*0301 doubled from 0.04 in type 1 diabetes under 4 years to 0.08 in over 11 years.
The inverse associations of the classical alleles with type 1 diabetes compared with IA-2A and GADA are further illustrated by our results. HLA-DRB1*03 is associated with risk of type 1 diabetes, and also of GADA positivity, but is negatively associated with IA-2A positivity. HLA-DRB1*04 is highly predisposing for type 1 diabetes, is only associated with GADA (negatively) in the presence of HLA-DRB1*01 or *13, and is positively associated with IA-2A. Both HLA-A*24 and HLA-B*39 have been shown to independently confer susceptibility to type 1 diabetes (10,11). Yet HLA-A*24 was negatively associated with IA-2A positivity (in agreement with another report [15]), whereas HLA-B*39 was not associated with either GADA or IA-2A independently of the class II effects. Consequently, as suggested previously (32,33), autoantibody positivity does not necessarily reflect what is occurring at the T-cell level.
Overall, given that there are these strong differences in the genetics of type 1 diabetes and of IA-2A and GADA, these data suggest that GADA and IA-2A may not be the first consequence of the autoimmune attack against β-cells (34), and their exact role in pathogenesis remains to be established.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Juvenile Diabetes Research Foundation International (JDRF), the Wellcome Trust, and the National Institute for Health Research Cambridge Biomedical Centre. The Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (079895). DNA control samples were prepared and provided by S. Ring, R. Jones, and M. and W. McArdle of the University of Bristol, D. Strachan of the University of London, and P. Burton of the University of Leicester. The authors acknowledge use of DNA from The U.K. Blood Services collection of Common Controls (UKBS collection), funded by the Wellcome Trust Grant 076113/C/04/Z, by the Wellcome Trust/JDRF Grant 061858, and by the National Institute for Health Research of England. The collection was established as part of the Wellcome Trust Case-Control Consortium. This research utilizes resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and JDRF and supported by U01-DK-062418. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of data is available from http://www.wtccc.org.uk/. This project was funded by Wellcome Trust under award 076113.
No potential conflicts of interest relevant to this article were reported.
J.M.M.H. researched and analyzed data and wrote the manuscript. H.S. helped with DNA preparation and autoantibody data. D.J.S. genotyped SNPs. N.M.W. performed data management. K.A.C. and P.J.B. measured autoantibodies. J.A.T. researched and analyzed data and wrote the manuscript. All authors reviewed and edited the manuscript.
The authors thank David Dunger and Barry Widmer of the University of Cambridge and the British Society for Paediatric Endocrinology and Diabetes for the type 1 diabetic case collection; P. Clarke, G. Coleman, S. Duley, D. Harrison, S. Hawkins, M. Maisuria, J. Denesha, and T. Mistry of the JDRF/Wellcome Trust Diabetes and Inflammation Laboratory, University of Cambridge, for preparation of DNA samples; and the U.K. Medical Research Council and Wellcome Trust, who funded the collection of DNA for the British 1958 Birth Cohort (MRC Grant G0000934, Wellcome Trust Grant 068545/Z/02). The authors also thank the patients and control subjects for participating.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0131/-/DC1.
REFERENCES
- 1.Borg H, Marcus C, Sjöblad S, Fernlund P, Sundkvist G. Insulin autoantibodies are of less value compared with islet antibodies in the clinical diagnosis of autoimmune type 1 diabetes in children older than 3 yr of age. Pediatr Diabetes 2002;3:149–154 [DOI] [PubMed] [Google Scholar]
- 2.Bilbao JR, Rica I, Vázquez JA, Busturia MA, Castaño L. Influence of sex and age at onset on autoantibodies against insulin, GAD65 and IA2 in recent onset type 1 diabetic patients. Horm Res 2000;54:181–185 [DOI] [PubMed] [Google Scholar]
- 3.Verge CF, Gianani R, Kawasaki E, et al. Number of autoantibodies (against insulin, GAD or ICA512/IA2) rather than particular autoantibody specificities determines risk of type I diabetes. J Autoimmun 1996;9:379–383 [DOI] [PubMed] [Google Scholar]
- 4.Bingley PJ, Christie MR, Bonifacio E, et al. Combined analysis of autoantibodies improves prediction of IDDM in islet cell antibody-positive relatives. Diabetes 1994;43:1304–1310 [DOI] [PubMed] [Google Scholar]
- 5.Achenbach P, Warncke K, Reiter J, et al. Stratification of type 1 diabetes risk on the basis of islet autoantibody characteristics. Diabetes 2004;53:384–392 [DOI] [PubMed] [Google Scholar]
- 6.Bingley PJ, Gale EA; European Nicotinamide Diabetes Intervention Trial (ENDIT) Group Progression to type 1 diabetes in islet cell antibody-positive relatives in the European Nicotinamide Diabetes Intervention Trial: the role of additional immune, genetic and metabolic markers of risk. Diabetologia 2006;49:881–890 [DOI] [PubMed] [Google Scholar]
- 7.Orban T, Sosenko JM, Cuthbertson D, et al. ; Diabetes Prevention Trial-Type 1 Study Group Pancreatic islet autoantibodies as predictors of type 1 diabetes in the Diabetes Prevention Trial-Type 1. Diabetes Care 2009;32:2269–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siljander HT, Simell S, Hekkala A, et al. Predictive characteristics of diabetes-associated autoantibodies among children with HLA-conferred disease susceptibility in the general population. Diabetes 2009;58:2835–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redondo MJ, Babu S, Zeidler A, et al. ; Diabetes Prevention Trial Type 1 Study Group Specific human leukocyte antigen DQ influence on expression of antiislet autoantibodies and progression to type 1 diabetes. J Clin Endocrinol Metab 2006;91:1705–1713 [DOI] [PubMed] [Google Scholar]
- 10.Howson JM, Walker NM, Clayton D, Todd JA; Type 1 Diabetes Genetics Consortium Confirmation of HLA class II independent type 1 diabetes associations in the major histocompatibility complex including HLA-B and HLA-A. Diabetes Obes Metab 2009;11(Suppl. 1):31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nejentsev S, Howson JM, Walker NM, et al. ; Wellcome Trust Case Control Consortium Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 2007;450:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietropaolo M, Becker DJ, LaPorte RE, et al. Progression to insulin-requiring diabetes in seronegative prediabetic subjects: the role of two HLA-DQ high-risk haplotypes. Diabetologia 2002;45:66–76 [DOI] [PubMed] [Google Scholar]
- 13.Graham J, Hagopian WA, Kockum I, et al. ; Diabetes Incidence in Sweden Study Group; Swedish Childhood Diabetes Study Group Genetic effects on age-dependent onset and islet cell autoantibody markers in type 1 diabetes. Diabetes 2002;51:1346–1355 [DOI] [PubMed] [Google Scholar]
- 14.Knip M, Kukko M, Kulmala P, et al. Humoral beta-cell autoimmunity in relation to HLA-defined disease susceptibility in preclinical and clinical type 1 diabetes. Am J Med Genet 2002;115:48–54 [DOI] [PubMed] [Google Scholar]
- 15.Qu HQ, Polychronakos C. The effect of the MHC locus on autoantibodies in type 1 diabetes. J Med Genet 2009;46:469–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kordonouri O, Hartmann R, Charpentier N, Knip M, Danne T, Ilonen J. Genetic risk markers related to diabetes-associated autoantibodies in young patients with type 1 diabetes in Berlin, Germany. Exp Clin Endocrinol Diabetes 2010;118:245–249 [DOI] [PubMed] [Google Scholar]
- 17.Williams AJ, Aitken RJ, Chandler MA, Gillespie KM, Lampasona V, Bingley PJ. Autoantibodies to islet antigen-2 are associated with HLA-DRB1*07 and DRB1*09 haplotypes as well as DRB1*04 at onset of type 1 diabetes: the possible role of HLA-DQA in autoimmunity to IA-2. Diabetologia 2008;51:1444–1448 [DOI] [PubMed] [Google Scholar]
- 18.Field SF, Nejentsev S, Walker NM, et al. Sequencing-based genotyping and association analysis of the MICA and MICB genes in type 1 diabetes. Diabetes 2008;57:1753–1756 [DOI] [PubMed] [Google Scholar]
- 19.Barrett JC, Clayton DG, Concannon P, et al. ; Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bingley PJ, Bonifacio E, Williams AJ, Genovese S, Bottazzo GF, Gale EA. Prediction of IDDM in the general population: strategies based on combinations of autoantibody markers. Diabetes 1997;46:1701–1710 [DOI] [PubMed] [Google Scholar]
- 22.Bingley PJ, Williams AJ, Gale EA. Optimized autoantibody-based risk assessment in family members. Implications for future intervention trials. Diabetes Care 1999;22:1796–1801 [DOI] [PubMed] [Google Scholar]
- 23.Törn C, Mueller PW, Schlosser M, Bonifacio E, Bingley PJ; Participating Laboratories Diabetes Antibody Standardization Program: evaluation of assays for autoantibodies to glutamic acid decarboxylase and islet antigen-2. Diabetologia 2008;51:846–852 [DOI] [PubMed] [Google Scholar]
- 24.R Development Core Team. R: A Language and Environment for Statistical Computing 2.11.1 ed. Vienna, Austria, R Foundation for Statistical Computing, 2008
- 25.Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet 2002;70:124–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen R, Gilliam L, Torn C, et al. ; Diabetes Incidence Study in Sweden group Islet cell autoantibody levels after the diagnosis of young adult diabetic patients. Diabet Med 2007;24:1221–1228 [DOI] [PubMed] [Google Scholar]
- 27.Vandewalle CL, Falorni A, Lernmark A, et al. Associations of GAD65- and IA-2- autoantibodies with genetic risk markers in new-onset IDDM patients and their siblings. The Belgian Diabetes Registry. Diabetes Care 1997;20:1547–1552 [DOI] [PubMed] [Google Scholar]
- 28.Koeleman BP, Lie BA, Undlien DE, et al. Genotype effects and epistasis in type 1 diabetes and HLA-DQ trans dimer associations with disease. Genes Immun 2004;5:381–388 [DOI] [PubMed] [Google Scholar]
- 29.Cucca F, Lampis R, Congia M, et al. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Hum Mol Genet 2001;10:2025–2037 [DOI] [PubMed] [Google Scholar]
- 30.Skowera A, Ellis RJ, Varela-Calviño R, et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J Clin Invest 2008;118:3390–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friese MA, Jakobsen KB, Friis L, et al. Opposing effects of HLA class I molecules in tuning autoreactive CD8+ T cells in multiple sclerosis. Nat Med 2008;14:1227–1235 [DOI] [PubMed] [Google Scholar]
- 32.Roep BO, Duinkerken G, Schreuder GM, Kolb H, de Vries RR, Martin S. HLA-associated inverse correlation between T cell and antibody responsiveness to islet autoantigen in recent-onset insulin-dependent diabetes mellitus. Eur J Immunol 1996;26:1285–1289 [DOI] [PubMed] [Google Scholar]
- 33.Huurman VA, Hilbrands R, Pinkse GG, et al. Cellular islet autoimmunity associates with clinical outcome of islet cell transplantation. PLoS ONE 2008;3:e2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oresic M, Simell S, Sysi-Aho M, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med 2008;205:2975–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.