Abstract
Tetraploidy and the depolyploidization of tetraploid cells may contribute to oncogenesis. Several mechanisms have evolved to avoid the generation, survival, proliferation and depolyploidization of tetraploids. Cells that illicitly survive these checkpoints are prone to chromosomal instability and aneuploidization. Along with their replication, tetraploids constantly undergo chromosomal rearrangements that eventually lead to pseudodiploidy by two non-exclusive mechanisms: (i) multipolar divisions and (ii) illicit bipolar divisions in the presence of improper microtubule-kinetochore attachments. Here, we describe the regulation and the molecular mechanisms that underlie such a ‘polyploidization–depolyploidization' cascade, while focusing on the role of oncogenes and tumor suppressor genes in tetraploidy-driven tumorigenesis. We speculate that the identification of signaling/metabolic cascades that are required for the survival of tetraploid or aneuploid (but not diploid) cancer cells may pave the way for the development of novel broad-spectrum anticancer agents.
Keywords: tetraploid, mitosis, cancer, aneuploidy, TP53
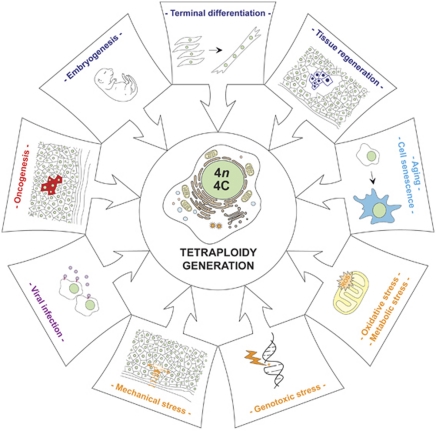
Diploidy, the condition of having two complete haploid sets of homologous chromosomes (2n), is the standard state for eukaryotic organisms that reproduce sexually. The accumulation of more than two chromosome sets, polyploidy (>2n), is common in nature, especially in its simplest form, tetraploidy (4n). In contrast to some plant species and animal taxa (e.g., insects), mammals do not tolerate germline polyploidization.1 However, shifts in the ploidy of somatic cells (endopolyploidization) participate in the developmental program of several tissues and organs (Figure 1).2 Thus, although the vast majority of mammalian cells are diploid, multinucleated and endopolyploid cells can be found at relatively high frequency (0.5–20%), especially in highly proliferating tissues like the placenta.3 Endopolyploidization, which sometimes constitutes a regulated mechanism for increasing cell size,4 has been documented in multiple cell types including megakaryocytes and neurons.4, 5
Figure 1.
Tetraploidy in physiological and pathological conditions. In several, evolutionarily distant organisms, tetraploid cells are implicated in a number of physiological processes, including embryogenesis as well as the terminal differentiation and the regeneration of specific tissues (e.g., muscles and the liver, respectively). Polyploidy has also been shown to arise in response to aging (cell senescence), to stress (including genotoxic, metabolic, oxidative and mechanical insults) and to infection by fusogenic (e.g., HIV-1) and oncogenic (e.g., human papillomavirus, HPV) viruses. Finally, tetraploidy has a critical role in oncogenesis. ROS, reactive oxygen species
In physiological conditions, endopolyploidization is frequently linked to the generation of terminally differentiated cells that cannot proliferate. Alternatively, endopolyploidization can constitute an adaptive response to stress (Figure 1).2 In some physiological scenarios (e.g., liver re-population by bone marrow-derived cells), polyploid intermediates generated by heterotypic cell-to-cell fusion can recover an apparently diploid genome and proliferate.6 Similarly, under specific conditions, the human pathogen Candida albicans can complete a life cycle that includes tetraploid intermediates but terminates with mating-competent diploid/near-to-diploid cells.7 Thus, the depolyploidization of tetraploids may also constitute a physiological process. However, the illicit polyploidization of proliferating cells has been associated with human diseases including cancer (Figure 1).8 In this context, polyploids, especially those arising from mitotic dysfunctions, are believed to contribute to tumorigenesis/cancer progression by favoring aneuploidy and chromosomal instability (CIN). This review explores the mechanisms of illicit polyploidization/depolyploidization.
Physiological and pathological generation of polyploids
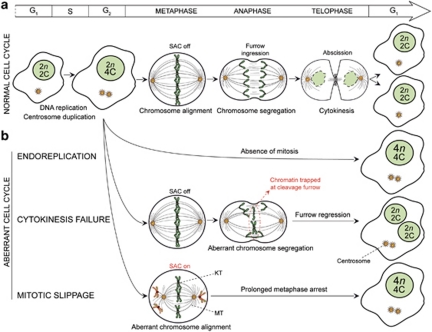
Several routes lead to polyploidization (Figures 2 and 3). Germline polyploidization, which is lethal (it accounts for 5–15% spontaneous abortions) can arise from: (i) gametes that failed to complete the reductional step of meiosis (normally leading to two daughter cells with an haploid DNA content); (ii) polyspermy (the fertilization of a single egg by more than one sperm); (iii) the fusion between the egg and one polar body (followed by normal fertilization); or (iv) events of mitotic failure or cell fusion occurring early during embryogenesis.9 On the contrary, somatic polyploidization often represents a programmed event resulting from: (i) cell-to-cell fusion, leading to multinucleated cells that are arrested in the G1 phase of the cell cycle;10 (ii) endoreplication, during which cells enter two consecutive rounds of DNA replication that are not separated by mitosis;2 or (iii) endomitosis, during which cells with duplicated chromosomes fail to undergo both nuclear (karyokinesis) and cytoplasmic (cytokinesis) division and abort mitosis.2 Endopolyploidization by cell-to-cell fusion has been documented in some cell types including myocytes.11 Endoreplication (whose most striking example is provided by trophoblastic giant cells that develop from the murine trophectoderm)3 exerts critical functions for normal development, efficient nutrient uptake/storage and differentiation.2 Endomitosis occurs in megakaryocytes and in some hepatocytes.2
Figure 2.
Mitotic dysfunctions as a generator of tetraploidy. Most unscheduled tetraploids are generated by the illicit progression along aberrant cell cycles. (a) Schematic representation of a normal cell cycle (mitotic prophase is omitted for clarity). (b) Diploid cells with normally replicated DNA (2n, 4C) and duplicated centrosomes can fail to undergo mitosis altogether (endoreplication) or just cytokinesis (cytokinesis failure), resulting in the generation of a mononucleated (4n, 4C) or binucleated (2 × (2n, 2C)) tetraploid cell, respectively. If viable, binucleated tetraploids will become mononucleated after one additional round of mitosis, when all chromosomes will align onto a single metaphase plate and will segregate into two daughter cells (not depicted). Alternatively, misattached chromosomes may trigger the activation of the spindle assembly checkpoint (SAC), which can lead to a prolonged metaphase arrest and eventually to the generation of a mononucleated (4n, 4C) tetraploid cell. Additional details in text. C, DNA content of a haploid nucleus; KT, kinetochore; MT, microtubule; n=number of chromosomes in the haploid set (23 for humans)
Figure 3.
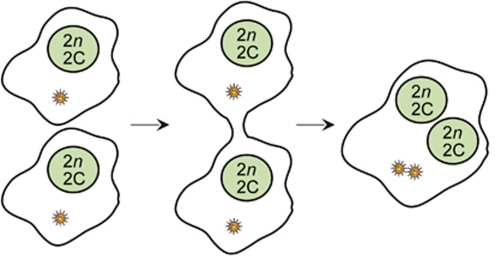
Cell-to-cell fusion as a generator of tetraploidy. In the presence of mechanical stress or viral fusogenic proteins, binucleated (2x 2n, 2C) tetraploids (as well as higher-order polyploids, not depicted) can derive from cell-to-cell fusion events. Additional details in text. C, DNA content of a haploid nucleus; n=number of chromosomes in the haploid set (23 for humans)
In injured or diseased tissues, somatic polyploidy may also be generated accidentally. Thus, macrophages fuse among each other during inflammation,12 as do hematopoietic cells with several cell types including hepatocytes and neurons in response to organ-specific or whole-body stress.13, 14 Oncogenic viruses can trigger illicit cell-to-cell fusion of somatic cells, contributing to cancer pathogenesis.15 Similarly, inflammation-triggered fusion between leukocytes and cancer cells reportedly creates tumor hybrids with metastatic potential.16 Unscheduled tetraploidization also arises in tumor cell precursors, most often on mitotic dysfunctions (Figure 2) including: (i) mitotic slippage (mitotic adaptation), a phenomenon by which cells that cannot proceed through mitosis due to continuous activation of the spindle assembly checkpoint (SAC) ‘slip' to the subsequent cell cycle in a tetraploid state17 and (ii) cytokinesis failure, which is provoked by cleavage furrow regression (due to chromatin bridges occluding the cleavage furrow or to defects in cytokinetic proteins) and results in the generation of binucleated cells.18
Chromosome non-disjunction (an anaphase error resulting in the segregation of both sister chromosomes to the same daughter cell) favors the generation of tetraploidy, rather than aneuploidy,19 indicating that accurate chromosome segregation is required for cytokinesis completion. Recently, a link between tetraploidy and pre-cancerous DNA lesions induced by oncogene activation20 and a novel pathway of endoreplication that is ignited by prolonged DNA damage signaling21 have been unveiled. Thus, there are multiple, independent pathways to polyploidization.
Preventing tetraploid accumulation
Cells developed several strategies to limit the generation, proliferation and/or survival of tetraploids. Pharmacological cytokinesis inhibition (for instance with cytochalasin) arrests the proliferation of non-transformed cells at a binucleated stage, through mechanisms that involve the oncosuppressors TP53, cyclin-dependent kinase inhibitor 1A (CDKN1A), CDKN2A and retinoblastoma (RB).22, 23 On the contrary, upon cytochalasin exposure, cells transformed by the Simian virus 40 (which inactivates both TP53 and RB) maintain their proliferative potential and generate aneuploids.24 Similarly, tetraploid cells deriving from cell-to-cell fusion propagate in culture in the absence of TP53.15 These observations have led some researchers to hypothesize the existence of a TP53- and RB-dependent ‘tetraploidy checkpoint' that block the proliferation and DNA re-replication of tetraploids by arresting them in a tetraploid G1 phase.22, 23 The existence of this checkpoint remains controversial, because TP53 activation might derive from the use of tetraploidy-inducing agents rather than from tetraploidy itself.25
Most tetraploids that are generated by cytokinesis or karyokinesis inhibition quickly undergo apoptosis.26 TP53 has a major role in the prevention of tetraploidy and its absence facilitates the generation/survival of tetraploid cancer cells,22, 26 as well as of tetraploid primary mouse mammary epithelial cells.27 These results suggest that the ‘tetraploidy checkpoint' might consist in the selective apoptotic removal of tetraploids. In support of this hypothesis, depletion of the pro-apoptotic factor BAX by RNA interference (RNAi) mimicked the permissive effect of TP53 deficiency on tetraploidization in vitro.26 Similarly, overexpression of the anti-apoptotic protein BCL-XL facilitated polyploidization in the absence of TP53.28 Both TP53 and RB are frequently absent or mutated in tumors, a situation that may provide newly generated tumor cells with an intrinsic potential to survive/proliferate.
The post-mitotic activation of TP53 responsible for the elimination of polyploids might be induced/influenced by DNA lesions accumulated during mitotic arrest.29 However, it remains controversial whether the DNA gets damaged during a prolonged mitotic blockage.30 Growing evidence indicates that functional SAC is required for the execution of TP53-dependent post-mitotic arrest.22, 31 This concept has been strengthened by the demonstration that SAC components like TTK/MPS1 directly phosphorylate TP53 on spindle damage and mitotic slippage, engaging the tetraploidy checkpoint.32 Thus, the absence/inhibition of SAC components favors the generation of tetraploids and allows for their survival/proliferation, thereby increasing CIN.
Mitotic deregulation as a generator of tetraploidy and tumorigenesis
Aneuploidy (the condition of having a number of chromosomes that deviates from, but that is not multiple of, the haploid number) represents the most common type of genomic instability in cancer. Tumors cells are characterized by a highly heterogeneous number of chromosomes with a dominance of aneuploid distributions. Often, cancer cells contain between 40 and 60 chromosomes, a condition that is referred to as ‘near-to-diploidy' or ‘pseudodiploidy'.33 Many tumors show an elevated rate of chromosome missegregation, provoking structural (translocations, deletions or inversions) and numerical (chromosome losses and gains) aberrations at each cell cycle, namely CIN.34 CIN results in continuous karyotypic rearrangements, favoring the generation of aneuploids and hence, the spontaneous evolution of tumors to malignancy.
At all tumor stages, a substantial number of cancer cells contains a tetraploid or near-to-tetraploid genome (‘large-scale aneuploidy'). Tetraploids have been documented in the early stages of colorectal, breast and cervical cancer.8, 33 Moreover, tetraploids are frequently found in pre-cancerous lesions including Barrett's esophagus.35 Bacteria belonging to the intestinal flora can induce tetraploidization, which has speculatively been linked to carcinogenesis.36 Chemical mutagenesis of tetraploid Tp53−/− mouse mammary epithelial cells in vivo leads to the generation of transplantable tumors, which exhibit high level of CIN/aneuploidy.27 Thus, primary tetraploids are more prone to transformation than diploids. Accordingly, some viruses including retroviruses have been shown to favor malignant transformation on cell-to-cell fusion, in vitro and in vivo.37 Chromosome non-disjunction does not directly trigger aneuploidy, but favors the generation of tetraploids that may become aneuploid on further divisions.19 This mechanism might constitute the most prominent source of aneuploidy in cancer.
The discovery of links between cell cycle regulators and SAC components has strengthened the hypothesis that tetraploidy might favor tumor progression. Inactivation of the RB pathway provokes aneuploidy by inducing a mitotic blockage followed by the generation of tetraploidy via mitotic slippage.38 Several mitotic checkpoint genes including MAD2L1 (a SAC component) are transcriptionally controlled by E2F (which is negatively regulated by RB), implying that their expression may increase when RB is inhibited.38 Aberrant expression of SAC components including MAD2L1, BUB1B and aurora kinase B (AURKB) can induce mitotic slippage-dependant tetraploidization.39 Accordingly, transgenic mice engineered for the inducible overexpression of Mad2l1 develop multiple tumors, and cells isolated from these animals display high CIN levels.40 Prolonged activation of the mitotic checkpoint may sustain tumorigenesis. Thus, the cancer-prone phenotype of Mad2l1 transgenic mice might result from Mad2l1-dependent hyperactivation of the mitotic checkpoint. SAC overactivation favors, indeed, the ‘mitotic arrest-mitotic errors-mitotic slippage' sequence that leads to tetraploidy, enhanced CIN and aneuploidy.40
The amplification of regulators of the mitotic progression like polo-like kinase 1 (PLK1) and aurora kinase A (AURKA) also generates polyploidy via mitotic dysfunction and cytokinesis failure.41 Mitotic kinases are frequently hyperactivated in human tumors and cancer cell lines, which might facilitate tumorigenesis/cancer progression by promoting CIN. In the murine mammary epithelium, the overexpression of AURKA, which controls several mitotic processes, increases CIN and induces tetraploidy before tumor formation,42 strengthening the idea that tetraploidy precedes malignant transformation. In ovarian cancer cells, RNAi-mediated depletion of AURKA limits genomic instability, centrosome amplification and in vivo tumorigenic potential.43 Thus, AURKA might function as an oncoprotein.
Similar results have been reported on the overexpression of other mitotic regulators like KIF11, a kinesin-related protein that allows for bipolar spindle assembly and elongation, and AURKB, which supervises several mitotic processes. In mice, KIF11 overexpression results in perturbations of mitotic spindle assembly, favoring genomic instability, tetraploidization and tumor development.44 Similarly, AURKB overexpression facilitates the generation of polyploids (presumably owing to an effect on the separation of sister chromatids), which form aggressive tumors in vivo.45 Thus, KIF11 and AURKB turn out to be bona fide oncoproteins, a notion that is further corroborated by the fact that KIF11 and AURKB are overexpressed in multiple malignancies.46
The downregulation of SAC proteins may also drive tumorigenesis. Bub1b haploinsufficient (Bub1b+/−) murine embryonic fibroblasts (MEFs) in culture spontaneously generates a high percentage of polyploids and Bub1b+/− mice are prone to develop cancer.47 Similarly, MEFs haploinsufficient for Plk4 (encoding a regulator of centriole duplication) are defective in cytokinesis, prone to genomic instability and display an elevated tumorigenic potential in vivo.48 Tetraploidy might account for the increased tumorigenicity of mammary epithelial cells subjected, in vivo, to the tissue-specific deletion of one copy of Tp53 or of Chek1 (coding an important regulator of cell cycle progression).49
These observations indicate that tetraploidy participates in oncogenesis by constituting an intermediate state between healthy diploidy and neoplastic aneuploidy.
The contribution of oncogenes and oncosuppressor genes to tetraploidization
A growing literature indicates that multiple oncosuppressor proteins actively repress tetraploidy (Table 1).
Table 1. Oncosuppressor proteins involved in the ‘polyploidization–depolyploidization' cascade.
| Name | Physiological function(s) | Role in oncogenesis | Implication in tetraploidy | Ref. |
|---|---|---|---|---|
| APC | Involved in cytoskeleton organization, cell migration, chromosomal segregation, transcriptional activation and apoptosis. | Gene defects cause FAP. ∼85% of sporadic colorectal tumors bear APC mutations. | Controls the expression of mitotic regulators. Its loss promotes the generation and survival of polyploids. | 50 |
| BRCA1 | Has a role in DSB DNA repair, transcription, recombination, cell cycle checkpoints and mitotic regulation. | Inherited mutations confer increased risk of developing breast or ovarian cancer. | Its absence favors the downregulation of SAC factors, centrosome amplification and genomic instability. | 56 |
| BRCA2 | Genome stabilizer that mediates DSB DNA repair, regulates cell cycle checkpoints, mitosis and cell division. | Required for the activation of (post) mitotic checkpoints. Limits centrosome amplification and genomic instability. | 52 | |
| CDKN1A | Controls G1 progression by inhibiting CDK2. Mediates TP53-dependent cell cycle arrest. Implicated in cell death, DNA repair and senescence. | Loss-of-function mutations are rare in human tumors. | Contributes to the genome stabilizing functions of TP53 by mediating the tetraploidy checkpoint. | 22,23 |
| CDKN2A | Negatively regulates cell cycle progression by inhibiting CDK4 and CDK6. Implicated in cell senescence. | Frequently inactivated in a wide range of human tumors | Involved in the tetraploidy checkpoint. Limits the proliferation and tumorigenic potential of polyploids. | 23 |
| FBW7 | Ubiquitin ligase, promotes the degradation of oncoproteins involved in cell growth and division (e.g., MYC). | Loss-of-function mutations occur in a variety of human tumors and cancer cell lines. | Controls SAC and G1 checkpoint by acting upstream of TP53. Prevents the proliferation of polyploids. | 53 |
| LATS1 | Ser/Thr kinase localized to the mitotic apparatus. Negative regulator of CDK1/cyclin complexes in early mitosis. | KO mice spontaneously develop tumors. Exerts antitumor effects in vivo. | Required for mitosis, SAC activation and tetraploidy checkpoint. | 55 |
| LATS2 | Centrosomal Ser/Thr kinase required for mitotic spindle formation. Interacts with AURKA and MDM2. | Downregulated in several tumors, and this correlates with increased tumor grade. | Binds and inactivates MDM2, thus triggering the tetraploidy checkpoint. | 30 |
| RB | Hypophosphorylated, it binds E2F1 and represses the transcription of S phase and mitosis regulators. Has a role in genomic stability and senescence. | Gene defects cause childhood RB. Genetic and epigenetic mutations are common to several types of cancer. | Represses the expression of inhibitors of the anaphase promoting complex (which are transactivated by E2F1), thereby limiting genomic instability. | 23,38 |
| PLK4 | Ser/Thr kinase implicated in centrosome duplication and in cell cycle regulation. | Plk4+/− mice spontaneously develop tumors. Deregulated in hepatocellular carcinoma. | Its haploinsufficiency favors centrosome amplification, tetraploidy and in vivo tumorigenicity. | 48 |
| TP53 | Master regulator of cell cycle, apoptosis, senescence, DNA repair, autophagy, metabolism, and genomic stability. | Mutated or epigenetically inactivate in more than 50% of all human tumors. | Limits the expansion of tetraploids and depolyploidization via MD. Implicated in the ‘non-diploidy intolerance'. | 23,26,72 and 81 |
Abbreviations: APC, adenomatous polyposis coli; AURKA, aurora kinase A; BASC, BRCA1-associated genome surveillance complex; CDK, cyclin-dependent kinase; CDKN cyclin-dependent kinase inhibitor; DSB, double-stranded breaks; FAP, familial adenomatous polyposis; FBW7, F-box and WD repeat domain containing 7; LATS, large tumor suppressor; MEFs, mouse embryonic fibroblasts; MD, multipolar division; MDM2, mouse double minute 2; PLK4, polo-like kinase 4; RB, retinoblastoma; SAC, spindle assembly checkpoint
APC is frequently mutated in colon carcinomas and is believed to drive colorectal oncogenesis. Both RNAi-mediated depletion in cultured cells and conditional knockout (with dominant negative mutations) of APC in vivo induce tetraploidization.50 This can result from SAC attenuation followed by mitotic slippage or from cytokinesis failure caused by disrupted spindle–cortical interactions. APC associates with MTs at the site of interaction between MTs and kinotechores (KTs), and might have a role in spindle dynamics and in the spindle checkpoint.51
The absence of BRCA2 and LATS2, two oncosuppressor proteins that regulate cytokinesis and the centrosomes cycle, induces polyploidization and allows for the proliferation of tetraploids, in vitro and in vivo.30, 52 Intriguingly, LATS2 is transactivated by TP53 during mitotic slippage and inhibits MDM2, the main negative regulator of TP53,30 constituting a feedforward loop that prevents the polyploid proliferation. Recently, the ubiquitin ligase FBW7 (which is mutated in numerous cancers) has been shown to regulate both the mitotic and tetraploidy checkpoints.53 FBW7 inhibits mitotic signal transducers including AURKA and reduces the activity of TP53,53 suggesting that FBW7 might suppress tetraploidy upstream of the LATS2-TP53 module. LATS1/hWARTS is an evolutionarily conserved kinase and a dynamic component of the mitotic apparatus.54 Inhibition of LATS1 induces prolonged mitotic arrest followed by abortive cell division and polyploidy.55 Moreover, expression of kinase-dead LATS1 facilitates the survival and proliferation of tetraploids generated as a consequence of deficient TP53 activation, thereby facilitating CIN.55 BRCA1, whose germline mutations predispose women to breast and ovarian cancers, is implicated in the control of mitotic progression and in the centrosome cycle. In mammary epithelial cells, deficient BRCA1 function leads to binucleation, abnormal centrosome amplification and tetraploidy.56
The expression of well-known oncogenes including MYC57 and AURKA42 stimulates tetraploidization by a mechanism involving TP53 inactivation (Table 2). E6, an oncogene encoded by human papillomavirus, also induces transformation although favoring the generation/proliferation of tetraploids.58 This may be linked to the capacity of E6 to trigger TP53 degradation, as proficient TP53 preserves genome integrity not only by strengthening the G1 tetraploidy checkpoint but also by repressing PLK1.58
Table 2. Oncoproteins involved in the ‘polyploidization–depolyploidization' cascade.
| Name | Physiological function(s) | Role in oncogenesis | Implication in tetraploidy | Ref. |
|---|---|---|---|---|
| AURKA | Mitotic kinase involved in centrosome separation, correct spindle polarity and chromosome segregation. | Overexpressed in multiple types of human cancer. | Favors centrosome amplification, genomic instability and tumorigenicity in vivo, in a TP53-inhibitable fashion. | 41,42 and 43 |
| AURKB | Kinase associated to the mitotic apparatus that supervises spindle polarity, chromosome segregation and cytokinesis. | Overexpressed in multiple cancers, correlating with advanced tumor grade. | AURKB overexpression (but also its depletion) stimulates the generation of polyploids and aggressive tumors in vivo. | 45 |
| E6 | Encoded by HPV. Binds to and promotes the degradation of TP53 and other transcription factors. | Immortalizes primary cells but is insufficient for oncogenesis. High-risk HPV strains are associated with cervical cancer. | Favors the generation and proliferation of tetraploids by upregulating PLK1 (which is normally repressed by TP53). | 58 |
| ID1 | HLH proteins that inhibits HLH transcription factors. Involved in cell growth, senescence and differentiation. Also localized to centrosomes. | Upregulated in several types of cancer. High ID1 levels are a negative prognostic indicator in breast cancer patients. | Stabilizes AURKA by inhibiting CDH1, a coactivator of the anaphase promoting complex. Favors the accumulation of extra centrosomes and tetraploidy. | 60 |
| KIF11 | Kinesin-like motor protein involved in chromosome positioning, centrosome separation and spindle bipolarity. | Overexpressed in some human neoplasms. | Transgenic overexpression inhibits centrosome separation and favors tetraploidy and in vivo tumorigenesis. | 44 |
| MADl1 | SAC component monitoring MT attachment and tension in mitosis. Induces metaphase arrest by inhibiting the anaphase promoting complex. | Overexpressed in many human tumors. In transgenic mice, accelerates Myc-induced oncogenesis and increases the spontaneous tumor development. | In the absence of RB, favors SAC overactivation and polyploidy. MADL1 depletion facilitates senescence-associated polyploidy. | 38,40 |
| MYC | Transcription factor controlling cell cycle progression, proliferation, differentiation, apoptosis, adhesion, and cell size. | Genetic/epigenetic alterations are common to several types of human tumors. | In the absence TP53, promotes the generation and survival of polyploids. Controls MAD2L1 expression. | 57 |
| PIM1 | CAMK that favors cell survival (via the upregulation of BCL-2) as well as the G1/S and G2/M cell cycle transitions. | Upregulated in many tumors. High expression levels correlate with poor prognosis. | Its overexpression favors the generation and survival of polyploids, which in turn can sustain tumorigenesis. | 59 |
| PLK1 | Ser/Thr kinase that regulates mitosis by activating the MPF and by degrading FBXO5. Involved in spindle elongation and cleavage furrow ingression. | Overexpressed in a variety of human tumors. High expression levels constitute a negative prognostic factor. | Its overexpression favors the accumulation of extra centrosomes and polyploidy, in a TP53-inhibitable fashion. Upregulated by HBX and E6. | 41,58 |
Abbreviations: AURK, aurora kinase; CAMK, calmodulin-dependent protein kinase; HBX, hepatitis B virus protein X; HLH, helix-loop-helix; HPV, human papillomavirus; MPF, maturation promoting factor; MT, microtubule; PLK1, polo-like kinase 1; SAC, spindle assembly checkpoint
Some additional oncogenes induce CIN and aneuploidy through polyploidization (Table 2). Overexpression of the tumorigenic kinase PIM1 gradually induces polyploidy in human prostate and mammary epithelial cells via cytokinesis failure.59 Overexpression of the potential oncogene ID1, a dominant negative inhibitor of basic helix-loop-helix transcription factors, induces supernumerary centrosomes and centrioles, cytokinesis failure and tetraploidization.60
Thus, tetraploidy can follow the inactivation of oncosuppressor proteins or the activation of oncoproteins.
Genomic instability of tetraploids
The mechanisms through which tetraploidy favors oncogenesis have not yet been entirely elucidated. Accumulating evidence indicates that during malignant transformation, polyploids constitute a metastable intermediate between diploidy and aneuploidy.8 Tetraploidy/polyploidy provokes severe geometric/physical constraints on the machineries that mediate chromosomal duplication, repair and segregation, intrinsically complicating the maintenance of a stable genome.8 Tetraploid human fibroblasts display a high rate of spontaneous DNA lesions, pointing to an increased activation of (and hence, an elevated dependency on) the DNA repair machinery.61 Notably, the efficiency of DNA repair is lower in tetraploids than in their diploid counterparts,62 which may further sustain genomic instability via gross chromosomal rearrangements.
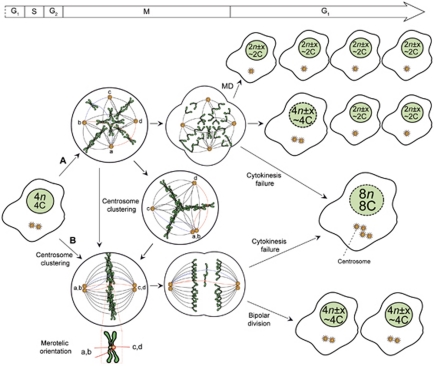
The most intuitive consequence of polyploidization is the dramatic increase in the complexity of the mitotic processes that manage and partition chromosomes (Figure 4). The high rates of segregation errors exhibited by newly formed tetraploids are due to the concomitant presence of supernumerary centrosomes (the microtubule organizing centers (MTOCs) of animal cells) and double chromosomes.63 Activated extra centrosomes can form supernumerary spindle poles, leading to a multipolar mitotic spindle and possibly to a multipolar division (MD). During MD, chromosomes are segregated in a multidirectional fashion and three or more daughter cells are generated (Figure 4).8 This process (which can be considered as a ‘reductional mitosis', as it actually reduces the genome of daughter cells) leads to the depolyploidization of tetraploid cells. An elegant study by Gisselsonn et al.64 clearly demonstrates that chromosomes are unequally segregated among daughter cells during MD, presumably constituting the major generator of aneuploidy and genomic instability in tetraploids.
Figure 4.
Aneuploidy generation by MD and centrosome clustering. In tetraploid cells, supernumerary centrosomes (here indicated as a–d) can drive MDs or can be clustered into two dominant poles (a, b and c, d) leading to a bipolar, yet error prone, division of the genetic material. Centrosome coalescence can be achieved directly or on a transient multipolar metaphase. (A) Most frequently, tetrapolar divisions result in the near-to-random segregation of chromosomes among typically unviable pseudodiploid (2n± × , ∼2C) daughter cells. However, as tetrapolar divisions (and MDs in general) are prone to mitotic defects, they can be aborted along one or more cleavage planes (cytokinesis failure), resulting in the generation of daughter cells with heterogeneous ploidy ranging from pseudodiploidy (2n± × , ∼2C) to octaploidy (8n, 8C) (B) Centrosome coalescence operates to limit MDs. Still, centrosome clustering is associated with a relatively high incidence of merotely (see insert), in turn favoring cytokinesis failure (and hence octoploidy 8n, 8C) or pseudotetraploidy (4n± × , ∼4C). In this scheme, dashed nuclei can be single o multiple, depending on the exact mechanism by which they were generated. Additional details in text. C, DNA content of a haploid nucleus; KT, kinetochore; MT, microtubule; n=number of chromosomes in the haploid set (23 for humans)
Polyploidy also complicates the regulation of the chromosome and centrosome cycles. In particular, polyploidy can compromise the coordination between DNA replication and centrosome duplication (both of which occur only once in a normal cell cycle), leading to the continuous emergence of chromosome and centrosome abnormalities, increased aberrant divisions and impaired inheritance of the genetic material.65
MDs are not the exclusive source of aneuploidy in polyploids. Tetraploids are known for their high propensity to lose one or more chromosomes and to accumulate structural chromosome aberrations. The levels of the components of the mitotic machinery do not necessarily increase with ploidy, implying that the interaction between these proteins may be compromised or imbalanced, leading to alterations in the geometry of the mitotic spindle.8 In budding yeast, tetraploidization is neither accompanied by changes in KT size nor by increased spindle length, leading to potential spindle irregularities.62
Alterations of the spindle geometry may lead to improper MT-KT attachments. For instance, in yeast, tetraploidy increases the frequency of syntelic chromosome attachments (a type of misattachment in which both sister KTs bind MTs that emanate from the same spindle pole),66 conferring a high level of genetic instability.62 In mammals, syntelic attachments efficiently trigger the SAC, causing a prolonged mitotic arrest that allows for error correction.66 Human tetraploid cancer cells usually possess intact and functional SAC,67 meaning that syntelic attachments are normally detected/corrected before the onset of anaphase to ensure faithful chromosome segregation. Thus, sintely does not represent a major source of aneuploidy/genomic instability in tetraploid cells. Tetraploids spontaneously activate the SAC more frequently than their diploid counterparts, because of a higher frequency of aberrant mitoses. Accordingly, inhibition/depletion of SAC components preferentially kills tetraploids.67
The SAC does not prevent mitotic progression in a permanent fashion, and can be circumvented though a poorly characterized process by which cells progress from metaphase to anaphase in spite of continuous SAC activation.17 In this setting, cells divide without correcting spindle and attachment errors, representing a potential mechanism of aneuploidization. Interestingly, tetraploid cells seem more prone to circumvent the SAC than diploids, as suggested by the localization of BUB1B on anaphase-lagging KTs (IV, unpublished observations).
In the presence of merotelic chromosome attachments (i.e., when a single KT binds MTs from both spindle poles)66 (Figure 4), chromosomes may align correctly to the metaphase plate.66 As all chromosomes are attached and under tension, the SAC is poorly sensitive to this scenario, which potentially results in aneuploidy.66 Importantly, multipolar spindles favor merotelic attachments68 and tetraploids have an intrinsic propensity to undergo MDs owing to supernumerary centrosomes. Thus, by favoring merotelic attachments, extra centrosomes may constitute an important source of aneuploidy/genomic instability for polyploids (Figure 4).
In conclusion, tetraploids (which are intrinsically hindered in the correct segregation of their genome) can generate an aneuploid offspring by two non-mutually exclusive mechanisms (Figure 4).
Multipolar mitoses induce the depolyploidization of tetraploid tumor cells
Supernumerary centrosome-driven MDs are common in malignant lesions and have been suspected to contribute to oncogenesis for more than a century.63 Two other histological features of malignant tissues, the heterogeneity in cell (‘anisocytosis') and nuclear (‘anisokaryosis') size, might also result from asymmetric cell divisions.63 Extra centrosomes have been detected in pre-invasive carcinomas of the cervix, breast and prostate, often correlating with aneuploidy, CIN and aggressiveness.65 Moreover, experimental induction of supernumerary centrosomes suffices to trigger oncogenesis.69 Thus, centrosome amplification is not a mere consequence of cancer, but frequently constitutes an oncogenic event. Still, RNAi-mediated depletion of the oncoprotein BCL3 has also been associated with centrosome amplification.70 This suggests that, in some instances, the amplification of centrosomes may constitute an epiphenomenon of oncogene/oncosuppressor gene deregulation.
Supernumerary centrosomes directly arise from polyploidization. Alternatively, centrosomes can be amplified on overduplication or centriole neogeneration.65 AURKA overexpression provokes both tetraploidy and centrosome amplification.71 Similarly, tetraploids that accumulate in the absence of TP53 contain supernumerary centrosomes.72 It remains unclear whether the absence of TP53 is permissive for the generation/survival of tetraploids with extra centrosomes or whether TP53 (which also localizes to centrosomes) directly influences the centrosome cycle.73 In tetraploid cancer cells, MDs require the absence of TP53 as well as the activation of MOS,72 an oncogenic kinase that is critical for oocyte meiosis. MOS favors aneuploidization via the depolyploidization of tetraploids,72 in line with the finding that, in lung cancer patients, MOS overexpression correlates with TP53 alterations, aneuploidy, anisocytosis, anisokaryosis and tumor aggressiveness.74 How TP53 and MOS functionally interact is obscure. As a possibility, the absence of TP53 may facilitate MOS upregulation. Alternatively, the absence of TP53 may be required for tolerating MOS activation-driven oncogenic stress, which would lead to proliferative arrest and cell death in TP53-proficient cells.
Thus, supernumerary centrosomes and whole-genome duplication might initiate tumorigenesis via a three-step mechanism (Figure 5): (i) extra centrosomes accumulate on tetraploidization; (ii) polyploids harboring supernumerary centrosomes bypass mitotic checkpoints and (iii) extra centrosomes of proliferating polyploids promote MDs, leading to cancer-associated aneuploidy.
Figure 5.
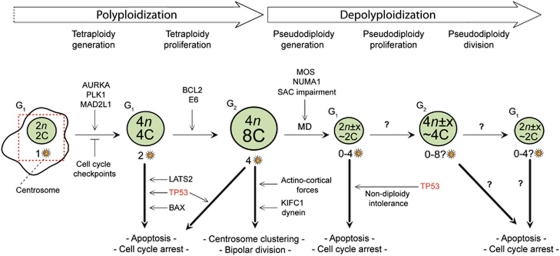
Regulation of the ‘polyploidization–depolyploidization' cascade. Diploid cells (2n, 2C) can become tetraploid (4n, 4C) as a consequence of endoreplication, mitotic errors and cell-to-cell fusion (see also Figure 2), a phenomenon that is favored by some oncogenes (e.g., aurora kinase A, AURKA) and prevented by many cell cycle checkpoints including the SAC. Freshly generated tetraploids normally undergo apoptosis or are permanently blocked in the G1 phase of the cell cycle, due to the activity of tumor suppressors including LATS2, TP53 and BAX. When such a tetraploidy control is deficient (for instance owing to the overexpression of the BAX antagonist BCL-2, when TP53 is inactivated by genetic or epigenetic events or in the presence of the viral oncoprotein E6), tetraploids illicitly proliferate and reach the G2 phase of the cell cycle. Depolyploidization most often occurs via MD, which is prevented by centrosome coalescence (involving action-cortical forces and motor protein like KIFC1). Alternatively, TP53 mediates the apoptotic elimination of tetraploid cells that might have illicitly reached the G2 phase of the cell cycle. Some oncogenes (e.g., MOS, NUMA1), dysfunctions of the SAC and/or of the TP53 system all favor the overcoming of centrosome coalescence, MD and hence depolyploidization, leading to the generation of aneuploid, most often pseudodiploid (2n± × , ∼2C), cells, which may contain 0–4 centrosomes. These aneuploid cells are normally unviable either because of the absence of centrosomes or to nullisomies and/or polysomies that are incompatible with life. Moreover, the proliferation of aneuploid cells is actively blocked by TP53, giving rise to the so-called ‘non-diploidy intolerance'. In rare instances (for instance when the TP53 system is inactive), pseudodiploids may bypass the apoptotic control and proliferate. Frequently, these cells display an increased tumorigenic potential and hence, activity support oncogenesis. Thicker arrows represent the most common events in the ‘polyploidization–depolyploidization' cascade. Additional details in text. C, DNA content of a haploid nucleus; KT, kinetochore; MT, microtubule; n=number of chromosomes in the haploid set (23 for humans)
Limiting MD
Several mechanisms normally prevent MD in the presence of extra centrosomes. Multipolar mitotic spindles can undergo bipolarization before the onset of anaphase, a mechanism named ‘centrosome clustering', ‘aggregation' or ‘coalescence' (Figure 4). Through coalescence, supernumerary centrosomes are clustered into two dominant poles, (at least theoretically) allowing for bipolar genome segregation.75, 76 Bipolar mitoses resulting from centrosome clustering are prone to segregation errors owing to incorrect, often merotelic, MT-KT attachments (Figure 4).68
Although in normal bipolar spindles, the merotelic orientation is thermodynamically unfavorable because of the geometry of sister KTs, in the presence of multipolar spindles, the probability that MTs enucleated from two spindle poles capture a single KT is increased.66 As merotelic attachments are poorly detected by the SAC, on centrosome clustering and the onset of a bipolar anaphase, merotelically attached chromatids segregate to the pole attached to a thicker MT bundle. However, if the tension generated by opposite bundles is equal, merotelically attached chromosomes persist at the spindle equator (so-called ‘anaphase-lagging chromosomes') until (i) they are randomly forced into one daughter cell by the ingression of the cleavage furrow66 or (ii) they are trapped in the furrow, resulting in cytokinesis failure (Figure 4).18
The machinery for correcting merotely is often deregulated in cancer cells.66 Thus, anaphase-lagging chromosomes may drive chromosome missegregation and induce aneuploidy through a mechanism that is dependent on extra centrosomes but MD-independent (Figure 4). Accordingly, merotely- and anaphase-lagging chromosomes are common in human cancer cell lines, especially in those that display high CIN levels.68
Centrosome extrusion and inactivation also ensure bipolar mitosis in the presence of supernumerary centrosomes. Centrosome extrusion involves the active expulsion of extra centrosomes within newly generated cytoplasts.75 During centrosome inactivation, supernumerary centrosomes cannot interact with MTs and hence, cannot act as MTOCs.69
The understanding of the molecular mechanisms accounting for centrosome clustering, extrusion and inactivation is partial. Centrosome coalescence relies on microtubule-associated proteins including ASPM,77 CKAP578 and NUMA1,76 as well as on motor proteins like dynein76 and KIFC1/HSET,69, 77 which contribute to mitotic spindle organization and centrosome positioning. Motor proteins crosslink MTs anchored at distinct centrosomes to generate the forces that are responsible for centrosome aggregation.79 Components of the chromosomal passenger complex like AURKB, BIRC5, CDCA8 and INCENP, proteins involved in the MT-KT attachment (e.g., NCD80 complex) and factors that ensure the cohesion between sister chromatids (e.g., CDCA5, SGOL1) also participate in centrosome aggregation.78 Thus, centrosome coalescence is a complex process whose regulation is intertwined with mitotic checkpoints and cell polarity/adhesion.77
MDs reportedly take longer than normal bipolar divisions,69 suggesting that MDs spontaneously activate SAC, in turn delaying anaphase onset and favoring centrosome coalescence. Moreover, the success of MDs is influenced by the interplay between spindle-intrinsic forces that operate on centrosomes and actin-dependent cortical forces operating on astral MTs and centrosomes.77 In this context, the poly(ADP-ribose) polymerizing enzyme tankyrase 1 has been suggested to provide the matrix for the anchorage of MT motors to MTs, whereas several proteins involved in the interaction between astral MTs and actin (including CLIP-170, dynein and myosin 10) would be responsible for the cortical forces that influence centrosome positioning.77 The distribution of such forces might also be influenced by the cell adhesion pattern, pulling the centrosomes toward the site of strongest adherence and influencing spindle polarity.77 Intriguing similarities between meiotic spindle poles (which lack centrosomes) and centrosome clustering led to the hypothesis that proteins involved in the generation/regulation of the forces that bundle MTs in acentrosomal spindles may also be implicated in centrosome aggregation.77
MOS depletion results in the conversion of MDs into bipolar mitoses, presumably via centrosome coalescence.72 Thus, MOS exerts oncogenic functions (at least partially) by inhibiting centrosome aggregation. Intriguingly, MOS can exacerbate CIN by favoring multipolar/reductional cell divisions (complete inhibition of centrosome clustering) or by increasing the incidence of merotelic attachments in (centrosome-clustered) bipolar mitoses (partial inhibition of centrosome clustering). The downstream target(s) of MOS have not been characterized yet. As a possibility, MOS may directly regulate centrosome separation.72 Alternatively, MOS may influence centrosome coalescence indirectly by acting on proteins that control the position of supernumerary centrosomes or by impairing SAC function.
Fate of pseudodiploid cells derived from MD
Time-lapse microscopic study has documented that cells generated by MD can undergo different fates. Most frequently, multipolar anaphases finish and produce three or more daughter cells (complete MD). Occasionally, MDs abort on cytokinesis failure along one or all division planes, for instance, due to incorrect MT-KT orientations or chromatin trapping in the cleavage furrow (Figure 4). For the same reason, higher-order multipolar spindles (containing more than four poles) normally fail to elicit cytokinesis and lead to the generation of binucleated cells or cells that contain one large nucleus.73
In most cases, the progeny of complete MDs is unviable,68, 72 presumably because of the lack of essential genes. Nullisomy (the absence of one specific chromosome) and polysomy (the presence of extra chromosome copies) are near-to-always lethal because they result in major defects in the assembly of multiprotein complexes and fatal linkage disequilibria. Thus, the standard fate of aneuploid cells generated from MD is to die during interphase or after one additional round of mitosis. Sometimes, these cells can proliferate very slowly, and are hence, outcompeted by their normal counterparts (Figure 4). In most cases, as aneuploidy compromises cell fitness, newly generated aneuploidy cells are not tolerated by the organism and undergo ‘non-diploid intolerance'.80 Some insights on the molecular cascade accounting for this phenomenon have recently been obtained. In particular, it appears that the intolerance for non-diploid genomes is regulated by a TP53-dependent mechanism involving CDKN1A.81 Accordingly, unstable TP53−/− tetraploids (which incur in progressive depolyploidization) generate tumors in vivo more rapidly than stable ones.72 Once formed, such tumors display an elevated rate of aneuploidy,72 suggesting that MDs have occurred and that some extent of ‘non-diploidy tolerance' has been acquired, in vivo. Thus, the absence of TP53 and tetraploidization cooperatively accelerate tumor progression by facilitating the generation (because of increased MD rates) and/or the survival (because of the acquisition of ‘non-diploid tolerance') of an aneuploid cancer cells.
In conclusion, TP53 preserves genomic stability by regulating the ‘diploidy–tetraploidy–pseudodiploidy' cascade at different levels (Figure 5): (i) it limits the generation/survival of tetraploid cells, (ii) it inhibits the depolyploidization of illicitly generated tetraploids and (iii) it restricts the proliferation of the rare pseudodiploid cells escaped from previous checkpoints.
Concluding remarks
Illicitly generated from healthy diploids, tetraploid cells can spontaneously revert to potentially tumorigenic aneuploids. Multiple mechanisms inhibit the ‘polyploidization–depolyploidization' cascade. First, multiple cell cycle checkpoints avoid the entry into/progression through mitosis of diploids showing unreplicated/damaged DNA or mitotic dysfunctions (limitation of tetraploid generation). Second, distinct cell cycle checkpoints block the proliferation of illicitly generated tetraploids (limitation of tetraploid proliferation). Third, most tetraploid cells spontaneously undergo apoptosis (limitation of tetraploid survival). Fourth, centrosome clustering (and perhaps other mechanisms) restricts the genomic instability of illicitly proliferating tetraploids (limitation of aneuploid generation). Fifth, hitherto poorly characterized pathways intercept illicitly generated, proliferating aneuploid cells (limitation of aneuploid survival/proliferation). Multiple oncogenes and oncosuppressor genes intervene in one or more of these checkpoints. The full comprehension of the ‘diploidy–tetraploidy–aneuploidy' cascade will yield cardinal insights into the mechanisms of oncogenesis and tumor progression and hence, generate ample opportunities for cancer prevention or therapy.
Acknowledgments
We thank Dr. Lugli Enrico (NIH, Bethesda) for fruitful discussions. GK is supported by the Ligue Nationale contre le Cancer (Equipes labellisée), Agence Nationale pour la Recherche (ANR), European Commission (Active p53, Apo-Sys, ChemoRes, ApopTrain), Fondation pour la Recherche Médicale (FRM), Institut National du Cancer (INCa) and Cancéropôle Ile-de-France. LG receives a post-doctoral fellowship from Apo-Sys. IV is supported by the Ligue Nationale contre le Cancer, LS by FRM.
Glossary
- AURK
aurora kinase
- CDKN
cyclin-dependent kinase inhibitor
- KT
kinetochore
- MD
multipolar division
- MEFs
murine embryonic fibroblasts
- MT
microtubule
- MTOC
microtubule organizing center
- RNAi
RNA interference
- PLK1
polo-like kinase 1
- SAC
spindle assembly checkpoint
- RB
retinoblastoma
The authors declare no conflict of interest.
Footnotes
Edited by RA Knight
References
- Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybina TG, Zybina EV. Cell reproduction and genome multiplication in the proliferative and invasive trophoblast cell populations of mammalian placenta. Cell Biol Int. 2005;29:1071–1083. doi: 10.1016/j.cellbi.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Morillo SM, Escoll P, de la Hera A, Frade JM. Somatic tetraploidy in specific chick retinal ganglion cells induced by nerve growth factor. Proc Natl Acad Sci USA. 2010;107:109–114. doi: 10.1073/pnas.0906121107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid K, Lu J, Zimmet JM, Jones MR. Roads to polyploidy: the megakaryocyte example. J Cell Physiol. 2002;190:7–20. doi: 10.1002/jcp.10035. [DOI] [PubMed] [Google Scholar]
- Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, Al-Dhalimy M, et al. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–901. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Ogle BM, Cascalho M, Platt JL. Biological implications of cell fusion. Nat Rev Mol Cell Biol. 2005;6:567–575. doi: 10.1038/nrm1678. [DOI] [PubMed] [Google Scholar]
- Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: when it takes more to make one. Dev Biol. 2010;341:66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helming L, Tomasello E, Kyriakides TR, Martinez FO, Takai T, Gordon S, et al. Essential role of DAP12 signaling in macrophage programming into a fusion-competent state. Sci Signal. 2008;1:ra11. doi: 10.1126/scisignal.1159665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson CB, Youssef S, Koleckar K, Holbrook C, Doyonnas R, Corbel SY, et al. Extensive fusion of haematopoietic cells with Purkinje neurons in response to chronic inflammation. Nat Cell Biol. 2008;10:575–583. doi: 10.1038/ncb1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren JM, Liuba K, Breitbach M, Stott S, Thoren L, Roell W, et al. Myeloid and lymphoid contribution to non-haematopoietic lineages through irradiation-induced heterotypic cell fusion. Nat Cell Biol. 2008;10:584–592. doi: 10.1038/ncb1721. [DOI] [PubMed] [Google Scholar]
- Duelli D, Lazebnik Y. Cell-to-cell fusion as a link between viruses and cancer. Nat Rev Cancer. 2007;7:968–976. doi: 10.1038/nrc2272. [DOI] [PubMed] [Google Scholar]
- Pawelek JM, Chakraborty AK. Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat Rev Cancer. 2008;8:377–386. doi: 10.1038/nrc2371. [DOI] [PubMed] [Google Scholar]
- Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- Ichijima Y, Yoshioka K, Yoshioka Y, Shinohe K, Fujimori H, Unno J, et al. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS One. 2010;5:e8821. doi: 10.1371/journal.pone.0008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- Wright WE, Hayflick L. Formation of anucleate and multinucleate cells in normal and SV 40 transformed WI-38 by cytochalasin B. Exp Cell Res. 1972;74:187–194. doi: 10.1016/0014-4827(72)90496-x. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a ‘tetraploidy checkpoint'. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Coquelle A, Vivet S, Vitale I, Kauffmann A, Dessen P, et al. Apoptosis regulation in tetraploid cancer cells. EMBO J. 2006;25:2584–2595. doi: 10.1038/sj.emboj.7601127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Boise LH, Thompson CB. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- Dalton WB, Yu B, Yang VW. p53 suppresses structural chromosome instability after mitotic arrest in human cells. Oncogene. 2010;29:1929–1940. doi: 10.1038/onc.2009.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Kienitz A, Hofmann I, Muller R, Bastians H. Crosstalk of the mitotic spindle assembly checkpoint with p53 to prevent polyploidy. Oncogene. 2004;23:6845–6853. doi: 10.1038/sj.onc.1207860. [DOI] [PubMed] [Google Scholar]
- Huang YF, Chang MD, Shieh SY. TTK/hMps1 mediates the p53-dependent postmitotic checkpoint by phosphorylating p53 at Thr18. Mol Cell Biol. 2009;29:2935–2944. doi: 10.1128/MCB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, F M.2010Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer . ( http://cgap.nci.nih.gov/Chromosomes/Mitelman ).
- Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining ‘chromosomal instability'. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Li X, Sanchez CA, Paulson TG, Blount PL, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004;64:7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68:9909–9917. doi: 10.1158/0008-5472.CAN-08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duelli DM, Padilla-Nash HM, Berman D, Murphy KM, Ried T, Lazebnik Y. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr Biol. 2007;17:431–437. doi: 10.1016/j.cub.2007.01.049. [DOI] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr Opin Genet Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhou YX, Qiao W, Tominaga Y, Ouchi M, Ouchi T, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–7158. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- Yang G, Chang B, Yang F, Guo X, Cai KQ, Xiao XS, et al. Aurora kinase A promotes ovarian tumorigenesis through dysregulation of the cell cycle and suppression of BRCA2. Clin Cancer Res. 2010;16:3171–3181. doi: 10.1158/1078-0432.CCR-09-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A, Morse HC, III, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- Nguyen HG, Makitalo M, Yang D, Chinnappan D, St Hilaire C, Ravid K. Deregulated Aurora-B induced tetraploidy promotes tumorigenesis. FASEB J. 2009;23:2741–2748. doi: 10.1096/fj.09-130963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang X, Yang Y, Li D, Ren H, Zhu Q, et al. Ectopic expression of the microtubule-dependent motor protein Eg5 promotes pancreatic tumourigenesis. J Pathol. 2010;221:221–228. doi: 10.1002/path.2706. [DOI] [PubMed] [Google Scholar]
- Rao CV, Yang YM, Swamy MV, Liu T, Fang Y, Mahmood R, et al. Colonic tumorigenesis in BubR1+/−ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci USA. 2005;102:4365–4370. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, et al. Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci USA. 2010;107:6888–6893. doi: 10.1073/pnas.0910941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishler T, Li YY, Wang RH, Kim HS, Sengupta K, Vassilopoulos A, et al. Genetic instability and mammary tumor formation in mice carrying mammary-specific disruption of Chk1 and p53. Oncogene. 2010;29:4007–4017. doi: 10.1038/onc.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell CM, Green RA, Kaplan KB. APC mutations lead to cytokinetic failures in vitro and tetraploid genotypes in Min mice. J Cell Biol. 2007;178:1109–1120. doi: 10.1083/jcb.200703186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke I. Cytoskeleton out of the cupboard: colon cancer and cytoskeletal changes induced by loss of APC. Nat Rev Cancer. 2006;6:967–974. doi: 10.1038/nrc2010. [DOI] [PubMed] [Google Scholar]
- Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- Finkin S, Aylon Y, Anzi S, Oren M, Shaulian E. Fbw7 regulates the activity of endoreduplication mediators and the p53 pathway to prevent drug-induced polyploidy. Oncogene. 2008;27:4411–4421. doi: 10.1038/onc.2008.77. [DOI] [PubMed] [Google Scholar]
- Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, et al. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000;149:1073–1086. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S, Hirota T, Morisaki T, Marumoto T, Hara T, Kuninaka S, et al. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 2004;23:5266–5274. doi: 10.1038/sj.onc.1207623. [DOI] [PubMed] [Google Scholar]
- Schlegel BP, Starita LM, Parvin JD. Overexpression of a protein fragment of RNA helicase A causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene. 2003;22:983–991. doi: 10.1038/sj.onc.1206195. [DOI] [PubMed] [Google Scholar]
- Yin XY, Grove L, Datta NS, Long MW, Prochownik EV. C-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene. 1999;18:1177–1184. doi: 10.1038/sj.onc.1202410. [DOI] [PubMed] [Google Scholar]
- Incassati A, Patel D, McCance DJ. Induction of tetraploidy through loss of p53 and upregulation of Plk1 by human papillomavirus type-16 E6. Oncogene. 2006;25:2444–2451. doi: 10.1038/sj.onc.1209276. [DOI] [PubMed] [Google Scholar]
- Roh M, Franco OE, Hayward SW, van der Meer R, Abdulkadir SA. A role for polyploidy in the tumorigenicity of Pim-1-expressing human prostate and mammary epithelial cells. PLoS One. 2008;3:e2572. doi: 10.1371/journal.pone.0002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man C, Rosa J, Yip YL, Cheung AL, Kwong YL, Doxsey SJ, et al. Id1 overexpression induces tetraploidization and multiple abnormal mitotic phenotypes by modulating aurora A. Mol Biol Cell. 2008;19:2389–2401. doi: 10.1091/mbc.E07-09-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe PH, Gonzalez-Barrera S, Rothstein R. More is not always better: the genetic constraints of polyploidy. Trends Genet. 2007;23:263–266. doi: 10.1016/j.tig.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Storchova Z, Breneman A, Cande J, Dunn J, Burbank K, O'Toole E, et al. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Hakanson U, Stoller P, Marti D, Jin Y, Rosengren AH, et al. When the genome plays dice: circumvention of the spindle assembly checkpoint and near-random chromosome segregation in multipolar cancer cell mitoses. PLoS One. 2008;3:e1871. doi: 10.1371/journal.pone.0001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyss D, Gergely F. Centrosome function in cancer: guilty or innocent. Trends Cell Biol. 2009;19:334–346. doi: 10.1016/j.tcb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Vitale I, Galluzzi L, Vivet S, Nanty L, Dessen P, Senovilla L, et al. Inhibition of Chk1 kills tetraploid tumor cells through a p53-dependent pathway. PLoS One. 2007;2:e1337. doi: 10.1371/journal.pone.0001337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora R, Espinosa M, Ceballos-Cancino G, Segura B, Maldonado V, Melendez-Zajgla J. Depletion of the oncoprotein Bcl-3 induces centrosome amplification and aneuploidy in cancer cells. Mol Cancer. 2010;9:223. doi: 10.1186/1476-4598-9-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Jemaa M, Michaud M, Galluzzi L, Kepp O, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–1284. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K. P53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim Biophys Acta. 2008;1786:15–23. doi: 10.1016/j.bbcan.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Zacharatos P, Mariatos G, Liloglou T, Kokotas S, Kastrinakis N, et al. Deregulated expression of c-mos in non-small cell lung carcinomas: relationship with p53 status, genomic instability, and tumor kinetics. Cancer Res. 2001;61:538–549. [PubMed] [Google Scholar]
- Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B, Maier B, Fuchs F, Chi J, Riffel P, Anderhub S, et al. Proteins required for centrosome clustering in cancer cells. Sci Transl Med. 2010;2:33ra38. doi: 10.1126/scitranslmed.3000915. [DOI] [PubMed] [Google Scholar]
- Acilan C, Saunders WS. A tale of too many centrosomes. Cell. 2008;134:572–575. doi: 10.1016/j.cell.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]