Abstract
OBJECTIVE
In contrast with childhood-onset type 1 diabetes, the genetics of autoimmune diabetes in adults are not well understood. We have therefore investigated the genetics of diabetes diagnosed in adults positive for autoantibodies.
RESEARCH DESIGN AND METHODS
GAD autoantibodies (GADAs), insulinoma-associated antigen-2 antibodies (IA-2As), and islet cell autoantibodies were measured at time of diagnosis. Autoantibody-positive diabetic subjects (n = 1,384) and population-based control subjects (n = 2,235) were genotyped at 20 childhood-onset type 1 diabetes loci and FCRL3, GAD2, TCF7L2, and FTO.
RESULTS
PTPN22 (1p13.2), STAT4 (2q32.2), CTLA4 (2q33.2), HLA (6p21), IL2RA (10p15.1), INS (11p15.5), ERBB3 (12q13.2), SH2B3 (12q24.12), and CLEC16A (16p13.13) were convincingly associated with autoimmune diabetes in adults (P ≤ 0.002), with consistent directions of effect as reported for pediatric type 1 diabetes. No evidence of an HLA-DRB1*03/HLA-DRB1*04 (DR3/4) genotype effect was obtained (P = 0.55), but it remained highly predisposing (odds ratio 26.22). DR3/4 was associated with a lower age at diagnosis of disease, as was DR4 (P = 4.67 × 10−6) but not DR3. DR3 was associated with GADA positivity (P = 6.03 × 10−6) but absence of IA-2A (P = 3.22 × 10−7). DR4 was associated with IA-2A positivity (P = 5.45 × 10−6).
CONCLUSIONS
Our results are consistent with the hypothesis that the genetics of autoimmune diabetes in adults and children are differentiated by only relatively few age-dependent genetic effects. The slower progression toward autoimmune insulin deficiency in adults is probably due to a lower genetic load overall combined with subtle variation in the HLA class II gene associations and autoreactivity.
The genetics of type 1 diabetes in children under 17 years of age (which is characterized by autoimmune destruction of the insulin-producing β-cells in the pancreatic islets and insulin deficiency) has been comprehensively studied, with over 50 susceptibility loci reported to date (1) (www.t1dbase.org). Investigation of the genetics of autoimmune diabetes in subjects who develop the disease as adults can elucidate the etiology of this late-onset autoimmunity and could impact its future treatment. However, most studies in adults have been confined to the study of a condition diagnosed as latent autoimmune diabetes in adults (LADA), a classification that is variable (2–4) (www.actionlada.org). In general, these case subjects are aged between 30 and 50 years and noninsulin dependent/not insulin treated at diagnosis and remain so for at least 3–6 months postdiagnosis and must be positive for GAD autoantibodies (GADAs), which are associated with type 1 diabetes (www.actionlada.org). A number of studies have investigated the association of the major histocompatibility complex (MHC) region, specifically the HLA class II genes HLA-DRB1 and HLA-DQB1, with LADA (5–7). They reported an age-at-diagnosis effect for the HLA-DRB1*03/HLA-DRB1*04 (DR3/4) genotype, with the frequency of DR3/4 lower in the older age-groups compared with that in the youngest age-groups, a finding that is also characteristic of pediatric type 1 diabetes. HLA class II susceptibility alone cannot account for type 1 diabetes development (8,9). However, in LADA cases to date, only one non-MHC gene has been shown to be convincingly associated: the −23 HphI INS (rs689) single nucleotide polymorphism (SNP) (10). This SNP is a near perfect proxy for the disease causal variable number tandem repeat (VNTR) polymorphism in the insulin (INS) gene in European ancestry populations. In that study, the 400 case subjects used were diagnosed with type 2 diabetes at an age at diagnosis between 25 and 68 years, were positive for GADA and/or insulinoma-associated antigen-2 autoantibodies (IA-2As), and did not require insulin during the first 3 months following diagnosis. The T allele had an odds ratio (OR) of 0.42 (95% CI 0.31–0.58), which is consistent with reports in childhood-onset type 1 diabetes and is associated with increased immune tolerance to insulin and its precursors (11). The authors concluded that the INS VNTR locus does not distinguish LADA and pediatric type 1 diabetes.
In the current study, we have taken a simpler, clinic-based approach to defining adult-onset autoimmune diabetes. To be included in the study, all diabetic case subjects were positive for one or more islet autoantibody, specifically, islet cell antibody (ICA), GADA, IA-2A, or insulin-specific autoantibodies (IAAs), and had glucose levels, oral glucose tolerance test, or HbA1c at a level diagnostic for diabetes. With these specific criteria for inclusion, the complexities associated with classification of diabetes in adults is avoided (2) and, as advocated by a number of authors in recent commentaries (3,4), the study of autoimmune diabetes may be more informative. Twenty type 1 diabetes–associated loci, selected based on effect size (OR ≥1.15, leading to power ≥0.7 at α = 0.05), potential age-at-diagnosis effects (IL2RA, IL2, and RNLS), and putative associations with thyroid peroxidize autoantibodies (STAT4, UBASH3A, IL2, BACH2, and CTLA4 [12,13]) were tested for association with autoimmune diabetes diagnosed in adults. The obesity gene, FTO, has been shown to be associated with LADA (14) and so was also tested for association in the autoimmune diabetic case subjects, as was the gene TCF7L2, which has the strongest effect of any locus in type 2 diabetes (15). Finally, FCRL3 is thought to be a general autoimmunity locus because of its association with several autoimmune diseases (16–19), so both FCRL3 and the gene GAD2 encoding the GAD antigen itself were also tested for association in these 1,212 adult-onset autoimmune diabetic case subjects. Hence, we have tested more loci in more adult-onset autoimmune diabetic case subjects than any other study has reported on to date.
RESEARCH DESIGN AND METHODS
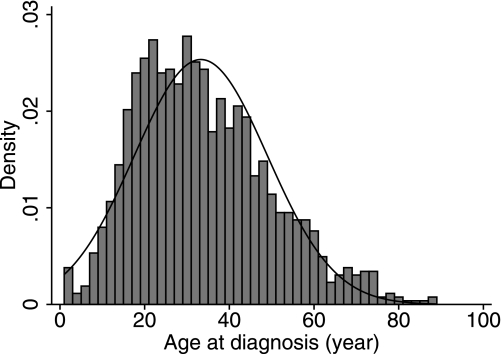
Diabetic case subjects were recruited at 14 diabetes practices in Ulm and the surrounding area, southwest Germany, as part of the Baden-Württemberg Consortium, Arbeitsgemeinschaft Diabetologie Baden-Württemberg - Erhebung Neu entdeckter Typ 1 Diabetiker (ADBW-END) Study Group. A total of 1,384 case subjects had autoimmune diabetes defined by glucose, oral glucose tolerance test, or HbA1c at a level diagnostic for diabetes according to World Health Organization criteria; in addition, at least one islet autoantibody had to be positive (ICA, GADA, IA-2A, or IAA). All case subjects were treated with insulin upon diagnosis of diabetes. One case subject was diagnosed within the 1st year of life and so was removed from all subsequent analyses. The remaining case subjects were aged between 3 and 89 years at diagnosis (the average age-at-diagnosis was 33.3 years [Fig. 1]). Of 1,212 case subjects diagnosed at age 17 years or over, 668 (55.4%) were male and 7 did not have sex information. A large proportion of case subjects diagnosed under age 18 years in Germany are seen by pediatricians and, hence, are underrepresented in our collection, thereby accounting for the high median age at diagnosis (31 years) of our case subjects.
FIG. 1.
Frequency histogram of age at diagnosis in the 1,384 German autoimmune diabetic case subjects. The curve represents the normal density.
Control sample subjects were recruited as part of a population-based, cross-sectional health survey in southwestern Germany. Of 12,475 inhabitants of an urban population, 4,000 were randomly selected by staff at the municipal registration office from the registry of inhabitants and invited to participate in the study. One hundred and seven were ineligible, having moved from the area. Subjects with glucose levels outside the normal range according to World Health Organization criteria and HbA1c levels ≥6.0% (using the Diabetes Control and Complications Trial/UK Prospective Diabetes Study assay as a reference method) were excluded. A total of 2,513 people aged 10–70 years, 47.4% (n = 1,192) of whom were male, participated in the study. The average age of control subjects was 39.7 years; 261 were 17 years of age or younger at the time of recruitment and were not analyzed.
Ethical agreement.
The study meets the international agreements of the World Medical Association Declaration of Helsinki 2000 about ethical principles for medical research involving human subjects and was approved by the ethics committee of the State Medical Chamber Baden-Württemberg. Written consent was obtained from every study participant.
Autoantibodies.
All case subjects were tested for serum autoantibodies to at least one of ICA, GADA, IA-2A, and IAA using established methodology (20,21). All samples were tested at a single laboratory (Ulm) in duplicate including positive and negative control standard sera, and 15% were then validated with 100% concordant. Diabetes Autoantibody Standardization Program (DASP) 2007 Workshop results for GAD65 was 95% specificity, 86% sensitivity. DASP 2007 Workshop results for IA-2ic was 99% specificity, 73% sensitivity. The 99th percentile, used as the cutoff for positivity, was calculated from 1,200 healthy school children for IAA (22) and from 2,000 healthy adults for GADA and IA-2A. Cutoff for positivity was >0.9 arbitrary units (AUs) for GADA, >0.75 for IA-2A, and >1 for IAA.
ICAs were measured by indirect immunofluorescence on cryosections of human pancreas after being incubated overnight at 4°C. The detection limit was 5 Juvenile Diabetes Foundation units (JDFUs). ICA levels ≥20 JDFU were considered positive. The assay achieved an analytical sensitivity and specificity of 100% in the 13th ICA Workshop in 1998.
Genotyping.
All SNPs were genotyped using the TaqMan 5′ nuclease assay (Applied Biosystems, Warrington, U.K.) according to the manufacturer’s protocol. Genotyping was performed blind to case-control status and double scored to minimize errors.
Tagging SNPs.
Tag SNPs were selected based on up to 6,205 British pediatric-onset type 1 diabetic case subjects and 5,325 British control subjects, described elsewhere (1,23). These case and control subjects were genotyped at 2,420 SNPs within the extended MHC region as part of the T1DGC genome-wide association study (GWAS) using the Illumina 550 K Infinium platform (1) and at 1,687 SNPs from the Wellcome Trust Case-Control Consortium (WTCCC) GWAS using the Affymetrix GeneChip Human Mapping 500 K Array set (23). The British pediatric-onset case and the British control subjects were genotyped at HLA-DRB1 using Dynal RELI SSO assays (Invitrogen, Paisley, U.K.) or Roche Molecular Systems SSO reverse dot blot technology.
The linkage disequilibrium (LD) measures, r2 and D′, between each SNP on the SNP chip platforms and the HLA-DRB1*03 allele and the HLA-DRB1*04 allele were calculated separately in case and control subjects using STATA (www.stata.com). The SNP in strongest LD with HLA-DRB1*03 was rs2187668 (r2 = 0.99 in case subjects and r2 = 0.87 in control subjects), and the one in strongest LD with HLA-DRB1*04 was rs660895 (r2 = 0.87 in case subjects and r2 = 0.74 in control subjects). For HLA-DRB1*15, rs9270986 was found to be in strongest LD (r2 = 0.97 in the subset of control subjects used by the WTCCC). However, when this SNP was genotyped using TaqMan in all 6,205 case and 5,325 control subjects with HLA-DRB1 genotypes, the LD dropped to r2 = 0.87 in control subjects and r2 = 0.52 in case subjects. Therefore, the next best SNP was used, rs9271366, which in the full sample set had r2 = 0.98 in control subjects and r2 = 1.00 in case subjects.
Statistics.
Association analyses were performed using STATA. All SNPs were in Hardy-Weinberg equilibrium in control subjects (P ≥ 0.06). Association with autoimmune diabetes was tested using disease outcome as the dependent variable and genotype as the independent variable in a logistic regression model. Sex was included as a covariate in the logistic model because of the different proportion of male and female subjects between the case and control groups (Supplementary Data). To test whether a multiplicative model was an appropriate approximation for the SNP association tests, a genotype effects model on 2 degrees of freedom (df), which does not assume a specific mode of inheritance, was compared with a multiplicative allelic effects model on 1 df using a likelihood ratio test. In the HLA region, rs2187668 and rs660895 were used to code the genotypes HLA-DRB1*03/03, 03/04, 03/X, 04/04, 04/X, and X/X. The 5 df genotype effects model was compared with a 2 df multiplicative allelic effects (DR3, DR4) model using a likelihood ratio test. Only samples genotyped at both loci were included in this test. Power calculations for association were performed using CaTS (24).
Association with autoantibody positivity was tested in the same manner as above, using logistic regression with autoantibody status as outcome variable, and included all necessary covariates (see results). No adjustments of P values to allow for multiple comparisons were made. However, the threshold of significance for association with adult-onset autoimmune diabetes, or autoantibody positivity, was lowered to P ≤ 0.002 to account for multiple testing.
Interaction with age at diagnosis was tested in a case-only linear regression model, with age at diagnosis as the outcome variable, and also in a multinomial logistic regression model with quartiles of the age-at-diagnosis distribution as the categorical outcome variable. The SNP allele, or genotypes, were used as the independent variables. Only the pediatric type 1 diabetes–associated regions were tested for age-at-diagnosis effects; hence, the threshold of significance used was raised to P ≤ 0.0025. The study had 80% power to detect effect sizes of 2.6 years per allele using α = 0.0025 and an allele frequency of 0.3 (equivalent to the DR3 allele). For α = 0.05, the study was powered to detect effects as low as 1.8 years per allele for a minor allele frequency of 0.3.
A genetic risk score (or “load”) was estimated using the predictors in a logistic regression model in which disease status was used as the outcome variable and all 25 type 1 diabetes–associated SNPs were included as independent variables. The scores were regressed on age at diagnosis to estimate the dependence of age at diagnosis on genetic risk.
RESULTS
HLA associations.
Case subjects under 17 years old at diagnosis (n = 172) were excluded from the SNP association tests because we were principally interested in the genetics of adult-onset autoimmune diabetes. The most associated SNPs, rs2187668 and rs660895 (P = 2.31 × 10−65 and 8.65 × 10−84, respectively, and P = 2.02 × 10−179 for the joint effect of both SNPs [Table 1]), were in the HLA class II region, which is the primary genetic locus for type 1 diabetes diagnosed in childhood. These SNPs are accurate proxies for the highly type 1 diabetes–predisposing alleles, HLA-DRB1*03 (DR3) and HLA-DRB1*04 (DR4) alleles (r2 >0.7 [see research design and methods and Supplementary Data]). Both SNPs had independent effects on risk of adult-onset autoimmune diabetes (the two SNPs added to each other in a logistic regression model, P < 1 × 10−98), and the minor alleles of both SNPs were strongly predisposing to adult-onset autoimmune diabetes (OR 5.36 for DR3 and 4.98 for DR4 [Table 1]). However, there was no evidence of a synergistic DR3/4 effect (P = 0.55); i.e., the DR3/3, DR3/4, and DR4/4 genotypes all had similar predisposing effects (the 95% CIs all overlapped [Table 1]). The X/X genotype was strongly protective, with an OR of 0.19 (95% CI 0.15–0.24) (using DR4/X as reference [Table 1]) but was more common than reported in British childhood-onset case subjects (Supplementary Table 1).
TABLE 1.
HLA associations in a maximum of 1,177 adult-onset autoimmune diabetic case subjects and 2,210 control subjects
| SNP (allele or genotype) |
N (frequency) |
OR [95% CI] | P | ||
|---|---|---|---|---|---|
| Case subjects | Control subjects | ||||
| rs2187668 (DR3)* | 585 (0.25) | 395 (0.09) | 3.37 [2.92–3.90] | 2.31 × 10−65 | |
| rs660895 (DR4)* | 868 (0.37) | 662 (0.16) | 3.34 [2.94–3.81] | 8.65 × 10−84 | |
| DR3† | 567 (0.25) | 380 (0.09) | 5.36 [4.53–6.34] | ||
| DR4† | 852 (0.37) | 656 (0.16) | 4.98 [4.23–5.77] | ||
| DRX† | 857 (0.38) | 3,138 (0.75) | 1.00 (ref.) | 2.02 × 10−179 | |
| 3/3‡ | 75 (0.07) | 15 (0.01) | 37.97 [21.26–67.81] | 7.22 [4.07–12.79] | |
| 3/4‡ | 222 (0.20) | 64 (0.03) | 26.22 [18.94–36.30] | 4.98 [3.65–6.80] | |
| 4/4‡ | 142 (0.12) | 47 (0.02) | 22.42 [15.48–32.48] | 4.26 [2.98–6.10] | |
| 4/X‡ | 346 (0.30) | 498 (0.24) | 5.26 [4.23–6.54] | 1.00 (ref.) | |
| 3/X‡ | 195 (0.17) | 286 (0.14) | 5.15 [4.02–6.60] | 0.98 [0.78–1.23] | |
| X/X‡ | 158 (0.14) | 1,177 (0.56) | 1.00 (ref.) | 0.19 [0.15–0.24] | |
The SNPs rs2187668 and rs660895 were used as proxies for HLA-DRB1*03 (DR3) and HLA-DRB1*04 (DR4) alleles, respectively, and to code for the DR3/4/X genotypes (see research design and methods). The P values reported are for a model that assumes multiplicative allelic effects either for the individual SNPs or for the joint effects of both DR3 and DR4.
*Individual SNP associations of rs2187668 and rs660895 use all samples genotyped at the test SNP.
†A joint effects model including both DR3 (rs2187668) and DR4 (rs660895). Only samples genotyped at both SNPs were included (see research design and methods).
‡The genotype effects model. Only samples genotyped at both SNPs were included (see research design and methods). No P value is reported for this model because a multiplicative allelic effects model was an appropriate approximation (P = 0.55). N, number of chromosomes or genotypes.
Non-HLA associations.
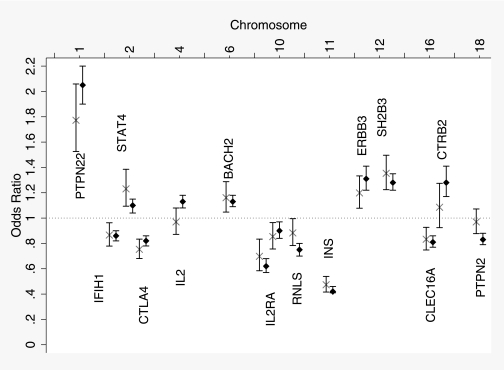
The strongest association outside of the MHC region was with INS (P = 1.97 × 10−32) (Table 2), followed by PTPN22 (1p13.2), which mirrors the strong associations of these genes in British pediatric-onset type 1 diabetes (Fig. 2). A further six candidate genes (regions), STAT4 (2q32.2), CTLA4 (2q33.2), IL2RA (10p15.1), ERBB3 (12q13.2), SH2B3 (12q24.12), and CLEC16A (16p13.13), were all convincingly associated with adult-onset autoimmune diabetes, with the same direction of effect as reported for British pediatric-onset type 1 diabetes (P ≤ 0.002) (Table 2; Fig. 2; Supplementary Table 2). The evidence of association at the remaining eleven loci (IFIH1 [2q24.2], IL2 [4q27], BACH2 [6q15], GLIS3 [9p24.2], RNLS [10q23.31], IL27 [16p11.2], CTRB2 [16q23.1], GSDMB [17q12], PTPN2 [18p11.21], CD226 [18q22.2], and UBASH3A [21q22.3]; P > 0.004) was not at a level required to be convincing (P ≤ 0.002). Three independent effects have been reported in type 1 diabetes at IL2RA, with rs2104286, rs12722495, and rs11594656 (25). The most associated SNP in adult-onset autoimmune diabetic case subjects was rs12722495 (P = 5.22 × 10−5) (Table 2). Once the effect of this SNP was accounted for, there was no further evidence of association at IL2RA (P > 0.06), probably owing to a lack of power (Table 2).
TABLE 2.
Associations at 24 non-HLA loci in a maximum of 1,196 autoimmune diabetic case subjects diagnosed at over 17 years of age and 2,215 control subjects
| Candidate gene (region), SNP |
N Chromosomes (MAF) |
OR [95% CI] | P | Power* |
||
|---|---|---|---|---|---|---|
| Case subjects | Control subjects | α = 0.001 | α = 0.05 | |||
| PTPN22 (1p13.2), rs2476601 (C>T) | 376 (0.16) | 431 (0.10) | 1.77 [1.53–2.06] | 8.03 × 10−14 | 1.00 | 1.00 |
| IFIH1 (2q24.2), rs1990760 (A>G) | 832 (0.36) | 1,643 (0.39) | 0.87 [0.78–0.96] | 0.0077 | 0.35 | 0.83 |
| STAT4 (2q32.2), rs7574865 (G>T) | 590 (0.25) | 938 (0.21) | 1.23 [1.09–1.39] | 5.93 × 10−4 | 0.04 | 0.35 |
| CTLA4 (2q33.2), rs3087243 (G>A) | 928(0.39) | 2,043 (0.46) | 0.75 [0.68–0.83] | 3.65 × 10−8 | 0.74 | 0.98 |
| IL2 (4q27), rs2069763 (G>T) | 786(0.34) | 1,484 (0.34) | 0.96 [0.86–1.08] | 0.51 | 0.17 | 0.64 |
| IL2 (4q27), rs2069762 (T>G) | 715 (0.30) | 1,366 (0.31) | 0.96 [0.86–1.08] | 0.51 | 0.12 | 0.57 |
| BACH2 (6q15), rs11755527 (C>G) | 1,144 (0.49) | 1,686 (0.45) | 1.16 [1.05–1.29] | 0.0045 | 0.20 | 0.68 |
| GLIS3 (9p24.2), rs7020673 (G>C) | 1,052 (0.44) | 2,083 (0.48) | 0.88 [0.79–0.97] | 0.012 | 0.23 | 0.72 |
| IL2RA (10p15.1), rs12722495 (A>G) | 190 (0.08) | 499 (0.11) | 0.70 [0.58–0.83] | 5.22 × 10−5 | 0.99 | 1.00 |
| IL2RA (10p15.1), rs2104286 (A>G) | 523 (0.22) | 1,083 (0.25) | 0.85 [0.76–0.96] | 0.031 | 0.07 | 0.44 |
| IL2RA (10p15.1), rs11594656 (T>A) | 576 (0.25) | 1,127 (0.27) | 0.92 [0.82–1.03] | 0.13 | 0.20 | 0.68 |
| RNLS (10q23.31), rs10509540 (T>C) | 530 (0.24) | 1,133 (0.26) | 0.88 [0.78–1.00] | 0.041 | 0.95 | 1.00 |
| INS (11p15.5), rs689 (A>T) | 382 (0.16) | 1,280 (0.29) | 0.47 [0.42–0.54] | 1.97 × 10−32 | 1.00 | 1.00 |
| ERBB3 (12q13.2), rs2292239 (C>A) | 846 (0.36) | 1,376 (0.32) | 1.20 [1.08–1.33] | 8.40 × 10−4 | 0.97 | 100 |
| SH2B3 (12q24.12), rs3184504 (A>G) | 1,030 (0.44) | 2,114 (0.48) | 1.35 [1.22–1.50] | 2.67 × 10−9 | 0.95 | 100 |
| CLEC16A (16p13.13), rs12708716 (A>G) | 763 (0.34) | 1,583 (0.38) | 0.83 [0.75–0.93] | 7.82 × 10−4 | 0.77 | 0.98 |
| IL27 (16p11.2), rs4788084 (G>A) | 939 (0.39) | 1,830 (0.42) | 0.90 [0.81–1.00] | 0.042 | 0.37 | 0.84 |
| CTRB2 (16q23.1), rs7202877 (T>G) | 271 (0.12) | 468 (0.10) | 1.09 [0.92–1.27] | 0.32 | 0.41 | 0.86 |
| GSDMB (17q12), rs2290400 (G>A) | 1,127 (0.49) | 2,115 (0.50) | 0.97 [0.87–1.07] | 0.051 | 0.30 | 0.79 |
| PTPN2 (18p11.21), rs478582 (T>C) | 1,027 (0.43) | 1,947 (0.44) | 0.96 [0.87–1.06] | 0.55 | 0.65 | 0.96 |
| PTPN2 (18p11.21), rs45450798 (G>C) | 390 (0.17) | 644 (0.15) | 1.15 [1.00–1.31] | 0.054 | 0.63 | 0.95 |
| CD226 (18q22.2), rs763361 (C>T) | 1,157 (0.49) | 2,178 (0.49) | 0.98 [0.89–1.08] | 0.68 | 0.25 | 0.74 |
| UBASH3A (21q22.3), rs3788013 (C>A) | 1,035 (0.44) | 1,853 (0.42) | 1.06 [0.96–1.17] | 0.26 | 0.19 | 0.68 |
| Obesity locus | ||||||
| FTO (16q12.2), rs9939609 (T>A)† | 953 (0.41) | 1,798 (0.42) | 0.96 [0.87–1.06] | 0.45 | 0.42 | 0.86 |
| Type 2 diabetes locus | ||||||
| TCF7L2 (10q25.2), rs12255372 (G>T) | 571 (0.27) | 1,147 (0.27) | 1.01 [0.89–1.13] | 0.93 | 0.99 | 0.99 |
| Autoantibody loci | ||||||
| GAD2 (10p12.1), rs2839671 (G>A) | 412 (0.18) | 776 (0.18) | 0.96 [0.84–1.09] | 0.50 | 0.13 | 0.59 |
| FCRL3 (1q23.1), rs7528684 (A>G) | 1,045 (0.44) | 1,919 (0.44) | 1.03 [0.93–1.14] | 0.58 | 0.30 | 0.79 |
MAF, minor allele frequency; number of chromosomes is listed for the minor allele.
*Power is calculated for the susceptible allele, assuming multiplicative allelic effects at all loci except INS. For all type 1 diabetes–associated loci, the effect estimates and minor allele frequencies used to calculate the power were taken from Smyth et al. (38), Fung et al. (39), and Barrett et al. (1) and are also available from www.t1dbase.org/page/regions. Note that if the association of a locus is reduced with increasing age at diagnosis, these power estimates could be inflated. For the TCF7L2 and FTO loci, the effect estimates as reported for type 2 diabetes were used (40,41). For the autoantibody loci, an effect estimate of 1.15, which is not unusual for type 1 diabetes, was used.
†rs9939609 is in LD with rs9931494, the SNP reported as associated with LADA (r2 = 0.91, D′ = 0.98 in Centre d'Etude du Polymorphisme Humaine samples) (14).
FIG. 2.
Comparison of effects of the minor allele in German autoimmune diabetic case subjects diagnosed between 17 and 89 years of age (X) and British pediatric-onset type 1 diabetic case subjects diagnosed before 17 years of age (♦). Effects in type 1 diabetic subjects are taken from Smyth et al. (38), Fung et al. (39), and Barrett et al. (1) and are also available from www.t1dbase.org/page/regions. Effects for all SNPs tested in Table 2 are given for British pediatric-onset type 1 diabetic case subjects diagnosed before 17 years of age in Supplementary Table 2.
No evidence of association with adult-onset autoimmune diabetes was obtained at TCF7L2 or FTO (P > 0.4) (Table 2), implying that our case series does not contain a sizeable number of type 2 diabetic case subjects because our study had 99% power to detect the effect at TCF7L2 (Table 2). Given the work by Saxena et al. (26), the subgroup of type 2 diabetic subjects with impaired insulin secretion does not appear to be represented. The lack of association at TCF7L2 suggests that it does not significantly affect risk of adult-onset autoimmune diabetes, which contrasts with another study of 197 adult-onset GADA-positive diabetic case subjects (27) but is consistent with findings in type 1 diabetic subjects (28). Neither FCRL3 nor GAD2 was associated with adult-onset autoimmune diabetes (P ≥ 0.5) (Table 2).
Age-at-diagnosis effects.
Evidence of an age-at-diagnosis effect was obtained with the DR4 SNP, rs660895 (P = 4.67 × 10−6) (Table 3), using all 1,384 autoimmune diabetic case subjects but not with the DR3 SNP, rs2187668 (P = 0.067) (Table 3). The average age at diagnosis of case subjects homozygous for DR4/4 (the G/G genotype of rs660895) was 30.0 years compared with 32.3 years for DR4/X (the A/G genotype at rs660895) and 35.8 years for DRX/X (the A/A genotype at rs660895). The same trend was not observed for the DR3 genotypes (Table 3). However, the known DR3/4 age-at-diagnosis effect was observed, having a frequency of 0.23 in case subjects diagnosed before age 22 years compared with 0.17 in case subjects diagnosed after age 43 years (Table 3). Most striking was the increase in frequency of the DRX/X genotype with increasing age at diagnosis. DRX/X was over twice as common in the >43-years age category (0.20) as in the 0- to 21-years category (0.08) (Table 3). The inverse was observed with the DR4/4 genotype (Table 3). The large increase (12%) in frequency of the X/X category in the oldest onset case subjects could not be attributed to the type 1 diabetes–protective HLA-DRB1*15 allele (rs9271366) alone, which was only 3% more frequent in the oldest case subjects compared with the youngest (Table 3). The G allele of the SNP rs9271366 is in LD with HLA-DRB1*15 (r2 ≥0.98) (see research design and methods).
TABLE 3.
Age at diagnosis in all 1,384 autoimmune diabetic case subjects, irrespective of age at diagnosis, by HLA genotype (rs9271366 being too rare to analyze for age-at-diagnosis effects by age category)
| Mean age at diagnosis (years) (SD) | Genotype counts (frequency) by age-group |
P | Pcat | ||||
|---|---|---|---|---|---|---|---|
| 0–21 years | 22–31 years | 32–43 years | ≥44 years | ||||
| rs660895 (DR4) | |||||||
| A/A (X/X) | 35.8 (16.4) | 97 (0.30) | 104 (0.33) | 130 (0.41) | 133 (0.44) | 4.67 × 10−6 | 2.58 × 10−4 |
| G/A (4/X) | 32.3 (15.5) | 172 (0.53) | 174 (0.54) | 146 (0.46) | 146 (0.48) | ||
| G/G (4/4) | 30.0 (14.7) | 54 (0.17) | 42 (0.13) | 41 (0.13) | 27 (0.09) | ||
| rs2187668 (DR3) | |||||||
| G/G (X/X) | 34.0 (16.3) | 187 (0.56) | 176 (0.55) | 177 (0.56) | 187 (0.61) | 0.067 | 0.51 |
| G/A (3/X) | 31.9 (15.3) | 128 (0.38) | 126 (0.39) | 118 (0.37) | 104 (0.34) | ||
| A/A (3/3) | 32.9 (13.0) | 18 (0.05) | 20 (0.06) | 21 (0.07) | 17 (0.06) | ||
| rs9271366 (DR15) | |||||||
| A/A (X/X) | 33.1 (15.9) | 320 (0.98) | 307 (0.96) | 298 (0.95) | 297 (0.96) | 0.13 | NA |
| A/G (15/X) | 36.7 (11.1) | 4 (0.01) | 12 (0.04) | 15 (0.05) | 13 (0.04) | ||
| G/G (15/15) | 37.3 (18.2) | 1 (0.00) | 0 (0.00) | 1 (0.00) | 1 (0.00) | ||
| 3/3 | 33.3 (12.9) | 16 (0.05) | 20 (0.06) | 21 (0.07) | 17 (0.06) | 1.78 × 10−7 | 0.0029 |
| 3/4 | 30.4 (14.4) | 72 (0.23) | 73 (0.23) | 57 (0.18) | 50 (0.17) | ||
| 4/4 | 30.0 (14.3) | 54 (0.17) | 41 (0.13) | 40 (0.13) | 27 (0.09) | ||
| 3/X | 33.9 (16.3) | 51 (0.16) | 49 (0.16) | 59 (0.19) | 50 (0.17) | ||
| 4/X | 33.4 (16.1) | 98 (0.31) | 99 (0.31) | 85 (0.28) | 92 (0.31) | ||
| X/X | 39.2 (17.3) | 26 (0.08) | 33 (0.10) | 47 (0.15) | 60 (0.20) | ||
NA, not analyzed. P, the P value for interaction of age at diagnosis as a continuous trait with genotype. Pcat, P value for interaction of genotype with the four age-groups corresponding to the quartiles of the age-at-diagnosis distribution.
Outside of the HLA, no convincing evidence of age-at-diagnosis effects was obtained (P > 0.0037) (Supplementary Table 3), with only suggestive evidence of an effect with rs478582 at PTPN2 (P = 0.0057), rs7202877 at CTRB2 (P = 0.0050), and rs2104286 at IL2RA (P = 0.0037) (Supplementary Table 3). Nevertheless, evidence that age at diagnosis was dependent on the overall genetic load attributable to the 25 type 1 diabetes–associated SNPs tested was obtained (P = 6.18 × 10−8). The genetic load in case subjects was inversely related to age at diagnosis (regression coefficient β = –2.1), such that the overall genetic risk was lower in case subjects diagnosed at an older age than in those diagnosed at a younger age.
Associations with autoantibodies.
Autoantibodies were measured at time of diagnosis in 83% of case subjects and postdiagnosis in the remaining 17% of case subjects. All case subjects were positive for one or more of the islet autoantibodies. ICA was measured in 1,270 samples, of which 59% were positive; IA-2A was measured in 933 case subjects, of whom 51% were positive; GADA was measured in 1,380 case subjects, of whom 83% were positive. Fifty-seven percent of our samples were positive for more than one autoantibody. Only 411 case subjects were GADA positive but negative for IA-2A, IAA, and ICA. GADA positivity was correlated with an older age at diagnosis, an earlier date of birth, and a shorter duration of disease (Supplementary Table 4). IA-2A positivity was pairwise correlated with a more recent date of birth and shorter duration of disease (Supplementary Table 4). However, age at diagnosis and date of birth together were used as covariates in the genetic association tests because they accounted for more variation in IA-2A positivity than date of birth and duration (Supplementary Table 4).
No evidence of association with IA-2A or GADA was obtained at the non-HLA type 1 diabetes–associated SNPs (P > 0.01). In contrast, both the DR3 and DR4 tag SNPs, rs2187668 and rs660895, were strongly associated with IA-2A (P = 3.22 × 10−7 and 5.45 × 10−6, respectively) (Table 4). Interestingly, their effects on IA-2A were in opposite directions, such that the DR3 allele was associated with IA-2A negativity and DR4 was associated with IA-2A positivity. The DR3/4 genotype effect with IA-2A was close to neutral; the DR3 and DR4 effects essentially cancelled each other out (Table 4).
TABLE 4.
Association of HLA class II with IA-2A in 904 autoimmune diabetic case subjects, using rs660895 and rs2187668 to model HLA-DR4 and -DR3, respectively
|
N (frequency) |
OR [95% CI] | P | ||
|---|---|---|---|---|
| IA-2A positive | IA-2A negative | |||
| rs660895 | ||||
| G (DR4)* | 390 (0.42) | 291 (0.33) | 1.62 [1.31–2.00] | 5.45 × 10−6 |
| rs2187668 | ||||
| A (DR3)† | 186 (0.20) | 260 (0.29) | 0.56 [0.44–0.70] | 3.22 × 10−7 |
| DR4‡ | 382 (0.42) | 286 (0.33) | 1.34 [1.06–1.70] | |
| DR3‡ | 184 (0.20) | 255 (0.30) | 0.64 [0.49–0.82] | 1.43 × 10−7 |
| 4/4‡ | 73 (0.16) | 42 (0.10) | 1.90 [1.09–3.29] | |
| 3/4‡ | 81 (0.18) | 96 (0.22) | 0.84 [0.52–1.36] | |
| 3/3‡ | 21 (0.05) | 34 (0.08) | 0.54 [0.28–1.06] | |
| 4/X‡ | 155 (0.34) | 106 (0.25) | 1.59 [1.02–2.50] | |
| 3/X‡ | 61 (0.14) | 91 (0.21) | 0.68 [0.41–1.12] | |
| X/X‡ | 61 (0.14) | 62 (0.14) | 1.00 (ref.) | |
Both DR3 and DR4 were required to model the association with IA-2A. The DR3/4/X genotype coding did not improve on the allele-specific model (P = 0.66).
*Uses all samples genotyped at rs660895.
†Uses all samples genotyped at rs2187668.
‡Uses samples genotyped at both rs660895 and rs2187668. N, number of chromosomes or genotypes.
The DR3 SNP, rs2187668, was associated with GADA positivity (P = 6.03 × 10−6) (Table 5). The DR4 SNP, rs660895, was not associated with GADA in a single locus model (P = 0.090) (Table 5) but did improve a model including DR3 (rs2187668) (P = 2.22 × 10−5). However, there was only a small difference in the frequency of DR4 in GADA positives and negatives, suggesting that perhaps it was the non-DR4 alleles that were associated with GADA positivity. SNPs rs2187668 and rs660895 were used to code the non-DR3 non-DR4 allele, DRX, which was found to be negatively associated with GADA positivity (OR 0.56 [95% CI 0.43–0.74]) (Table 5). Analysis of the genotypes revealed that while DR4/4 was associated with GADA positivity (OR 2.57) (Table 5), both DR4/X and DRX/X were negatively associated with GADA positivity (OR 0.53 and 0.27, respectively, using DR3/4 as reference) (Table 5).
TABLE 5.
Association of HLA class II with GADA, using rs660895 and rs2187668, to model HLA-DR4 and -DR3 (in 1,327 and 1,342 autoimmune diabetic case subjects, respectively)
| SNP |
N (frequency) |
OR [95% CI] | P | ||
|---|---|---|---|---|---|
| GADA positive | GADA negative | ||||
| rs2187668 | |||||
| A (DR3)* | 586 (0.26) | 76 (0.17) | 1.90 [1.42–2.55] | 6.03 × 10−6 | |
| rs660895 | |||||
| G (DR4)* | 847 (0.38) | 168 (0.36) | 1.23 [0.97–1.57] | 0.090 | |
| DR3† | 569 (0.26) | 72 (0.16) | 1.43 [1.02–1.99] | 2.53 [1.82–3.51] | |
| DR4† | 833 (0.39) | 166 (0.38) | 1.00 (ref.) | 1.78 [1.36–2.33] | |
| DRX† | 754 (0.35) | 202 (0.46) | 0.56 [0.43–0.74] | 1.00 (ref.) | 5.31 × 10−9 |
| 4/4 | 147 (0.14) | 22 (0.10) | 2.57 [1.44–4.59] | 1.37 [0.72–2.59] | |
| 3/X | 194 (0.18) | 23 (0.10) | 2.37 [1.38–4.09] | 1.26 [0.69–2.32] | |
| 3/3 | 74 (0.07) | 6 (0.03) | 3.27 [1.25–8.61] | 1.74 [0.64–4.76] | |
| 3/4 | 227 (0.21) | 37 (0.17) | 1.88 [1.18–2.98] | 1.00 (ref.) | |
| 4/X | 312 (0.29) | 85 (0.39) | 1.00 (ref.) | 0.53 [0.34–0.85] | |
| X/X | 124 (0.12) | 47 (0.21) | 0.51 [0.32–0.81] | 0.27 [0.16–0.47] | |
*Single SNP analyses use all samples genotyped at the test SNP.
†Uses the DR3 and DR4 SNPs to generate a non-DR3, non-DR4 allele, DRX, and to analyze the class II locus HLA-DRB1 as a single locus including alleles using two alleles in the model and using the third as reference. Only samples genotyped at both SNPs (rs2187668 and rs660895) were included in this analysis. The evidence that the DR3/4/X genotype coding was a more appropriate model than the combined DR3 and DR4 allelic effects model was unconvincing (P = 0.024). N, number of chromosomes or genotypes.
FCRL3 (rs7528684) was found to be associated with IA-2A (P = 9.16 × 10−4) (Table 6). The minor (G) allele was negatively associated with IA-2A positivity (OR 0.73) (Table 6). Neither the FCRL3 SNP (rs7528684) nor the GAD2 SNP (rs2839671) was associated convincingly with GADA positivity (P = 0.012 and 0.17, respectively).
TABLE 6.
Association of rs7528684 (A>G) in FCRL3 (1q23.1) with IA-2A positivity in 903 autoimmune diabetic case subjects
| Allele or genotype |
N (frequency) |
OR [95% CI] | P | ||
|---|---|---|---|---|---|
| IA-2A positive | IA-2A negative | ||||
| G | 386 (0.42) | 450 (0.50) | 0.73 [0.60–0.88] | 9.16 × 10−4 | |
| A/A | 161 (0.35) | 116 (0.26) | 1.00 (ref.) | 1.44 [1.05–1.97] | |
| A/G | 210 (0.46) | 212 (0.48) | 0.70 [0.51–0.96] | 1.00 (ref.) | |
| G/G | 88 (0.19) | 116 (0.26) | 0.53 [0.37–0.78] | 0.77 [0.54–1.09] | |
N, number of chromosomes or genotypes.
DISCUSSION
In contrast with smaller studies of adult-onset autoimmune diabetes or LADA that ascertained case subjects from type 2 diabetes clinics, this study, for the first time, has gathered together a large collection of samples from diabetic patients of all ages who are autoantibody positive at the time of diagnosis and tested them for association with previously confirmed pediatric type 1 diabetes–associated genetic loci. We obtained evidence for nine susceptibility loci, including HLA, INS, PTPN22, and IL2RA, which are shared in common between autoimmune diabetes diagnosed in adults and type 1 diabetes diagnosed in children. With more samples and improved power, the number of shared susceptibility loci will increase. For instance, nominal evidence of association at P < 0.01 was also obtained for IFIH1 (2q24.2) and BACH2 (6q15) with the same direction of effect as reported for pediatric type 1 diabetes (Table 2; Supplementary Table 3; Fig. 2). We were unable to account for any population substructure because the cohort has not been genotyped on a GWAS SNP chip. However, given that all case and control subjects were of white European ancestry, were recruited from a single region in Germany, and that evidence of association obtained in adult-onset autoimmune diabetes was consistent with the effects found with childhood-onset type 1 diabetes, confounding due to population effects should be minimal. A proportion of our case subjects is likely to fulfill the diagnostic criteria for LADA, which is thought to be an autoimmune form of diabetes. However, these case subjects could not be identified in our cohort because all case subjects were treated with insulin upon diagnosis of diabetes based on clinical grounds and not autoantibody status of the patient, of which the clinicians were unaware.
Given the known age dependence of the DR3/4 genotype association (5–7), the lack of evidence of a DR3/4 synergistic effect in our adult-onset autoimmune diabetic case subjects, despite >90% power assuming the same effect size as in childhood-onset type 1 diabetes, was unsurprising. DR4/4, which has not previously been shown to have an age-at-diagnosis effect in autoimmune diabetes, had a reduced frequency in case subjects with an older age at diagnosis. The converse increase in frequency of the DRX/X genotype that was observed (Table 3) was consistent with earlier reports that the HLA class II alleles, which confer protection for pediatric-onset type 1 diabetes, are more common in older-onset autoimmune diabetes (6). Other HLA-DRB1 or HLA-DQB1 alleles or genotypes, in addition to the protective HLA-DRB1*15 allele, must have elevated frequencies in adult-onset autoimmune diabetes, but full HLA genotyping is required to determine which one(s).
Outside the HLA region, RNLS, rs10509540, and the IL2 SNP rs2069763 have been shown to have an age-at-diagnosis effect in pediatric-onset type 1 diabetes (J.M.M.H., J.A.T., unpublished data), with the minor allele conferring less susceptibility in older-onset case subjects. This direction of effect is consistent with our findings in the HLA. Owing to the age-at-diagnosis interaction, the effect size in adult-onset autoimmune diabetes is likely to be smaller than that in pediatric type 1 diabetes. Hence, using the effect size as reported in pediatric type 1 diabetes to calculate power may exaggerate the power of our study to detect an effect in adult-onset autoimmune diabetes. No effect of rs10509540, RNLS (10q23.31), was detected in adult-onset autoimmune diabetes, despite good power (80%) to detect an effect of OR 0.88, which is smaller than the effect reported in childhood-onset type 1 diabetes (OR 0.75 [95% CI 0.70–0.80]) (1). Consequently, in addition to the HLA class II alleles, RNLS (rs10509540) distinguishes pediatric-onset type 1 diabetes from adult-onset autoimmune diabetes because it either has an effect size significantly smaller than in childhood-onset type 1 diabetes or it is not associated in adult-onset autoimmune diabetes. In fact, all of the loci with possible age-at-diagnosis effects, HLA-DR3/4, HLA-DR4/4, RNLS, IL2, PTPN2, CTRB2, and IL2RA, appear to have a significantly smaller effect in adult-onset autoimmune diabetes than in childhood-onset diabetes (Table 3; Fig. 2). Three of the loci with possible age-at-diagnosis effects in autoimmune diabetes, PTPN2, IL2RA, and IL2, are all involved in interleukin-2 (IL-2) cytokine’s essential role in maintaining immune tolerance (9,29). These results are mirrored by findings regarding the IL-2 gene in susceptibility to autoimmune diabetes in the NOD mouse (9). Genetic deficiencies in the IL-2 pathway, in concert with HLA class II genes, are clearly a major driver for young-onset type 1 diabetes. The immune system is known to alter significantly over time, and these changes could imply that the role of the IL-2 pathway in loss of tolerance to islet antigens and susceptibility to autoimmune diabetes wanes with age (30). Importantly, IL-2 production and signaling, an essential requirement for immune-tolerating function of T regulatory cells, is the major causal effect outside the MHC in the NOD mouse model of autoimmune diabetes and here, too, the dependency on IL-2 decreases with increasing age (31,32).
The inverse association of DR3 and DR4 with IA-2A in our autoimmune diabetic case subjects is consistent with that observed in British pediatric-onset type 1 diabetes (OR 0.56 and 0.56, respectively, for DR3 and OR 1.62 and 1.75, respectively, for DR4) (33). However, despite an increase in frequency in adult-onset autoimmune diabetes compared with British pediatric-onset type 1 diabetes, DRX/X is consistently neutral for IA-2A in both groups. In contrast, the DRX/X group is not associated with GADA positivity in British childhood-onset type 1 diabetes (33) but was negatively associated with GADA positivity in adult-onset autoimmune diabetes. Having accounted for age-at-diagnosis effects, we found that DR4/4 and DR3/4, which are associated with a younger age at diagnosis, were associated with GADA positivity in our autoimmune diabetic case subjects (Table 5), a trait associated with older age at diagnosis (Supplementary Table 4). In concordance with this, neither DR4/4 nor DR3/4 was associated with GADA positivity in British childhood-onset type 1 diabetes (33). Others have reported, in a much smaller sample set, that there is an age-at-diagnosis dependence of the HLA-DQB1*0302 (which is in high LD with DR4) GADA association (34). These results together could indicate a change over time in autoantigenic targets and autoreactive T-cell repertoires.
The FCRL3 SNP, rs7528684 (A>G), has been shown to affect FCRL3 expression on B cells through nuclear factor-κB binding (16). It has also been shown to be associated with several autoimmune diseases (16–19) but not with type 1 diabetes (35,36). Despite not being associated in our adult-onset autoimmune diabetic case subjects, rs7528684 in FCRL3 was associated convincingly with IA-2A positivity, an effect also found in type 1 diabetes diagnosed before age 17 years (13). However, in contrast to Graves disease and rheumatoid arthritis, in which the G/G genotype confers susceptibility, G/G was associated with absence of IA-2A and A/A with IA-2A positivity in our case subjects.
Type 1 diabetes incidence is predicted to double in children by 2020 owing to undefined environmental changes (37). These children carry the most diabetogenic combinations of susceptibility alleles. Consequently, lower frequencies of high-risk alleles and genotypes are observed in older case subjects because individuals carrying the most strongly predisposing alleles in the population will have progressed to autoimmune insulin deficiency at a younger age. Although we have not scanned the entire genome for association with autoimmune diabetes diagnosed in adults, we propose that the genetic architecture will be almost identical to pediatric diabetes but with a lower genetic load and a different history of environmental exposure.
ACKNOWLEDGMENTS
This work was funded by the Juvenile Diabetes Research Foundation International, the Wellcome Trust, and the National Institute for Health Research Cambridge Biomedical Centre. The Cambridge Institute for Medical Research is in receipt of a Wellcome Trust Strategic Award (079895). B.O.B. was supported by Deutsche Forschungsgemeinschaft SFB 518, GrK1041, and Baden-Württemberg State (Centre of Excellence “Metabolic Disorders”).
No potential conflicts of interest relevant to this article were reported.
J.M.M.H. analyzed and interpreted data and wrote the manuscript. S.R. tested samples for autoantibodies and collected and edited phenotype data. D.J.S. performed all genotyping. B.O.B. designed the ADBW study, collected samples, and edited the manuscript. The ADBW-END Study Group collected all samples. J.A.T. conceived the study, interpreted data, and edited the manuscript.
The authors thank all the patients and control subjects for their participation. The authors also thank H. Stevens, P. Clarke, G. Coleman, S. Duley, D. Harrison, S. Hawkins, M. Maisuria, and T. Mistry of the University of Cambridge for preparation of DNA samples.
APPENDIX
Members of the ADBW-END Study Group (in alphabetical order): G. Aldinger, J. Aufschild, J. Bechtold, G. Becker, W. Beischer, R. Bickel, R. Blagieva, B.O. Boehm (Chairperson), J. Brückel, S. Claudi-Boehm, A. Dapp, T. Eiermann, R. Ermerling, J. Gloyer, T. Haak, M. Haenle, W. Hecker, R. Holl (Chairperson), A. Holland, S. Höpfer, S. Jung, W. Junginger, G. Jütting, T. Käser, J. Kieferle, W. Koenig, W. Kratzer, P. Kristen, P. Kühnl, A. Kurkhaus, G. Limberg, B. Lippmann-Grob, R. Lobmann, U. Maas, B. Manfras, S. Merger, K.A. Müller, T. Nikolaus, W.-B. Offensperger, H. Renner, C. Rosak, S. Rosinger, S. Schilling, G.H. Schreiber, A. Schuler, G.J. Schwinn, E. Siegel, R. Sing, E. Spohr, M. Standop, G. Trischler, U. Weickert, G. Wildemann-Gilbert, and D. Wörner.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0364/-/DC1.
A complete list of the members of the ADBW-END Study Group can be found in the appendix.
REFERENCES
- 1.Barrett JC, Clayton DG, Concannon P, et al. ; Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leslie RD. Predicting adult-onset autoimmune diabetes: clarity from complexity. Diabetes 2010;59:330–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolandsson O, Palmer JP. Latent autoimmune diabetes in adults (LADA) is dead: long live autoimmune diabetes! Diabetologia 2010;53:1250–1253 [DOI] [PubMed] [Google Scholar]
- 4.Steck AK, Eisenbarth GS. Genetic similarities between latent autoimmune diabetes and type 1 and type 2 diabetes. Diabetes 2008;57:1160–1162 [DOI] [PubMed] [Google Scholar]
- 5.Caillat-Zucman S, Garchon HJ, Timsit J, et al. Age-dependent HLA genetic heterogeneity of type 1 insulin-dependent diabetes mellitus. J Clin Invest 1992;90:2242–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai M, Zeggini E, Horton VA, et al. An association analysis of the HLA gene region in latent autoimmune diabetes in adults. Diabetologia 2007;50:68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton V, Stratton I, Bottazzo GF, et al. ; UK Prospective Diabetes Study (UKPDS) Group Genetic heterogeneity of autoimmune diabetes: age of presentation in adults is influenced by HLA DRB1 and DQB1 genotypes (UKPDS 43). Diabetologia 1999;42:608–616 [DOI] [PubMed] [Google Scholar]
- 8.Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet 2009;5:e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todd JA. Etiology of type 1 diabetes. Immunity 2010;32:457–467 [DOI] [PubMed] [Google Scholar]
- 10.Desai M, Zeggini E, Horton VA, et al. The variable number of tandem repeats upstream of the insulin gene is a susceptibility locus for latent autoimmune diabetes in adults. Diabetes 2006;55:1890–1894 [DOI] [PubMed] [Google Scholar]
- 11.Durinovic-Belló I, Wu RP, Gersuk VH, Sanda S, Shilling HG, Nepom GT. Insulin gene VNTR genotype associates with frequency and phenotype of the autoimmune response to proinsulin. Genes Immun 2010;11:188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howson JM, Dunger DB, Nutland S, Stevens H, Wicker LS, Todd JA. A type 1 diabetes subgroup with a female bias is characterised by failure in tolerance to thyroid peroxidase at an early age and a strong association with the cytotoxic T-lymphocyte-associated antigen-4 gene. Diabetologia 2007;50:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plagnol V, Howson JMM, Smyth DJ, et al. Genetic analysis of autoantibody positivity in Graves' disease and type 1 diabetes. PLoS Genet 2011;7:e1002216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettersen E, Skorpen F, Kvaløy K, Midthjell K, Grill V. Genetic heterogeneity in latent autoimmune diabetes is linked to various degrees of autoimmune activity: results from the Nord-Trøndelag Health Study. Diabetes 2010;59:302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voight BF, Scott LJ, Steinthorsdottir V, et al. ; MAGIC investigators; GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kochi Y, Yamada R, Suzuki A, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet 2005;37:478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owen CJ, Kelly H, Eden JA, Merriman ME, Pearce SH, Merriman TR. Analysis of the Fc receptor-like-3 (FCRL3) locus in Caucasians with autoimmune disorders suggests a complex pattern of disease association. J Clin Endocrinol Metab 2007;92:1106–1111 [DOI] [PubMed] [Google Scholar]
- 18.Simmonds MJ, Heward JM, Carr-Smith J, Foxall H, Franklyn JA, Gough SC. Contribution of single nucleotide polymorphisms within FCRL3 and MAP3K7IP2 to the pathogenesis of Graves’ disease. J Clin Endocrinol Metab 2006;91:1056–1061 [DOI] [PubMed] [Google Scholar]
- 19.Burton PR, Clayton DG, Cardon LR, et al. ; Wellcome Trust Case Control Consortium; Australo-Anglo-American Spondylitis Consortium (TASC); Biologics in RA Genetics and Genomics Study Syndicate (BRAGGS) Steering Committee; Breast Cancer Susceptibility Collaboration (UK) Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 2007;39:1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehm BO, Manfras B, Seissler J, et al. Epidemiology and immunogenetic background of islet cell antibody–positive nondiabetic schoolchildren: Ulm-Frankfurt population study. Diabetes 1991;40:1435–1439 [DOI] [PubMed] [Google Scholar]
- 21.Powell M, Prentice L, Asawa T, et al. Glutamic acid decarboxylase autoantibody assay using 125I-labelled recombinant GAD65 produced in yeast. Clin Chim Acta 1996;256:175–188 [DOI] [PubMed] [Google Scholar]
- 22.Endl J, Rosinger S, Schwarz B, et al. Coexpression of CD25 and OX40 (CD134) receptors delineates autoreactive T-cells in type 1 diabetes. Diabetes 2006;55:50–60 [PubMed] [Google Scholar]
- 23.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006;38:209–213 [DOI] [PubMed] [Google Scholar]
- 25.Dendrou CA, Plagnol V, Fung E, et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet 2009;41:1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxena R, Gianniny L, Burtt NP, et al. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes 2006;55:2890–2895 [DOI] [PubMed] [Google Scholar]
- 27.Bakhtadze E, Cervin C, Lindholm E, et al. Common variants in the TCF7L2 gene help to differentiate autoimmune from non-autoimmune diabetes in young (15-34 years) but not in middle-aged (40-59 years) diabetic patients. Diabetologia 2008;51:2224–2232 [DOI] [PubMed] [Google Scholar]
- 28.Raj SM, Howson JMM, Walker NM, et al. No association of multiple type 2 diabetes loci with type 1 diabetes. Diabetologia 2009;52:2109–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long SA, Cerosaletti K, Wan JY, Ho JC, Tatum M, Wei S, et al. An autoimmune-associated variant in PTPN2 reveals an impairment of IL-2R signaling in CD4(+) T cells. Genes Immun 2011;12:116–125 [DOI] [PMC free article] [PubMed]
- 30.Maue AC, Yager EJ, Swain SL, Woodland DL, Blackman MA, Haynes L. T-cell immunosenescence: lessons learned from mouse models of aging. Trends Immunol 2009;30:301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamanouchi J, Rainbow D, Serra P, et al. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet 2007;39:329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long SA, Cerosaletti K, Bollyky PL, et al. Defects in IL-2R signaling contribute to diminished maintenance of FOXP3 expression in CD4(+)CD25(+) regulatory T-cells of type 1 diabetic subjects. Diabetes 2010;59:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howson JMM, Stevens H, Smyth DJ, et al. Evidence that HLA class I and II associations with type 1 diabetes, autoantibodies to GAD and autoantibodies to IA-2, are distinct. Diabetes 2011;60:2635–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandewalle CL, Falorni A, Lernmark A, et al. Associations of GAD65- and IA-2- autoantibodies with genetic risk markers in new-onset IDDM patients and their siblings. The Belgian Diabetes Registry. Diabetes Care 1997;20:1547–1552 [DOI] [PubMed] [Google Scholar]
- 35.Duchatelet S, Caillat-Zucman S, Dubois-Laforgue D, Blanc H, Timsit J, Julier C. FCRL3 -169CT functional polymorphism in type 1 diabetes and autoimmunity traits. Biomed Pharmacother 2008;62:153–157 [DOI] [PubMed] [Google Scholar]
- 36.Turunen JA, Wessman M, Kilpikari R, Parkkonen M, Forsblom C, Groop PH; FinnDiane Study Group The functional variant -169C/T in the FCRL3 gene does not increase susceptibility to type 1 diabetes. Diabet Med 2006;23:925–927 [DOI] [PubMed] [Google Scholar]
- 37.Patterson CC, Dahlquist GG, Gyürüs E, Green A, Soltész G; EURODIAB Study Group Incidence trends for childhood type 1 diabetes in Europe during 1989-2003 and predicted new cases 2005-20: a multicentre prospective registration study. Lancet 2009;373:2027–2033 [DOI] [PubMed] [Google Scholar]
- 38.Smyth DJ, Plagnol V, Walker NM, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 2008;359:2767–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fung EY, Smyth DJ, Howson JM, et al. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun 2009;10:188–191 [DOI] [PubMed] [Google Scholar]
- 40.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–323 [DOI] [PubMed] [Google Scholar]
- 41.Zeggini E, Weedon MN, Lindgren CM, et al. ; Wellcome Trust Case Control Consortium (WTCCC) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]