Abstract
The pathological processes of neurodegenerative disorders such as Alzheimer's and Parkinson's diseases engender synaptic and neuronal cell damage. While mild oxidative and nitrosative (nitric oxide (NO)-related) stress mediates normal neuronal signaling, excessive accumulation of these free radicals is linked to neuronal cell injury or death. In neurons, N-methyl--aspartate (NMDA) receptor (NMDAR) activation and subsequent Ca2+ influx can induce the generation of NO via neuronal NO synthase. Emerging evidence has demonstrated that S-nitrosylation, representing covalent reaction of an NO group with a critical protein thiol, mediates the vast majority of NO signaling. Analogous to phosphorylation and other posttranslational modifications, S-nitrosylation can regulate the biological activity of many proteins. Here, we discuss recent studies that implicate neuropathogenic roles of S-nitrosylation in protein misfolding, mitochondrial dysfunction, synaptic injury, and eventual neuronal loss. Among a growing number of S-nitrosylated proteins that contribute to disease pathogenesis, in this review we focus on S-nitrosylated protein-disulfide isomerase (forming SNO-PDI) and dynamin-related protein 1 (forming SNO-Drp1). Furthermore, we describe drugs, such as memantine and newer derivatives of this compound that can prevent both hyperactivation of extrasynaptic NMDARs as well as downstream pathways that lead to nitrosative stress, synaptic damage, and neuronal loss.
Keywords: NMDA receptor, S-nitrosylation, misfolded protein, mitochondrial dysfunction, neurodegeneration
Excessive generation of reactive oxygen and nitrogen species (ROS/RNS), including superoxide anion (O2•−) and nitric oxide (NO•), contributes to neuronal cell injury and death in neurodegenerative diseases.1, 2, 3 Under basal conditions, mild activation of synaptic N-methyl--aspartate (NMDA)-type glutamate receptors (NMDARs) can result in physiological ROS and RNS production, which mediate normal signaling to support neuronal function and survival. However, under neurodegenerative conditions, overactivation of extrasynaptic NMDARs causes excessive influx of Ca2+ ions, generating neurotoxic levels of ROS and RNS (Figure 1).4 For instance, upon hyperactivation of NMDARs, NADPH oxidase (NOX) and mitochondrial respiration produce free radicals, principally superoxide O2•−.5 Increased Ca2+ influx also activates neuronal NO synthase (nNOS) to generate NO. In addition to nNOS, inducible NOS (iNOS) in glial cells can produce significant amounts of NO independent of NMDAR activation. Initially, the biological action of NO was thought to be mediated principally via guanylate cyclase activation and cyclic guanosine-3′,5′-monophosphate (cGMP) production. However, beginning with our own work in this area, over the past decade and a half, S-nitrosylation, a covalent reaction of an NO group with a reactive cysteine thiol on target proteins, has emerged as the principal mechanism exerting NO bioactivity.6 The formation of S-nitrosoproteins (SNO-Ps) generally regulates protein function either allosterically or by direct modification of an active site cysteine.6, 7 Collaboration between the groups of Stuart Lipton and Jonathan Stamler initially discovered and characterized this biochemical process on the NMDAR, showing that NO inhibits excessive NMDAR activity via S-nitrosylation.7 Currently, nearly a thousand proteins have been identified as SNO-Ps,8 supporting the notion that NO exerts most of its biological activity via S-nitrosylation, although the specific function of most of SNO-Ps merits further investigation (Figure 2). Analogous to phosphorylation, Lipton and Stamler coined the term ‘S-nitrosylation,' indicating a biological effect of the chemical reaction of S-nitrosation.9 Importantly, S-nitrosylation can mediate either protective or neurotoxic effects depending on the action of the target protein. Moreover, we have very recently shown that transnitrosylation (transfer of the NO group) between proteins appears to be a prominent mechanism of NO signaling that is just emerging in importance.
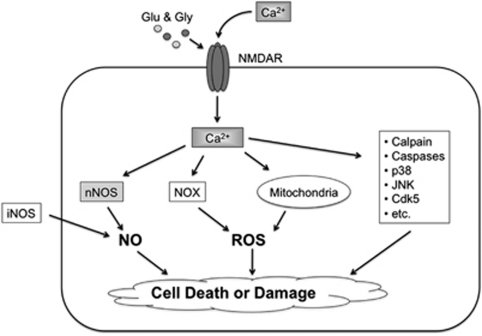
Figure 1.
Excessive stimulation of NMDARs triggers intracellular signaling pathways leading to synaptic damage and neuronal cell loss. Hyperactivation of NMDARs by glutamate (Glu) and glycine (Gly) induces excessive Ca2+ influx and activation of nNOS leading to NO production. Ca2+ signaling also promotes ROS generation via activation of NADPH oxidase and dysfunctional mitochondria. iNOS, which is predominantly present in astrocytes, can produce toxic levels of NO, representing NMDAR-independent NO synthesis. In addition to producing RNS and ROS, NMDAR/Ca2+ signaling influences the activity of many other proteins (examples are listed at right), contributing to synaptic dysfunction and neuronal death
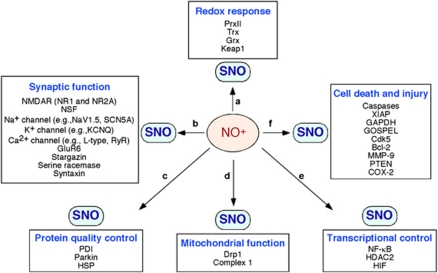
Figure 2.
Representative examples of S-nitrosylated proteins that can regulate neuronal function. NO generated in the nervous system modulates the activity of various proteins via S-nitrosylation (or further oxidation), controlling (a) redox responses, (b) synaptic function, (c) protein quality control, (d) mitochondrial function, (e) transcriptional regulation, and (f) cell death and injury. Depending on the levels of NO, S-nitrosylation of these proteins can mediate either neuroprotective or neurodestructive pathways
In recent work, we and others have shown that S-nitrosylation and subsequent further oxidation of critical cysteine residues can lead to protein misfolding and mitochondrial dysfunction, both of which are characteristic features of neurodegenerative diseases. Misfolded proteins form aggregates in many neurodegenerative diseases, and soluble oligomers of these aberrantly folded proteins are thought to adversely affect cell function by interfering with normal cellular processes or initiating cell death signaling pathways.10 As examples, α-synuclein and synphilin-1 in Parkinson's disease (PD), and amyloid-β (Aβ) and tau in Alzheimer's disease (AD) form toxic oligomers of non-native secondary structures. Interestingly, many lines of evidence suggest that the formation of larger aggregates may be an attempt of the cell to wall off these toxic proteins.11 In addition, neurodegenerative brains often manifest mitochondrial dysfunction, including accumulation of mutations in mitochondrial DNA, increased generation of ROS, and decreased respiration activity. These mitochondrial abnormalities may be engendered from defects in mitochondrial dynamics, which is controlled by fission and fusion of mitochondria. In this review, we discuss specific examples highlighting (i) the critical role of S-nitrosylation of an endoplasmic reticulum (ER) chaperone, protein-disulfide isomerase (PDI), in accumulation of misfolded proteins in neurodegenerative diseases (Figure 3),12 and (ii) the contribution of S-nitrosylated dynamin-related protein 1 (Drp1) to the pathological fragmentation of mitochondria and consequent loss of synapses in neurodegenerative disorders (Figure 4).13
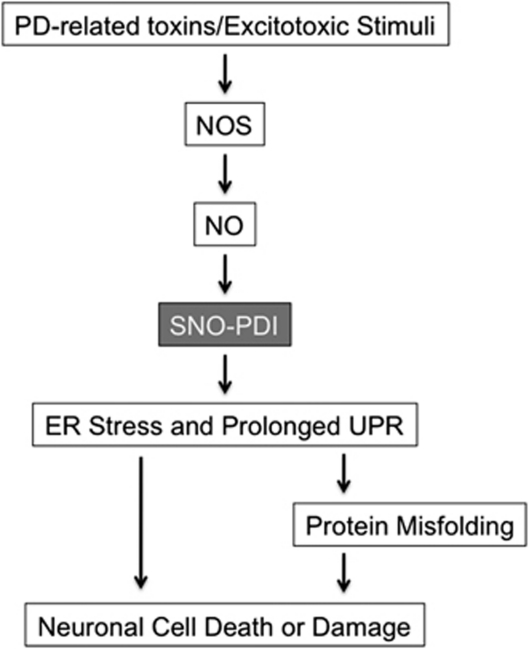
Figure 3.
Possible signaling pathways whereby SNO-PDI contributes to protein misfolding and neuronal damage. In PD, mitochondrial dysfunction caused by pesticides, herbicides, or other environmental toxins can trigger NO and ROS production, possibly via mitochondrial pathways and the NMDAR/Ca2+ cascade.12, 31, 94, 95 NO produced by NOS reacts with sulfhydryl groups of PDI to form SNO-PDI, inhibiting its isomerase and chaperone activities. SNO-PDI formation causes ER stress and a prolonged UPR, and thereby contributes to neuronal cell injury, in part, by triggering accumulation of misfolded proteins
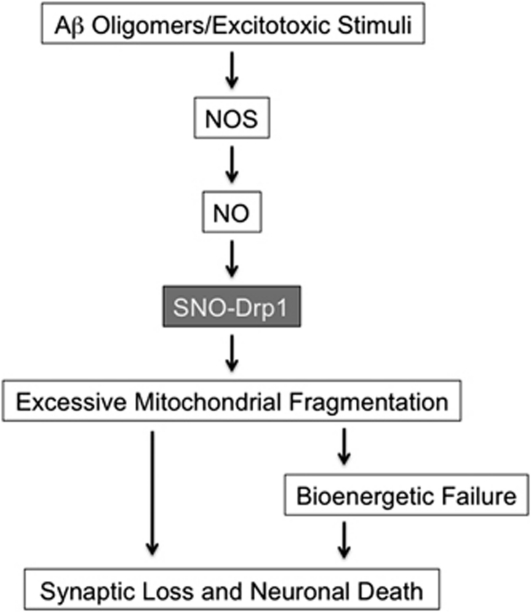
Figure 4.
Possible signaling pathways that S-nitrosylated Drp1 contribute to excessive mitochondrial fission and synaptic damage. Soluble oligomers of Aβ peptide, thought to be a key mediator in AD pathogenesis, can facilitate neuronal NO and ROS production in both NMDAR-dependent and -independent manners.96, 97, 98 S-Nitrosylation of Drp1 (forming SNO-Drp1) causes excessive mitochondrial fragmentation in neurodegenerative conditions. Compromised mitochondrial dynamics may result in the impairment of bioenergetics, and thus contribute to synaptic damage and eventually neuronal cell death
Generation of NO in Neuronal Cells
Three subtypes of NOS exist in the nervous system; the two constitutive forms of NOS – nNOS and endothelial NOS (eNOS) – take their names from the cell type in which they were first found, and can be activated by cell Ca2+. The name of the third subtype – iNOS – indicates that expression of the enzyme is induced by acute inflammatory stimuli. For example, activated glial cells may produce neurotoxic amounts of NO via iNOS expression in various neurodegenerative diseases. All three isoforms are widely distributed in the brain and possibly involved in the process of neurodegeneration.
In neurons, nNOS is not only enriched in postsynaptic structures but also localized in the cytoplasm. At synapses, nNOS and other intracellular signaling molecules are coupled to NMDARs via PSD-95 protein complexes.14 Upon glutamate stimulation of the NMDAR, Ca2+ enters into the cytoplasm through the receptor's ion channel. In conjunction with the Ca2+ binding protein, calmodulin, Ca2+ influx then triggers the activation of nNOS to generate NO from the amino acid -arginine (Figure 1).11, 14 Moderate levels of NO, produced under physiological conditions, trigger many normal intracellular signaling pathways. In contrast, overstimulation of NMDARs and resultant excessive Ca2+ influx promote pathological signaling, contributing to cell injury and death via production of toxic amounts of NO and other free radicals, as well as other enzymatic processes (Figure 1).2, 7, 15, 16, 17 Such hyperexcitation of NMDARs has a role in a variety of neurological disorders ranging from acute hypoxic-ischemic brain injury to chronic neurodegenerative diseases. Severe overstimulation of excitatory receptors can cause necrotic cell death, whereas less fulminant or chronic overstimulation can cause apoptotic or other forms of cell death.16, 17, 18 Consistent with this notion, disruption of either (a) the NMDAR/PSD-95 complex or (b) PSD-95/nNOS interaction decreases glutamate-induced neurotoxicity. These results suggest that the NMDAR/NO pathway potentially represents a therapeutic target for a number of neurodegenerative diseases.19, 20
In addition to synaptic NMDARs, extrasynaptic sites also contain functional NMDARs. In general, synaptic NMDAR activity signals to molecular pathways promoting neuronal survival, for example, by enhancing transcriptional expression of antioxidative enzymes.21 Conversely, excessive activation of extrasynaptic NMDAR mediates molecular pathways that trigger neurotoxicity associated with protein misfolding and mitochondrial dysfunction (Figure 5).22 Recently, extrasynaptic (in addition to synaptic) NMDARs were reported to be associated with PSD-95,23 consistent with the notion that nNOS/NO toxicity can have a role in cell death pathways triggered by extrasynaptic NMDARs.
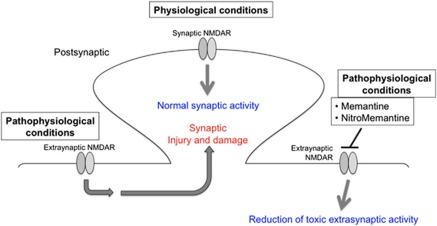
Figure 5.
‘UFO'-type neuroprotective drugs, like memantine and NitroMemantine, preferentially block extrasynaptic NMDARs. Physiological synaptic activity of the NMDAR is required for neurotransmission and neuronal cell survival. In contrast, excessive activation of extrasynaptic NMDARs induces synaptic injury and neuronal loss, and is often associated with the accumulation of misfolded proteins as well as mitochondrial dysfunction. Conventional NMDAR antagonists, such as MK-801, completely block all receptor activity, including physiological synaptic activity, and thus result in severe side effects and clinical intolerability. Memantine and the newer NitroMemantine drugs preferentially block excessive (pathological) extrasynaptic NMDA receptor activity, while relatively sparing normal (physiological) synaptic activity89, 90
S-Nitrosylation Signaling Pathways in the Nervous System
NO participates in cellular signaling pathways that regulate broad aspects of brain function, including synaptic plasticity, normal development, and neuronal cell death.15 These effects were thought to be largely achieved by activation of guanylate cyclase to form cGMP, but, as mentioned above, emerging evidence suggests that a more prominent reaction of NO is S-nitrosylation of regulatory protein thiol groups.4, 6, 7 S-Nitrosylation is the covalent addition of an NO group to a cysteine thiol/sulfhydryl (RSH or, more properly, thiolate anion, RS−) to form an S-nitrosothiol derivative (R-SNO). Such regulatory modifications are broadly found in mammalian, plant, and even in microbial proteins. One mechanism for the specificity of S-nitrosylation involves the presence of a SNO motif adjacent to the target cysteine. We and our colleagues initially found that a consensus motif of nucleophilic residues (generally an acid and a base) surrounds a critical cysteine, increasing the susceptibility of the sulfhydryl to S-nitrosylation.6 Recent studies, involving proteomics, bioinformatics, and structural analyses, have confirmed this initial finding by demonstrating that acidic and/or basic amino acids reside generally within 6–8 Å of the S-nitrosylated cysteine residue.24, 25 The process of SNO-P formation is counterbalanced by denitrosylation via enzymes such as thioredoxin/thioredoxin reductase, class III alcohol dehydrogenase, PDI, intracellular glutathione, or other mechanisms.26 Recently, we and others have reported an additional S-nitrosylation pathway that requires protein-protein transnitrosylation.27, 28, 29 Transnitrosylation reactions may propagate many NO-mediated cellular signaling pathways via production of either a denitrosylated protein (by donating an NO moiety to a second protein), or a SNO-P (by accepting an NO group from another protein), or both of these processes. As stated above, accumulating evidence now suggests that S-nitrosylation is analogous to phosphorylation in regulating the biological activity of many proteins.6, 7, 12, 30, 31, 32, 33 However, the chemistry of NO remains complex. NO is often a good ‘leaving group', resulting in further oxidation of the thiol to a disulfide bond between neighboring (vicinal) cysteine residues. Alternatively, as NO ‘leaves' for another reaction partner, the remaining thiol group can react with ROS to yield sulfenic (-SOH), sulfinic (-SO2H), or sulfonic (-SO3H) acid derivatives on the cysteine residue of the protein.12, 31, 34 S-Nitrosylation may also possibly produce a nitroxyl disulfide, in which the NO group is shared by proximate cysteine thiols.35
At low (physiological) levels, NO can mediate neuroprotective functions. For example, our group first identified the physiological relevance of S-nitrosylation by showing that NO reacts with the NMDAR to downregulate its excessive activity. Under excitotoxic conditions, S-nitrosylation of NMDARs can thus provide neuroprotective affects.7 Specifically, we found that NO can S-nitrosylate five different cysteine residues on extracellular domains of NR1 and NR2 subunits of the NMDAR, but cysteine residue no. 399 (Cys399) on the NR2A subunit mediates ≥90% of the NO's effect under most conditions.36 From crystal structure models and electrophysiological experiments, we further found that S-nitrosylation of Cys399 may induce a conformational change in the receptor protein that enhances glutamate and Zn2+ binding to the receptor. The enhanced binding of glutamate and Zn2+ in turn causes the receptor to desensitize and, consequently, the ion channel to close.37 Interestingly, we also found that the NMDAR becomes more sensitive to inhibition by S-nitrosylation, when oxygen tension is lowered to the levels observed in normal brain (10–20 Torr) as opposed to ambient air.38 In addition to the NMDAR, activity of other membrane proteins, including specific subtypes of potassium channels, sodium channels, and calcium channels, as well as ryanodine receptors, are regulated by S-nitrosylation (Figure 2).39, 40, 41, 42 In some cases, S-nitrosylation of these ion channels may contribute to neurodegenerative conditions.
In contrast, high levels of NO are thought to stimulate neurotoxic pathways. Increased NO rapidly reacts with superoxide anion, generated from both mitochondrial sources and non-mitochondrial sources (e.g. NOX), to form the very toxic product, peroxynitrite (ONOO−).5, 7 Moreover, we and other colleagues have also found that excessive (pathophysiological) production of oxidative/nitrosative stress contributes to neuronal cell death through S-nitrosylation of a number of targets, including MMP-9, cyclooxygenase-2, N-ethylmaleimide sensitive factor, and GAPDH (Figure 2).7, 32, 34, 43, 44 For instance, Solomon Snyder's group has extensively characterized the pathological consequence of SNO-GAPDH. S-Nitrosylation of GAPDH augments its binding to Siah, which possesses a nuclear localization signal, and thus precipitates the translocation of the GAPDH/Siah complex into the nucleus. In the nucleus, SNO-GAPDH influences many signaling molecules, such as p300/CBP, SIRT1, and HDAC2, to initiate apoptotic cascades.27, 32, 45 Interestingly, a cytosolic protein, GOSPEL, or the monoamine oxidase inhibitor R-(-)-deprenyl (deprenyl) appeared to exhibit neuroprotective effects at least in part by inhibiting the GAPDH-Siah interaction.46, 47 Collectively, these studies suggest a potential contribution of SNO-GAPDH-mediated neuronal cell death in neurodegenerative disease pathogenesis.
In addition, recent evidence suggests that the presence of excessive NO-related species may have a significant role in the mechanism of protein misfolding. However, until recently little was known regarding the underlying molecular and pathogenic events whereby NO contributes to the formation of aggregates such as amyloid plaques in AD or Lewy bodies in PD. We and others recently presented pathophysiological and chemical evidence that S-nitrosylation regulates the isomerase and chaperone activities of PDI,12 contributing to protein misfolding and neurotoxicity in models of neurodegenerative disorders.
Furthermore, increased nitrosative stress can elicit dysfunction of mitochondrial dynamics (fission and fusion events), resulting in the generation of excessive mitochondrial fragmentation.48, 49 Although the exact mechanism for this remains enigmatic, our recent findings have shed light on the molecular events underlying this relationship, particularly in AD. Specifically, we have recently discovered (patho) physiological and chemical evidence that S-nitrosylation increases the GTPase activity of the mitochondrial fission protein, Drp1, consequently precipitating mitochondrial fragmentation. We found that excessive mitochondrial fragmentation is triggered in this manner by oligomeric Aβ peptide, via increasing NO production, and results in bioenergetic impairment, synaptic damage, and eventually frank neuronal loss in models of AD.13
S-Nitrosylated PDI Mediates Protein Misfolding and Neurotoxicity in Models of Neurodegenerative Diseases
Healthy neurons generally do not accumulate protein aggregates, suggesting that the appearance of such structures is a response to pathological stresses. Considerable evidence suggests that misfolded or otherwise abnormal proteins are produced even in healthy cells. The difference can largely be accounted by cellular mechanisms for quality control, such as (1) molecular chaperones that promote proper folding of proteins and reduce toxic aggregation, (2) the ubiquitin-proteasome system (UPS) that targets misfolded proteins for degradation, (3) the unfolded protein response (UPR) that upregulates ER-resident chaperones to ameliorate the accumulation of unfolded proteins, and (4) lysosomal-autophagic degradation that is responsible for the removal of large protein complexes. For instance, a reduction in molecular chaperone or proteasome activity under pathological conditions can result in deposition and accumulation of aberrant proteins either within or outside of cells in the brain. In this manner, several mutations in molecular chaperones or UPS-associated enzymes are known to contribute to neurodegeneration.10 For example, a reduction in proteasome activity has been reported in the substantia nigra of PD patients,50 and overexpression of the molecular chaperone HSP70 prevented neurodegeneration in in vivo animal models of PD.51 Aggregated proteins were first considered to be pathogenic. Subsequent evidence suggested, however, that macroscopic aggregates are ‘an attempt' by the cell to sequester aberrant proteins, while, in contrast, soluble (micro-) oligomers of such proteins are the most likely toxic forms.11 Generally, the molecular quality control machinery of the cell can detoxify both large and small toxic aggregates; however, it is perhaps worth noting that excessive or persistent activation of quality control systems, especially the UPR and autophagy, may contribute to cell death, in part, via generation of ROS/RNS.
We recently reported that S-nitrosylation of PDI (forming SNO-PDI) disrupts normal protein folding and its neuroprotective role, particularly in PD conditions.12 In the ER, PDI normally facilitates proper protein folding by introducing disulfide bonds into proteins (oxidation), breaking disulfide bonds (reduction), and catalyzing thiol/disulfide exchange (isomerization), thus facilitating disulfide bond formation, rearrangement reactions, and structural stability of the mature protein.52 In this regard, PDI also has molecular chaperone activity capable of stabilizing the correct folding of substrate proteins. Increased expression of a PDI homologue, PDIp, in neuronal cells under the conditions mimicking PD, has suggested the possible contribution of PDIp to neuronal survival.53 In many neurodegenerative disorders and in cerebral ischemia, the accumulation of immature and denatured proteins results in ER dysfunction,53 and upregulation of PDI represents an adaptive response promoting protein refolding that may offer some degree of neuronal protection.53
We have recently reported that excessive NO, as well as rotenone exposure, which is known to lead to PD, can lead to S-nitrosylation of the active-site thiols of PDI, inhibiting its isomerase and chaperone activities.12 Formation of SNO-PDI led to accumulation of misfolded and polyubiquitinated proteins, resulted in prolonged UPR activation, and thus participated in persistent ER stress. Consequently, S-nitrosylation of PDI prevented the ability of PDI to attenuate neuronal cell death triggered by ER stress, resulted in misfolded proteins, and contributed to proteasome dysfunction. Also, we found that PDI was S-nitrosylated in the brains of virtually all cases of sporadic AD and PD that were available to us for study. Taken together, these results suggest that SNO-PDI can mediate protein misfolding and consequently neuronal cell injury or death (Figure 3).
Interestingly, when misfolding stress is very severe, despite its well-characterized neuroprotective function, PDI can trigger caspase-dependent neuronal apoptosis pathways.54 In a recent report, newly discovered inhibitors of PDI revealed that PDI accumulates at mitochondrial-associated ER membranes, increasing the formation of disulfide-linked Bak oligomers, thus promoting mitochondrial outer membrane permeabilization-mediated apoptotic signaling. This finding suggests that PDI is a bifunctional protein that can protect or enhance cell death depending on the cellular circumstances. For instance, PDI can provide a first line of defense by correcting protein misfolding, but under severe neurodegenerative conditions, PDI can contribute to neuronal cell death.54 An alternative view might be that PDI is S-nitrosylated under conditions of severe ER stress and thus it is the posttranslational modification of PDI that triggers its neurodestructive role. Further studies therefore are required to investigate whether SNO-PDI is localized in a mitochondrial fraction and thus effects this neurotoxic activity of PDI. To date, we have not found detectable levels of SNO-PDI in normal aged brain, but only in disease states, suggesting that this posttranslational modification may represent a therapeutically approachable target as it apparently occurs only in degenerative conditions.12, 30, 31 In summary, our findings suggest that S-nitrosylation of these and similar proteins may represent a key mechanism contributing to neurodegenerative conditions.
S-Nitrosylation of Drp1 Mediates Mitochondrial Fission and Neurotoxicity in Cell Models of AD
NO has also been reported to regulate mitochondrial function. Under physiological conditions, the NO-cGMP pathway induces mitochondrial biogenesis through peroxisome proliferator-activated receptor γ coactivator 1α.55 In contrast, increased nitrosative stress can result in defects in mitochondrial function. For example, S-nitrosylation affects mitochondrial respiration by reversibly inhibiting complexes I and IV,56, 57, 58 and formation of a SNO-complex I has been implicated in PD.59, 60 Mitochondria thus compromised will release ROS, and this in turn could contribute to brain aging and/or pathological conditions associated with neurodegenerative diseases.
Neurons are particularly vulnerable to mitochondrial defects because they require high levels of energy for maintenance of synapses, survival, and specialized functions. In models of PD, pesticides, herbicides, and other environmental toxins, such as MPTP and rotenone, specifically inhibit mitochondrial complex I, generating excessive ROS/RNS, and thus recapitulating many features of sporadic PD, including degeneration of dopaminergic neurons, overproduction and aggregation of α-synuclein, accumulation of Lewy body intraneuronal inclusions, and impairment of behavioral functions.1, 12, 30, 31, 61 Also, in AD models, accumulation of Aβ or tau proteins can induce deficits in the mitochondrial respiratory pathway.62
In healthy neurons, the physiological processes of mitochondrial fission and fusion can counteract mild mitochondrial defects. For example, mitochondrial biogenesis, an event that produces new mitochondria as a consequence of mitochondrial fission, can provide high concentrations of ATP at the synapse, preventing a loss of synaptic transmission and structure.63, 64, 65 The fission/fusion machinery proteins are known to maintain mitochondrial integrity and insure that new mitochondria are generated at critical locations. These proteins include Drp1 and Fis1, acting as fission proteins, and Mitofusin (Mfn) and Opa1, operating as fusion proteins.66 In both familial and sporadic neurodegenerative conditions, abnormal mitochondria regularly appears in the brain as a result of dysfunction in the fission/fusion machinery. For instance, genetic mutations in Mfn2, OPA1, or Drp1 disturb mitochondrial dynamics, causing neurological defects such as certain forms of peripheral neuropathy and optic atrophy.67, 68, 69, 70 These fission/fusion proteins are widely expressed in human tissues, but as defects preferentially affect nervous tissue, clearly neurons are particularly sensitive to mitochondrial dysfunction.
Additionally, analyses of autopsy and biopsy samples reveal that mitochondria isolated from sporadic AD brains exhibit diminished respiratory capacity,71 and AD neurons contain a number of mitochondria with fractured cristae.72, 73 Moreover, electron-microscopic studies have found an increase in mitochondrial fragmentation in human AD brains.74, 75 In cell-based models, Aβ production results in the appearance of fragmented and abnormally distributed mitochondria,48, 76 suggesting that Aβ (probably in the form of soluble oligomers) may trigger excessive mitochondrial fission in AD patients. Pathological forms of tau may also contribute to mitochondrial fragmentation in AD brains as expression of caspase-cleaved tau-induced mitochondrial fission in a calcineurin-dependent manner.77 Finally, it should be noted that dysfunction in mitochondrial dynamics has been associated with PD as well as AD, and these findings have been reviewed in detail elsewhere.78, 79
In several neurodegenerative diseases, in particular AD, synaptic damage in the hippocampus and cerebral cortex is thought to account for cognitive decline. Emerging evidence suggests that disruption of mitochondrial distribution and bioenergetics in synaptic structures, in part due to abnormal mitochondrial dynamics, can contribute to synaptic loss.80 Additionally, degenerating AD brains contain aberrant accumulations of misfolded, aggregated proteins – Aβ and tau – which can adversely affect neuronal connectivity and plasticity, and eventually trigger cell death signaling pathways leading to neurodegeneration. Recently, our group reported that S-nitrosylation of Drp1 at Cys644 mediates Aβ-induced disruption of mitochondrial dynamics, contributing to synaptic injury and neuronal damage, as described below.13
Drp1 includes four distinct structural domains: an N-terminal GTPase domain, a dynamin-like middle domain, an insert B domain, and a C-terminal GTPase effector domain (GED) domain. Cys644 resides within the GED of Drp1, which influences both GTPase activity and oligomer formation of Drp1.81, 82, 83 S-Nitrosylation of Drp1 (forming SNO-Drp1) induces formation of Drp1 dimers, which function as building blocks for tetramers and higher order structures of Drp1, and activates Drp1 GTPase activity; however, we found that mutation of Cys644 to Ala (C644A) abrogates these effects of NO.
We further demonstrated that exposure to oligomeric Aβ peptide results in formation of SNO-Drp1 in cell culture models. Moreover, we and our colleagues have observed that Drp1 is S-nitrosylated in the brains of virtually all cases of sporadic AD that we examined.13, 75 In order to determine the consequences of S-nitrosylated Drp1 in neurons, we exposed cultured cerebrocortical neurons to the physiological NO donor, S-nitrosocysteine, or to Aβ oligomers and found that both induced SNO-Drp1 formation and led to the accumulation of excessively fragmented mitochondria (Aβ was already known to induce the generation of NO in neurons). Finally, in response to Aβ, SNO-Drp1-induced mitochondrial fragmentation caused synaptic damage, an early characteristic feature of AD, and subsequently apoptotic neuronal cell death. Importantly, blockade of Drp1 nitrosylation (using the Drp1(C644A) mutant) prevented Aβ-mediated mitochondrial fission, synaptic loss, and neuronal cell death, suggesting that the posttranslational modification (S-nitrosylation) of Drp1 contributes to the pathogenesis of AD. Thus, SNO-Drp1 may represent a potential new therapeutic target for protecting neurons and their synapses in sporadic AD.
Multiple groups, including our own, have now reported S-nitrosylation and subsequent activation of dynamin family members, including Drp1.13, 75, 84, 85 In contrast, a single group has recently raised some concerns regarding the work, in particular related to the ability of S-nitrosylation to influence formation of dimers or higher order structures of Drp1 and to increase its GTPase activity.86 It should be noted, however, that in this contradictory report the authors used recombinant Drp1 that was already oxidized, and was thus already maximally dimerized or oligomerized.86 Such artifactual oxidation can occur, for example, if recombinant protein is prepared and kept in ambient air for a long period. Importantly, when a large portion of Drp1 is already oxidized and thus activated, it would prevent NO/S-nitrosylation from stimulating Drp1 activity. Under physiological conditions, NO can activate Drp1 via S-nitrosylation of cysteine 644 because Drp1 exists in a non-oxidized form (see Nakamura et al.87 for a detailed discussion on these topics). Also, the contrary group was able to show only a small increase in the SNO-Drp1 levels under AD conditions,86 despite the fact that at least two independent groups have encountered markedly elevated levels of SNO-Drp1 in AD brains.13, 75 This discrepancy is apparently due to the very high background observed in the control brains of the Bossy–Wetzel study. Conceivably, the use of an extremely large amount of brain lysate could account for such high background levels of nitrosylation under basal conditions, preventing them from detecting elevated SNO-Drp1 levels in AD brains.
Redox-Mediated Posttranslational Modifications may Mimic the Phenotype Caused by Rare Genetic Mutations Encoding Mitochondrial Fission and Fusion Proteins
Rare hereditary mutations in the genes encoding mitochondrial fission and fusion proteins can cause neurological diseases, including dominant optic atrophy and Charcot–Marie Tooth disease. Our recent studies as well as the work of other groups now suggests that posttranslational redox modification, such as S-nitrosylation or sulfonation, of these proteins can contribute to altered mitochondrial dynamics, protein misfolding, and neuronal dysfunction, in some sense mimicking or lowering the threshold for the effect of these rare genetic mutations. It is our feeling that this general mechanism may in fact represent a common etiology for sporadic cases of many neurodegenerative conditions, which represent the vast majority of affected patients.
Memantine Provides Neuroprotection Against NMDAR-Induced Excessive Ca2+ Influx and Oxidative/Nitrosative Stress in Cell-Based Models
Oxidative and nitrosative stress mediate, at least in part, glutamate-induced neuronal cell injury and death, and several antioxidant molecules have been reported to protect neurons against such assaults.18, 88 One mechanism that could potentially curtail excessive Ca2+ influx, and the resultant formation of neurotoxic ROS and RNS is inhibition of NMDAR hyperactivation. However, high-affinity NMDAR antagonists (e.g. MK-801) block virtually all NMDAR activity, including the physiological synaptic activity that is required for neuronal signaling and survival, and therefore are not clinically tolerated. In contrast, we have demonstrated that the adamantane derivative, memantine, preferentially blocks excessive (pathological/extrasynaptic) NMDAR activity while relatively sparing normal (physiological/synaptic) activity.89, 90 Memantine effectively blocks only excessively open channels through an uncompetitive mechanism of action in conjunction with a relatively fast off-rate, resulting in relatively low affinity for the NMDAR. Despite its low affinity for the NMDAR, memantine is still relatively selective for this receptor. As the action of an uncompetitive antagonist is contingent upon previous activation of the receptor by the agonist (i.e. glutamate), a fixed low dose of memantine blocks higher concentrations of agonist to a relatively greater degree than lower concentrations of agonist; hence, in an apparent paradox, we showed that a given concentration of memantine provides greater protection when more glutamate is present. The relatively fast off-rate of memantine is a major contributor to the drug's low affinity for the NMDAR as well as its clinical tolerability because this property insures that once excessive activity is normalized, the drug will leave the channel and not disrupt subsequent physiological neurotransmission. Thus, the critical features of memantine's mode of action are its Uncompetitive mechanism and Fast Off-rate, constituting what we have termed a ‘UFO' drug – a drug that is present at its site of inhibitory action only when you need it and then quickly disappears. A number of studies in vitro and in animal models of neurodegenerative diseases and stroke have shown that memantine protects neurons from NMDAR-mediated excitotoxic damage.91 In light of three successful human phase 3 clinical trials, the FDA approved memantine for the treatment of moderate-to-severe AD, and the drug is being tested in a number of other neurodegenerative conditions.
Recently, we developed an improved derivative of memantine based upon a combinatorial drug combining memantine and the active moiety of nitroglycerin (–NOx, where x=1 or 2). These new drugs, called NitroMemantines, use memantine to target the NO group to the nitrosylation sites of the NMDAR.3, 92 Because memantine, as an uncompetitive antagonist preferentially enters overactive NMDAR-coupled channels, it serves to bring NO directly to the NMDAR where it helps to curtail excessive activity, while avoiding other reactions of NO that could potentially be neurodestructive. Preliminary studies show that NitroMemantines are more effective neuroprotectants in vitro and in animal models than memantine and at lower concentrations.3, 93 Though much research remains to be done on these second generation NitroMemantine drugs, the combination of memantine with an NO group has created a new improved class of UFO drugs that should prove to be both clinically tolerated and neuroprotective.
Conclusions
Excessive production of ROS and RNS is thought to be a contributing factor to the sporadic forms of neurodegenerative diseases. NMDAR hyperactivation, protein misfolding, and mitochondrial dysfunction all contribute to neuronal damage and synaptic loss via excessive nitrosative and oxidative stress. Here, we have described a molecular mechanism involving S-nitrosylation of PDI and Drp1 that links free radical production, protein misfolding, and abnormal mitochondrial dynamics to neuronal cell injury in neurodegenerative disorders. These findings have important implications for new drug discovery efforts targeting aberrant S-nitrosylation pathways, and suggest that elucidation of additional S-nitrosylation pathways should help us attain a more complete understanding of the molecular processes involved in NO-mediated neurodegeneration. Furthermore, our findings highlight the importance of carefully characterizing SNO-proteins that may become candidate targets for future treatments of neurological disorders.
Acknowledgments
This work was supported in part by NIH grants P01 ES016738, P01 HD29587, P30 NS057096, R01 EY05477, R01 EY09024, the American Parkinson's Disease Association, San Diego Chapter, and an Ellison Senior Scholars Award in Aging (to S.A.L.).
Glossary
- Aβ
amyloid-β
- AD
Alzheimer's disease
- cGMP
cyclic guanosine-3′,5′-monophosphate
- Drp1
dynamin-related protein 1
- ER
endoplasmic reticulum
- GED
GTPase effector domain
- Mfn
Mitofusin
- NMDA
N-methyl--aspartate
- nNOS
neuronal NO synthase
- NO
nitric oxide
- NOX
NADPH oxidase
- PD
Parkinson's disease
- PDI
protein disulfide isomerase
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SNO-Ps
S-nitrosoproteins
- UPR
unfolded protein response
- UPS
ubiquitin-proteasome system
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Beal MF. Experimental models of Parkinson's disease. Nat Rev Neurosci. 2001;2:325–334. doi: 10.1038/35072550. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Rosenberg PA. Excitatory amino acids as a final common pathway for neurologic disorders. N Engl J Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]
- Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- Garthwaite J, Charles SL, Chess-Williams R. Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature. 1988;336:385–388. doi: 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- Brennan AM, Suh SW, Won SJ, Narasimhan P, Kauppinen TM, Lee H, et al. NADPH oxidase is the primary source of superoxide induced by NMDA receptor activation. Nat Neurosci. 2009;12:857–863. doi: 10.1038/nn.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, et al. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, et al. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci USA. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, et al. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, et al. S-Nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler R, Xiong Z, Lu WY, Hafner M, MacDonald JF, Tymianski M. Specific coupling of NMDA receptor activation to nitric oxide neurotoxicity by PSD-95 protein. Science. 1999;284:1845–1848. doi: 10.1126/science.284.5421.1845. [DOI] [PubMed] [Google Scholar]
- Dawson VL, Dawson TM, London ED, Bredt DS, Snyder SH. Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures. Proc Natl Acad Sci USA. 1991;88:6368–6371. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfoco E, Krainc D, Ankarcrona M, Nicotera P, Lipton SA. Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-D-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc Natl Acad Sci USA. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd SL, Tenneti L, Lishnak T, Lipton SA. Mitochondrial and extramitochondrial apoptotic signaling pathways in cerebrocortical neurons. Proc Natl Acad Sci USA. 2000;97:6161–6166. doi: 10.1073/pnas.100121097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankarcrona M, Dypbukt JM, Bonfoco E, Zhivotovsky B, Orrenius S, Lipton SA, et al. Glutamate-induced neuronal death: a succession of necrosis or apoptosis depending on mitochondrial function. Neuron. 1995;15:961–973. doi: 10.1016/0896-6273(95)90186-8. [DOI] [PubMed] [Google Scholar]
- Aarts M, Liu Y, Liu L, Besshoh S, Arundine M, Gurd JW, et al. Treatment of ischemic brain damage by perturbing NMDA receptor- PSD-95 protein interactions. Science. 2002;298:846–850. doi: 10.1126/science.1072873. [DOI] [PubMed] [Google Scholar]
- Zhou L, Li F, Xu HB, Luo CX, Wu HY, Zhu MM, et al. Treatment of cerebral ischemia by disrupting ischemia-induced interaction of nNOS with PSD-95. Nat Med. 2010;16:1439–1443. doi: 10.1038/nm.2245. [DOI] [PubMed] [Google Scholar]
- Papadia S, Soriano FX, Leveille F, Martel MA, Dakin KA, Hansen HH, et al. Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat Neurosci. 2008;11:476–487. doi: 10.1038/nn2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto SI, Pouladi MA, Talantova M, Yao D, Xia P, Ehrnhoefer DE, et al. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat Med. 2009;15:1407–1413. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Hua F, Yi Z, Zhou A, Ge L, et al. Organization of NMDA receptors at extrasynaptic locations. Neuroscience. 2010;167:68–87. doi: 10.1016/j.neuroscience.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, et al. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci USA. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar M, Forrester MT, Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, et al. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, et al. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, et al. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Lamas S, Fang FC. Nitrosylation. the prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, et al. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- Houk KN, Hietbrink BN, Bartberger MD, McCarren PR, Choi BY, Voyksner RD, et al. Nitroxyl disulfides, novel intermediates in transnitrosation reactions. J Am Chem Soc. 2003;125:6972–6976. doi: 10.1021/ja029655l. [DOI] [PubMed] [Google Scholar]
- Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HS, et al. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Takahashi H, Zhang D, Li W, Godzik A, et al. Cysteine regulation of protein function–as exemplified by NMDA-receptor modulation. Trends Neurosci. 2002;25:474–480. doi: 10.1016/s0166-2236(02)02245-2. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Shin Y, Cho SJ, Zago WM, Nakamura T, Gu Z, et al. Hypoxia enhances S-nitrosylation-mediated NMDA receptor inhibition via a thiol oxygen sensor motif. Neuron. 2007;53:53–64. doi: 10.1016/j.neuron.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Zoga V, Kimura M, Liang MY, Wu HE, Gemes G, et al. Nitric oxide activates ATP-sensitive potassium channels in mammalian sensory neurons: action by direct S-nitrosylation. Mol Pain. 2009;5:12. doi: 10.1186/1744-8069-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renganathan M, Cummins TR, Waxman SG. Nitric oxide blocks fast, slow, and persistent Na+ channels in C-type DRG neurons by S-nitrosylation. J Neurophysiol. 2002;87:761–775. doi: 10.1152/jn.00369.2001. [DOI] [PubMed] [Google Scholar]
- Tjong YW, Jian K, Li M, Chen M, Gao TM, Fung ML. Elevated endogenous nitric oxide increases Ca2+ flux via L-type Ca2+ channels by S-nitrosylation in rat hippocampal neurons during severe hypoxia and in vitro ischemia. Free Radic Biol Med. 2007;42:52–63. doi: 10.1016/j.freeradbiomed.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Eu JP, Sun J, Xu L, Stamler JS, Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- Huang Y, Man HY, Sekine-Aizawa Y, Han Y, Juluri K, Luo H, et al. S-nitrosylation of N-ethylmaleimide sensitive factor mediates surface expression of AMPA receptors. Neuron. 2005;46:533–540. doi: 10.1016/j.neuron.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Tian J, Kim SF, Hester L, Snyder SH. S-nitrosylation/activation of COX-2 mediates NMDA neurotoxicity. Proc Natl Acad Sci USA. 2008;105:10537–10540. doi: 10.1073/pnas.0804852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Thomas B, Cascio MB, Bae BI, Hester LD, Dawson VL, et al. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc Natl Acad Sci USA. 2006;103:3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N, Hara MR, Ahmad AS, Cascio MB, Kamiya A, Ehmsen JT, et al. GOSPEL: a neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron. 2009;63:81–91. doi: 10.1016/j.neuron.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, et al. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Lipton SA. Nitric oxide signaling regulates mitochondrial number and function. Cell Death Differ. 2003;10:757–760. doi: 10.1038/sj.cdd.4401244. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Perl DP, Brownell AL, Olanow CW. Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson's disease. Ann Neurol. 2004;56:149–162. doi: 10.1002/ana.20186. [DOI] [PubMed] [Google Scholar]
- Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- Lyles MM, Gilbert HF. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry. 1991;30:613–619. doi: 10.1021/bi00217a004. [DOI] [PubMed] [Google Scholar]
- Conn KJ, Gao W, McKee A, Lan MS, Ullman MD, Eisenhauer PB, et al. Identification of the protein disulfide isomerase family member PDIp in experimental Parkinson's disease and Lewy body pathology. Brain Res. 2004;1022:164–172. doi: 10.1016/j.brainres.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, et al. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 2010;6:900–906. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, et al. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- Cleeter MW, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AH. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci USA. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm CC, Moore K, Murphy MP. Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: implications for the interaction of nitric oxide with mitochondria. J Biol Chem. 2006;281:10056–10065. doi: 10.1074/jbc.M512203200. [DOI] [PubMed] [Google Scholar]
- Chinta SJ, Andersen JK. Reversible inhibition of mitochondrial complex I activity following chronic dopaminergic glutathione depletion in vitro: implications for Parkinson's disease. Free Radic Biol Med. 2006;41:1442–1448. doi: 10.1016/j.freeradbiomed.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Hsu M, Srinivas B, Kumar J, Subramanian R, Andersen J. Glutathione depletion resulting in selective mitochondrial complex I inhibition in dopaminergic cells is via an NO-mediated pathway not involving peroxynitrite: implications for Parkinson's disease. J Neurochem. 2005;92:1091–1103. doi: 10.1111/j.1471-4159.2004.02929.x. [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Muller WE, Eckert A, Kurz C, Eckert GP, Leuner K. Mitochondrial dysfunction: common final pathway in brain aging and Alzheimer's disease–therapeutic aspects. Mol Neurobiol. 2010;41:159–171. doi: 10.1007/s12035-010-8141-5. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Critical dependence of neurons on mitochondrial dynamics. Curr Opin Cell Biol. 2006;18:453–459. doi: 10.1016/j.ceb.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Li H, Chen Y, Jones AF, Sanger RH, Collis LP, Flannery R, et al. Bcl-xL induces Drp1-dependent synapse formation in cultured hippocampal neurons. Proc Natl Acad Sci USA. 2008;105:2169–2174. doi: 10.1073/pnas.0711647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- Kijima K, Numakura C, Izumino H, Umetsu K, Nezu A, Shiiki T, et al. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum Genet. 2005;116:23–27. doi: 10.1007/s00439-004-1199-2. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Parker WD, Jr, Parks J, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer's disease brain. Neurology. 1994;44:1090–1096. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, et al. Mitochondrial abnormalities in Alzheimer's disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Mao P, Calkins MJ, Cornea A, Reddy AP, Murphy MP, et al. Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer's disease neurons. J Alzheimers Dis. 20 (Suppl 2:S609–S631. doi: 10.3233/JAD-2010-100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloyannis SJ. Mitochondrial alterations in Alzheimer's disease. J Alzheimers Dis. 2006;9:119–126. doi: 10.3233/jad-2006-9204. [DOI] [PubMed] [Google Scholar]
- Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, et al. Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, et al. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci USA. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla RA, Matthews-Roberson TA, Dolan PJ, Johnson GV. Caspase-cleaved tau expression results in mitochondrial dysfunction in cortical neurons. Implications for the pathogenesis of Alzheimer disease. J Biol Chem. 2009;284:18754–18766. doi: 10.1074/jbc.M808908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laar VS, Berman SB. Mitochondrial dynamics in Parkinson's disease. Exp Neurol. 2009;218:247–256. doi: 10.1016/j.expneurol.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Zheng L, Perry G, Smith MA, Zhu X. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer's disease. J Neurochem. 2009;109 (Suppl 1:153–159. doi: 10.1111/j.1471-4159.2009.05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- Zhu PP, Patterson A, Stadler J, Seeburg DP, Sheng M, Blackstone C. Intra- and intermolecular domain interactions of the C-terminal GTPase effector domain of the multimeric dynamin-like GTPase Drp1. J Biol Chem. 2004;279:35967–35974. doi: 10.1074/jbc.M404105200. [DOI] [PubMed] [Google Scholar]
- Pitts KR, McNiven MA, Yoon Y. Mitochondria-specific function of the dynamin family protein DLP1 is mediated by its C-terminal domains. J Biol Chem. 2004;279:50286–50294. doi: 10.1074/jbc.M405531200. [DOI] [PubMed] [Google Scholar]
- Kang-Decker N, Cao S, Chatterjee S, Yao J, Egan LJ, Semela D, et al. Nitric oxide promotes endothelial cell survival signaling through S-nitrosylation and activation of dynamin-2. J Cell Sci. 2007;120:492–501. doi: 10.1242/jcs.03361. [DOI] [PubMed] [Google Scholar]
- Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci USA. 2006;103:1295–1300. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy B, Petrilli A, Klinglmayr E, Chen J, Lutz-Meindl U, Knott AB, et al. S-Nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer's disease. J Alzheimers Dis. 2010;20 (Suppl 2:S513–S526. doi: 10.3233/JAD-2010-100552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Cieplak P, Cho DH, Godzik A, Lipton SA. S-nitrosylation of Drp1 links excessive mitochondrial fission to neuronal injury in neurodegeneration. Mitochondrion. 2010;10:573–578. doi: 10.1016/j.mito.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TH, Schnaar RL, Coyle JT. Immature cortical neurons are uniquely sensitive to glutamate toxicity by inhibition of cystine uptake. Faseb J. 1990;4:1624–1633. [PubMed] [Google Scholar]
- Okamoto S, Pouladi MA, Talantova M, Yao D, Xia P, Ehrnhoefer DE, et al. Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingin. Nat Med. 2009;15:1407–1413. doi: 10.1038/nm.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Pellegrini JW, Aggarwal SK, Lei SZ, Warach S, Jensen FE, et al. Open-channel block of N-methyl-D-aspartate (NMDA) responses by memantine: therapeutic advantage against NMDA receptor-mediated neurotoxicity. J Neurosci. 1992;12:4427–4436. doi: 10.1523/JNEUROSCI.12-11-04427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Chen HS, Zhang D, Lipton SA. Memantine preferentially blocks extrasynaptic over synaptic NMDA receptor currents in hippocampal autapses. J Neurosci. 2010;30:11246–11250. doi: 10.1523/JNEUROSCI.2488-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton SA. Pathologically activated therapeutics for neuroprotection. Nat Rev Neurosci. 2007;8:803–808. doi: 10.1038/nrn2229. [DOI] [PubMed] [Google Scholar]
- Wang Y, Eu J, Washburn M, Gong T, Chen HS, James WL, et al. The pharmacology of aminoadamantane nitrates. Curr Alzheimer Res. 2006;3:201–204. doi: 10.2174/156720506777632808. [DOI] [PubMed] [Google Scholar]
- Beal MF. Excitotoxicity and nitric oxide in Parkinson's disease pathogenesis. Ann Neurol. 1998;44:S110–S114. doi: 10.1002/ana.410440716. [DOI] [PubMed] [Google Scholar]
- Gupta SP, Patel S, Yadav S, Singh AK, Singh S, Singh MP. Involvement of nitric oxide in maneb- and paraquat-induced Parkinson's disease phenotype in mouse: is there any link with lipid peroxidation. Neurochem Res. 2010;35:1206–1213. doi: 10.1007/s11064-010-0176-5. [DOI] [PubMed] [Google Scholar]
- Akama KT, Van Eldik LJ. Beta-amyloid stimulation of inducible nitric-oxide synthase in astrocytes is interleukin-1beta- and tumor necrosis factor-alpha (TNFalpha)-dependent, and involves a TNFalpha receptor-associated factor- and NFkappaB-inducing kinase-dependent signaling mechanism. J Biol Chem. 2000;275:7918–7924. doi: 10.1074/jbc.275.11.7918. [DOI] [PubMed] [Google Scholar]
- Li S, Hong S, Shepardson NE, Walsh DM, Shankar GM, Selkoe D. Soluble oligomers of amyloid beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron. 2009;62:788–801. doi: 10.1016/j.neuron.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon DT, Rogers SD, Ghilardi JR, Finke MP, Cleary JP, O'Hare E, et al. Fibrillar beta-amyloid induces microglial phagocytosis, expression of inducible nitric oxide synthase, and loss of a select population of neurons in the rat CNS in vivo. J Neurosci. 1998;18:2161–2173. doi: 10.1523/JNEUROSCI.18-06-02161.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]