Abstract
Wig-1 is a transcriptional target of the tumor suppressor p53 and encodes an unusual zinc-finger protein involved in post-transcriptional gene regulation. Wig-1 is expressed in all cell types investigated so far, with the highest levels in the brain, and is enriched in stem cells as compared with more differentiated cells of the same lineage. Wig-1 binds to both long double-stranded (ds) RNA and short microRNA-like dsRNA. We have shown that Wig-1 acts in a positive feedback loop that stabilizes p53 mRNA through an AU-rich element (ARE) in the p53 3′untranslated region. Our preliminary data indicate a more general effect of Wig-1 on ARE-containing mRNA. Here we shall summarize current knowledge about Wig-1 and discuss possible implications on p53 function and other cellular processes.
Keywords: Wig-1, p53, AU-rich elements, mRNA regulation
The p53 tumor suppressor protects us from cancer by sensing and reacting to a wide range of cellular stress factors, including DNA damage and oncogene activation. The p53 activation can result in various outcomes depending on stress agent and cellular context, the classical p53 stress responses being cell cycle arrest and apoptosis.1, 2 The importance of p53-mediated tumor suppression is demonstrated by the high frequency of p53 mutations (about 50%) in human tumors,3 (http://www-p53.iarc.fr; http://p53.free.fr). p53 orchestrates its responses mainly through its activity as transcription factor, regulating transcription of target genes that carry out a plethora of functions in the cell.1, 2 For this reason, delineating functions of p53 target genes is crucial in order to fully understand the function of p53 itself.
Structural and Functional Characteristics of the Wig-1 Protein
Wig-1 (also known as PAG608 or ZMAT3) is a direct transcriptional target gene of p53 that was discovered more than 10 years ago. Mouse Wig-1 was identified in our laboratory in J3D mouse T lymphoma cells carrying a temperature sensitive Val135 mutant p53 construct (tsp53). This construct is expressed as mutant p53 at 37 °C but temperature shift to 32 °C induces wild-type p53 expression, triggering cell cycle arrest and apoptosis. Wig-1 was identified by differential display analysis as an mRNA expressed at 32 °C but not at 37 °C.4, 5 The laboratory of Moshe Oren simultaneously identified Wig-1 (which they named PAG608 – a name that subsequently has been used to indicate the rat ortholog of Wig-1) using a similar approach in the mouse myeloid leukemia line LTR6.6 We and others later cloned human Wig-1 and confirmed that it is a p53 target gene.7, 8
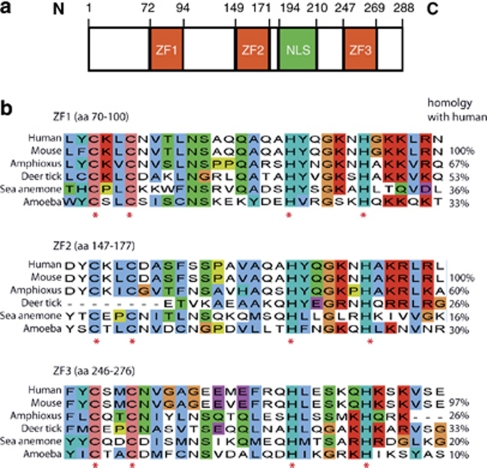
Human Wig-1 maps to chromosome 3q26.32. The Wig-1 protein exists in two isoforms differing by one amino acid only, and the Wig-1 open reading frame (ORF) translates into proteins of 288 or 289 amino acids. The ORF is followed by a very long 3′ untranslated region (UTR) with three polyA sites; usage of the most distal gives rise to an 8 kb long 3′UTR. The Wig-1 protein contains a nuclear localization signal and three zinc-fingers of the Cys2His2 type (Figure 1a). The zinc-fingers are unusual in their inter-histidine distance within the zinc-fingers (five amino acids instead of the normal three to four) and their long linkers between the zinc-fingers (56–75 amino acids compared with six to eight as in most other zinc-finger proteins). The unusual zinc-finger structure is shared with a small group of double-stranded RNA (dsRNA) binding proteins that lack consensus dsRNA-binding motifs. It has been suggested that members of this group of proteins that bind dsRNA through such widely spaced zinc-fingers may show greater versatility in binding specificity, strength and span.9, 10 The most well-studied member of this dsRNA-binding protein-group is JAZ, which binds to the dsRNA nuclear export receptor Exportin-5,11 and positively regulates p53 transcriptional activity by binding to the p53 protein.12 Like JAZ, Wig-1 is an RNA binding protein. It binds to long dsRNA of more than 50 bp,13 and can also bind to single-stranded RNA and to RNA-DNA hybrids, although dsRNA is the preferred target.13 Both the first and second zinc-fingers of Wig-1 are necessary for binding to dsRNA in living cells.13, 14 Wig-1 also binds to shorter RNA of ∼21 bp, but only those that resemble microRNA (miRNA) in that they have a two-nucleotide 3′ overhang. Interestingly, Wig-1 levels are decreased in embryonic stem cells (ESCs) null for Dicer, an RNAse III enzyme crucial for miRNA maturation,15 indicating that Wig-1 may have a function related to miRNA.
Figure 1.
Wig-1 protein structure and conservation during evolution. (a) Wig-1 protein structure. ZF indicates zinc-finger and NLS nuclear localization signal. N and C indicates N and C terminus, respectively, and numbers indicate amino acid position in the 288 amino-acid Wig-1 protein. (b) Conservation of the Wig-1 zinc-fingers (shown with a few surrounding residues) down to amoeba. Asterisks indicate the two cysteines and two histidines of the Wig-1 Cys2His2 zinc-fingers
Wig-1 is highly conserved from fish to human, especially with regard to the zinc-fingers that are almost completely conserved. Human and mouse Wig-1 show 87% protein identity with perfectly conserved zinc-fingers except for one amino-acid substitution in the third zinc-finger. The distance between the zinc-fingers are also conserved.16 Further analysis of species conservation has revealed that Wig-1 is in fact conserved throughout evolution all the way from amoeba, the ancestors of which separated from the human ancestors in evolution about 1.5 billion years ago, at the very beginning of eukaryotes.17 Again the zinc-fingers show significant conservation – the first and second zinc-fingers are ∼30% identical between human and amoeba (Figure 1b), whereas the overall identity is 22% (http://www.uniprot.org).
Wig-1 Expression
According to our studies, Wig-1 is expressed in all tissues, with the highest expression in brain (ref. 4, our unpublished results). These findings are supported by expression data available at BioGPS (http://biogps.gnf.org), showing basal Wig-1 levels in all tissues with higher levels in brain – especially in the amygdala and prefrontal cortex – and smooth muscle, cardiac myocytes, and adipocytes (Table 1, see Table 2 for Wig-1 expression in cancer cell lines). A role for Wig-1 in brain is further supported by the fact that PAG608, the rat ortholog of Wig-1, is constitutively expressed at relatively high levels in various regions of the rat nervous system, and is induced in the nervous system by a number of stress agents, including ischemia,18, 19 treatment with methamphetamine,20 onset of disease in a model of ALS,21 and by L-DOPA in a Parkinson model.22 Some studies also suggest pro-apoptotic functions of PAG608 in stressed brain.18, 23 Taken together, these data indicate a role for Wig-1/PAG608 in stress responses and pathological conditions in the nervous system.
Table 1. Wig-1 expression in selected human tissues, adopted from http://biogps.gnf.org.
| Tissue | Wig-1 expression relative to median |
|---|---|
| Kidney | 0.78 |
| Adipocyte | 3.00 |
| Uterus | 0.83 |
| Pancreas | 1.02 |
| Smooth muscle | 4.31 |
| Cardiomyocyte | 3.58 |
| Lung | 1.00 |
| Whole Brain | 3.73 |
| Amygdala | 3.06 |
| Prerontal cortex | 2.83 |
| Parietal lobe | 1.73 |
| Whole blood | 0.98 |
| Skin | 0.87 |
| Liver | 1.34 |
| Heart | 1.04 |
Table 2. Wig-1 expression in selected human cell lines, adopted from http://biogps.gnf.org.
| Cell line | Tissue of origin | Wig-1 expression relative to median |
|---|---|---|
| HCT116 | Colorectal carcinoma | 0.68 |
| HeLa | Cervical carcinoma | 1.45 |
| HT1080 | Fibrosarcoma | 27.8 |
| Jurkat | Acute T-cell leukemia | 1.47 |
| SHSY-5Y | Neuroblastoma | 3.43 |
| U2OS | Osteosarcoma | 1.97 |
| SKOV-3 | Ovarian carcinoma | 0.43 |
| HT-29 | Colorectal adenocarcinoma | 0.36 |
| HEK293 | Transformed human embryonic kidney cells | 0.78 |
| UACC62 | Melanoma | 25.4 |
| SKMEL5 | Melanoma | 13.7 |
| LNCAP | Prostate adenocarcinoma | 4.2 |
| MCF7 | Breast adenocarcinoma | 0.93 |
| T3M4 | Pancreatic adenocarcinoma | 0.30 |
| SN12C | Renal carcinoma | 0.25 |
Then how is Wig-1 expression regulated? Human Wig-1 has a functional, perfect consensus p53 response element in intron 1 (M Wilhelm, unpublished results) and mouse Wig-1 contains two functional, albeit not perfect, p53 response elements in the promoter region.5 That Wig-1 is a bona fide p53 target gene is confirmed by a number of studies 4, 5, 6, 7 and is further supported by microarray data showing Wig-1 upregulation after p53 activation by the small molecule RITA.24 In addition, Wig-1 is induced after DNA damage in vivo in hematopoietic stem cells after treatment with the chemotherapeutic drug 5-fluoro-uracil.25
Yet it is clear that p53 is not the only transcription factor that regulates Wig-1 expression, as Wig-1 is expressed at robust levels also in cells lacking p53. Our database searches (using pscan: http://159.149.109.9/pscan/ and genomatix: http://www.genomatix.de) identified between 45 and 130 putative transcription factor-binding sites in the Wig-1 promoter, depending on choice of database and search parameters. This suggests that Wig-1 is subjected to tight regulation by multiple factors. As expected, p53 is among the top ranked transcription factor candidates that may bind both human and mouse wig-1 promoters, according to Pscan (http://159.149.109.9/pscan/). Pscan also identified a number of other transcription factors with various functions and expression patterns (Table 3). These factors include myogenic factor 5, which is involved in specification and differentiation of the muscle lineage,26 fifth ewing variant, thought to be involved in the transcription of genes of the human serotonergic system and to have a role in early brain development,27 spleen focus forming virus (SFFV) proviral integration oncogene (SPI1) and hepatic leukemia factor, involved in normal and malignant hematopoiesis,28, 29 spermatogenic leucine zipper 1 (spz)1, a sperm specific growth promoting transcription factor,30, 31 POU class 5 homeobox 1 (Pou5f1) and SRY (sex-determining region Y)-box 2 (Sox2), both essential for the maintenance of ESCs,32 and Nr2e3, nuclear receptor subfamily 2, group E, member 3, which is a photoreceptor-specific nuclear receptor.33 This spectrum of transcription factors suggests an intricate regulation of Wig-1 expression in a variety of tissues (muscle, brain, lens, testis, hematopoietic cells, and ESCs) in accordance with previous observation of ubiquitous Wig-1 expression. In addition, according to Pscan, Wig-1 can both be activated by the tumor suppressor p53 and a number of transcription factors that promote cell growth and/or have oncogenic features (e.g., spz1, Pou5f1, and SPI1), indicating that Wig-1 expression may be induced under a variety of different cellular circumstances. Moreover, it is interesting to note that several of the potential Wig-1-regulating transcription factors are crucial for embryonic stem cell survival, such as Pou5f1 and Sox2. We will return to this subject below.
Table 3. Top putative transcription factors binding to the human and mouse Wig-1 promoters with P-values according to Pscan (http://159.149.109.9/pscan).
| Transcription factor | P-value |
|---|---|
| Myf5 | 0.0004 |
| FEV | 0.004 |
| SPI1 | 0.004 |
| Spz1 | 0.012 |
| Pou5f1 | 0.017 |
| GABPA | 0.022 |
| TP53 | 0.029 |
| Nr2e3 | 0.038 |
| Sox2 | 0.053 |
| HLF | 0.054 |
p53 is indicated in bold
To further study upstream regulatory cues with potential impact on Wig-1 expression, we performed gene ontology classifications (http://www.pantherdb.org) on the list of putative transcription factors (the most extensive list was chosen for this purpose). According to this analysis, the putative Wig-1-regulating transcription factors are involved in a number of important growth promoting signaling pathways (Table 4). Other top pathways identified are consistent with a role for Wig-1 in the p53 network, such as oxidative stress response, apoptosis signaling, and p53 feedback loops (Table 4). From this pathway analysis, we can deduce that the p53 network is indeed an important upstream Wig-1 activator, but so are a number of other prominent cell signaling networks – all in all, there are many conditions and signals that will lead to Wig-1 activation. This is in agreement with the fact that Wig-1 is expressed in all cells tested so far, including both tumor cell lines and primary cells from a number of different tissues and regardless of p53 status. However, induction of Wig-1 levels after DNA damage requires functional p53 (our unpublished results).
Table 4. Pathway analysis of putative transcription factors binding to the Wig-1 promoter using Panther (http://www.pantherdb.org) with P-values for human and mouse Wig-1 promoter.
| Pathway | P-value human | P-value mouse |
|---|---|---|
| Interleukin signaling | 3.08E-17 | 3.57E-16 |
| Insulin/IGF-protein kinase B | 4.14E-10 | 5.93E-09 |
| TGF-beta signaling | 3.13E-09 | 2.98E-08 |
| PI3 kinase | 4.67E-09 | 5.21E-08 |
| PDGF signaling | 8.01E-09 | 5.71E-09 |
| Oxidative stress | 1.47E-07 | 1.18E-07 |
| Ras | 1.34E-05 | 1.11E-05 |
| Toll-receptor signaling | 5.43E-05 | 4.66E-05 |
| Apoptosis signaling | 1.52E-04 | 1.27E-04 |
| Wnt signaling | 2.16E-04 | 1.69E-04 |
| Inflammation mediated by chemokines and cytokines | 4.99E-04 | 1.98E-03 |
| p53 pathway feedback loops 2 | 4.67E-03 | 4.27E-03 |
The pathway ‘unclassified' was omitted
Another interesting question is if Wig-1 expression is altered in tumor cells. Wig-1 is located on chromosome 3q26.32 within a region that is amplified in many tumors, including head and neck, breast, ovarian, cervical, prostate, esophageal, nasopharyngeal and lung squamous cell carcinomas, as well as in acute myeloid leukemia.34, 35, 36, 37, 38, 39, 40 This suggests that Wig-1 is amplified in a number of these tumors, an assumption supported by one report on overexpression of Wig-1 in lung cancer.7 However, the chromosomal region where Wig-1 resides harbors several genes with relevance for cancer, such as telomerase RNA component and B-cell CLL/lymphoma 6,41, 42 and so it is unclear whether Wig-1 is driving gene amplification or simply is co-amplified as a passenger gene. In addition, the indications that Wig-1 can potentially be transcriptionally upregulated by both tumor suppressors, for example, p53 and transcription factors with oncogenic activity (see above), along with the fact that both overexpression and knockdown of Wig-1 interferes with cell growth (see below), argues against a clear-cut role for Wig-1 either as a tumor suppressor gene or oncogene. In addition, both increased and decreased Wig-1 expression in tumors has been reported (Table 5).
Table 5. Comparison of Wig-1 in tumor versus normal tissues.
| Experiment | Wig-1 ratio tumor versus normal | Reference |
|---|---|---|
| Papillary thyroid carcinoma versus normal | 3.36 | GeoProfiles GDS1732 |
| Squamous cell carcinoma versus normal | 1.00 | 65 |
| Invasive lobular breast cancer versus normal | 0.79 | 66 |
| Invasive ductal breast cancer versus control | 0.91 | 66 |
Several studies have demonstrated increased expression of Wig-1 in stem cells, suggesting a role in stem cell maintenance. One study compared genes enriched in both haematopoietic, neuronal and ESCs (as compared with the corresponding differentiated cell types), and concluded that Wig-1 is upregulated in all three stem cell compartments.43 Another study showed increased expression of Wig-1 in haematopoietic stem cells (HSCs) null for BMI1 polycomb ring finger oncogene (Bmi-1), a factor necessary for HSC renewal. In the absence of Bmi-1 the pool of adult HSCs was lost, coinciding with Wig-1 upregulation.44 Whether this increased expression of Wig-1 promoted the loss of HSCs or if it occurred as a compensatory mechanism in the absence of Bmi-1 remains unclear. Furthermore, Wig-1 expression decreases during erythropoiesis, in accordance with a function for Wig-1 in the HSCs and haematopoietic progenitor cells.45 In addition, microarray studies support a role for Wig-1 in germ cells and ESCs. For example, one array study46 showed Wig-1 induction in spermatogonial stem cells following stimulation with Glial cell line-derived neurotrophic factor (GDNF) and GDNF-family receptor α-1. In another study,47 cumulus oocyte complexes isolated from human chorionic gonadotropin-treated ovaries showed increased Wig-1 expression compared with untreated oocytes. These array studies indicate that Wig-1 may be induced in germ cells in response to growth factors. In accordance with a role for Wig-1 in germ cells, Wig-1 is expressed in mouse oocytes before fertilization and maintained and somewhat increased until the E4.5 embryo.48 Wig-1 expression in pre-implantation embryos is further confirmed by additional array data.49, 50 A role for Wig-1 in ESC maintenance is strengthened by the observation that Wig-1 knockdown using short hairpin RNA (shRNA) reduced the stem cell phenotype in an shRNA screen to find novel ESC regulators.51 Furthermore, the two essential ESC transcription factors Pou5f1 and Sox2 are potential Wig-1 regulators (see above). In addition, classification of the putative Wig-1-regulating transcription factors using Panther (see above) revealed enrichment for these factors in biological processes such as reproduction and development, strengthening the idea that Wig-1 has a critical role or roles in ESCs and embryonic development. For an overview of Wig-1 enrichment in stem cells, see Table 6.
Table 6. Enrichment of Wig-1 mRNA in stem cells.
| Experiment | Wig-1 enrichment in stem cell | Species | Reference: |
|---|---|---|---|
| HSC versus differentiated tissue | 1.55 | Mouse | 43 |
| NSC versus differentiated tissue | 1.80 | Mouse | 43 |
| ESC versus differentiated tissue | 3.76 | Mouse | 43 |
| Neural crest stem cells versus mature Schwann cells | 1.58 | Mouse | 67 |
| Day 1 (stem cell) versus day 11 (differentiated tissue) after induction of erythropoiesis | 1.78 | Human | 45 |
Wig-1 Function
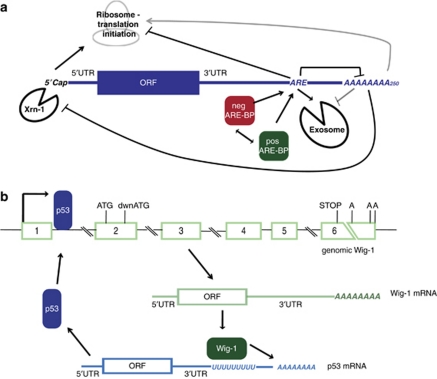
Our previous work showed that ectopically expressed Wig-1 inhibits cell growth modestly in long-term assays without any major impact on cell cycle distribution or cell death. In addition, Wig-1 knockdown using small interfering RNA leads to decreased cell viability.52 Hence, both too much and too little Wig-1 has a negative effect on cell viability, indicating Wig-1 levels must be controlled and maintained within an appropriate range. In order to identify Wig-1 partner proteins, we immunoprecipitated FLAG-tagged Wig-1 and identified co-precipitating proteins by mass spectrometry. We found that RNA helicase A and heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2/B1) are bound to Wig-1 in an RNA-dependent fashion.52 These two proteins are multifunctional RNA binding proteins, and therefore it is difficult to deduce a function of Wig-1 solely from these RNA dependent interactions. However, in accordance with a role for hnRNPA2/B1 in regulation of mRNA stability, we recently showed that Wig-1 can positively regulate p53 mRNA. Wig-1 binds to and stabilizes p53 mRNA via an U-rich region in the p53 3′UTR, forming a positive regulatory feedback loop. U-rich regions are a subgroup to the AU-rich elements (AREs) present in the 3′UTRs of certain mRNAs. The AREs are elements involved in regulation mRNA stability and translation, and consist of the element AUUUA in various constellations or of exclusively U-rich regions.53, 54 They are present in 5–8 % of the transcriptome, mostly in mRNAs of genes whose expression is precisely controlled, including genes encoding proteins that regulate cell growth or the cellular response to external factors, for example, c-Myc, N-Myc, cyclins, interferons, p53 and p21.53, 54 AREs are generally considered as negative regulators of gene expression. They can mediate mRNA degradation both by increasing deadenylation, which is the first and rate-limiting step of mRNA degradation, and by enhancing subsequent decay of the mRNA. AREs can also decrease translation efficiency (Figure 2a). Most factors that bind and regulate AREs affect the target mRNA negatively, by promoting degradation or inhibiting translation. However, there are also primarily positive ARE regulators such as the well known and ubiquitously expressed human antigen R (HuR) and ARE regulators that can act either positively or negatively, depending on the mRNA in question.53, 55
Figure 2.
ARE-mediated regulation of mRNA. (a) Mechanisms for ARE-mediated regulation of mRNA stability and translation efficiency. AREs are regions of the pentamer AUUUA in various constellations or U-rich regions present in the 3′UTR of mRNAs. In general AREs promote mRNA decay by increasing deadenylation, the first and rate-limiting step of mRNA degradation. When the polyA tail is shortened beyond a certain limit mRNA degradation commences, either 3′ to 5′ through the exosome or 5′ to 3′ mediated by Xrn-1. AREs can also regulate translation. Proteins that bind to AREs (ARE-BPs) are usually negative regulators carrying out the ARE effects on mRNA stability and translation, but there are also positive ARE-binding factors that prevent mRNA degradation and/or enhance translation. (b) Wig-1 stabilizes the p53 mRNA in a positive feedback loop in which Wig-1 transcription is induced by p53, and Wig-1 in turn binds to the U-rich region in the 3′UTR of p53 mRNA and protects it from deadenylation, thereby increasing p53 mRNA and protein levels. The p53 recognizes a perfect p53-binding motif in intron 1 of the human Wig-1 gene variant 1 (nt 1662-1681; NCBI reference sequence NC_000003.11). Note that most introns and exon 6 are not shown to scale (indicated by broken lines). The main Wig-1 ATG and a downstream ATG that gives rise to a shorter Wig-1 protein isoform, as well as the stop codon and the three polyA sites (‘A'), are also indicated. The U-rich region in p53 mRNA to which Wig-1 binds is located at nt 2094-2156 (AC NM_000546.4)
ARE-binding proteins (ARE-BPs) are important nodes of regulation, as they can potentially control the levels of many different mRNAs in the cell.54, 56 The 3′UTR of p53 harbors one U-rich region (18 continuous Us) and one additional ARE,57, 58 both of which are targeted for regulation by several ARE-BPs.59 Wig-1 binds to the U-rich region in the 3′UTR of p53 mRNA and stabilizes p53 mRNA by preventing its deadenylation (Figure 2b). This enhances p53 protein levels and potentiates the p53 response to DNA damage.60 These results are consistent with data demonstrating that inhibition of expression of the Wig-1 ortholog PAG608 using antisense RNA led to attenuated p53-dependent methamphetamine-induced cell death.20 Most ARE-BPs regulate multiple mRNAs54, 56, and our preliminary results indicate that Wig-1 also can target other ARE-containing mRNAs apart from p53.60
Critical Unanswered Questions
The fact that the p53 target Wig-1 has a role in post-transcriptional gene regulation suggests a whole new level of gene regulation orchestrated by p53, mediated by its downstream target Wig-1. This way, p53 might potentially regulate a different set of targets at the mRNA level in addition to its direct transcriptional targets. Moreover, it is conceivable that p53 could fine-tune the regulation of some or many of its transcriptional targets by additional regulation at the level of their mRNA stability and/or translation. However, a number of questions remain to be answered in order to achieve a more complete understanding of the role of Wig-1 in post-transcriptional gene regulation and in cell physiology. First, it is crucial to identify additional Wig-1 targets and elucidate exactly which cellular responses Wig-1 activation is associated with. Most mRNAs regulated by AREs correspond to proteins involved in immediate early responses – many of them are growth-promoting proteins such as cyclins, c-Jun, c-Fos, and c-Myc.54 We have identified several tentative Wig-1 targets apart from p53, including proteins normally associated with growth promotion.60 We are currently investigating global effects of Wig-1 on mRNA expression through microarray analysis in order to obtain a genome-wide picture of cellular processes that are regulated by Wig-1.
Another pressing task is the investigation of mechanisms behind Wig-1 function in ARE-mediated post-transcriptional gene regulation. What specific RNA motifs or structures are bound and regulated by Wig-1? Does Wig-1 target specificity change with cell type and/or stress induction? Also, we may have to look beyond ARE-mediated regulation – there are probably other, additional mechanisms of action for Wig-1. ARE-mediated regulation takes place in the cytoplasm, whereas Wig-1 mainly localizes to the nucleus and shuttles between nucleus and cytoplasm60 features that are shared with the positive ARE regulator HuR.61 It is possible that Wig-1, like HuR, is involved in nuclear processes in addition to its cytoplasmic function in ARE-mediated regulation. Such processes could include mRNA transport, processing, and splicing. The notion that Wig-1 has an important role in the nucleus is supported by the observed increase in Wig-1 levels in both the nucleus and the cytoplasm after stress.60 In addition to a likely nuclear role of Wig-1, we have shown that Wig-1 can bind to miRNA-like short RNA in vitro,14 suggesting that Wig-1 is somehow involved in miRNA-mediated regulation. There are several examples of cross talk between miRNA- and ARE-mediated mechanisms. Wig-1 may be one of the factors linking these two pathways together. However, we observed that Wig-1-mediated regulation of p53 mRNA was unaffected in cells with significantly reduced miRNA levels due to lack of Dicer, indicating that Wig-1 can regulate p53 mRNA independently of miRNA presence. Nonetheless, it is possible that Wig-1 has other important miRNA-related functions that are related or unrelated to its role in ARE-mediated mRNA regulation.
One critical question is the exact role of Wig-1 in the p53 stress response. Human Wig-1 has a perfect p53 binding site in its promoter, a feature shared with the cell cycle arrest-inducing p53 target p21. Moreover, Wig-1 was one of the most strongly upregulated genes in γ-radiated mouse embryonic fibroblasts from knock-in mice carrying a p53 mutant that can only activate cell cycle arrest-related genes.62 This suggests that p53-mediated induction of Wig-1 is related to cell cycle arrest, and that Wig-1 somehow fine-tunes or potentiates this response, although ectopically expressed Wig-1 by itself does not cause any major changes in cell cycle distribution.52
Further studies are also required to investigate the possible role of Wig-1 in tumor development. It will be interesting to examine and compare levels of Wig-1 in tumors that carry wild-type or mutant p53. Assuming that Wig-1 regulates mutant as well as wild-type p53, increased Wig-1 levels, for example via amplification of the 3q26.3–27 region (see above), could in theory lead to a further increase of mutant p53 mRNA and protein levels, which may promote tumorigenesis through mutant p53 gain-of-function mechanisms.63 Moreover, it will be important to investigate the expression of novel Wig-1 targets in tumors in relation to Wig-1 expression.
The fact that Wig-1 is expressed at significant levels also in cells lacking p53 clearly suggests that Wig-1 also can have p53 independent functions. In line with this assumption, Wig-1 can potentially be transactivated by a range of transcription factors. Further, a number of reports indicate a role for Wig-1 in stem cells. This is consistent with our preliminary results indicating that homozygous knockout of Wig-1 in mice causes early embryonic lethality. As all cells investigated to date – and all organisms as far back as the amoeba – express Wig-1, it seems reasonable to conclude that Wig-1 is required for cell survival, with a particularly important role in stem cell maintenance.
Concluding Remarks
The complexity of the p53 network is constantly increasing. It is now clear that p53 can have many other functions in the cell than induction of cell cycle arrest and apoptosis in response to stress. For example, p53 is implicated in regulation of metabolism, and low basal levels of p53 may actually promote cell survival.2, 64 In this context, the intriguing p53 target Wig-1 may fit right in. Wig-1 is a target gene not directly involved in induction of cell cycle arrest or apoptosis – instead, evidence suggests that it regulates gene expression in a post-translational manner and modulates the p53 response towards cell cycle arrest or apoptosis or other biological outcomes. Thus, Wig-1 may have a profound impact on cell fate. Moreover, as Wig-1 appears to be well conserved in all eukaryotic cells, further studies of its regulation and activities might provide important information about fundamental cellular processes. Clearly, our understanding of Wig-1 function and biological significance is still only at an early stage, and much remains to be discovered. We can expect exciting progress and many surprises from Wig-1 in the future!
Acknowledgments
We are supported by the Swedish Cancer Society (Cancerfonden), the Swedish Medical Research Council (VR), the Konung Gustaf V Jubilee Fund and Karolinska Institutet. We thank David P Lane for valuable advice on bioinformatic analysis of evolutionary conservation.
Glossary
- ARE
AU-rich element
- UTR
untranslated region
- dsRNA
double-stranded RNA
- ORF
open reading frame
- SPI1
spleen focus forming virus (SFFV) proviral integration oncogene
- spz1
spermatogenic leucine zipper 1
- Pou5f1
POU class 5 homeobox 1
- Sox2
SRY (sex-determining region Y)-box 2
- ESCs
embryonic stem cells
- Bcl-6
B-cell CLL/lymphoma 6
- HSCs
haematopoietic stem cells
- RITA
reactivation of p53 and induction of tumor cell apoptosis
- ALS
amyotrophic lateral sclerosis
- L-DOPA
levo-Dihydroxyphenylalanine
- Bmi-1
BMI1 polycomb ring finger oncogene
- GDNF
glial cell-derived neurotrophic factor
- shRNA
short hairpin RNA
- hnRNPA2/B1
heterogeneous nuclear ribonucleoprotein A2/B1
- HuR
human antigen R
- ARE-BPs
ARE-binding proteins
- miRNA
microRNA
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Varmeh-Ziaie S, Okan I, Wang Y, Magnusson KP, Warthoe P, Strauss M, et al. Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene. 1997;15:2699–2704. doi: 10.1038/sj.onc.1201454. [DOI] [PubMed] [Google Scholar]
- Wilhelm MT, Mendez-Vidal C, Wiman KG. Identification of functional p53-binding motifs in the mouse wig-1 promoter. FEBS Lett. 2002;524:69–72. doi: 10.1016/s0014-5793(02)03004-1. [DOI] [PubMed] [Google Scholar]
- Israeli D, Tessler E, Haupt Y, Elkeles A, Wilder S, Amson R, et al. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmeh-Ziaie S, Ichimura K, Yang F, Rabbits P, Collins VP. Cloning and chromosomal localization of human WIG-1/PAG608 and demonstration of amplification with increased expression in primary squamous cell carcinoma of the lung. Cancer Lett. 2001;174:179–187. doi: 10.1016/s0304-3835(01)00699-1. [DOI] [PubMed] [Google Scholar]
- Hellborg F, Qian W, Mendez-Vidal C, Asker C, Kost-Alimova M, Wilhelm M, et al. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene. 2001;20:5466–5474. doi: 10.1038/sj.onc.1204722. [DOI] [PubMed] [Google Scholar]
- Reuter G, Giarre M, Farah J, Gausz J, Spierer A, Spierer P. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature. 1990;344:219–223. doi: 10.1038/344219a0. [DOI] [PubMed] [Google Scholar]
- Fasano L, Roder L, Core N, Alexandre E, Vola C, Jacq B, et al. The gene teashirt is required for the development of Drosophila embryonic trunk segments and encodes a protein with widely spaced zinc finger motifs. Cell. 1991;64:63–79. doi: 10.1016/0092-8674(91)90209-h. [DOI] [PubMed] [Google Scholar]
- Chen T, Brownawell AM, Macara IG. Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol Cell Biol. 2004;24:6608–6619. doi: 10.1128/MCB.24.15.6608-6619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Wu S, Su X, May WS. JAZ mediates G1 cell-cycle arrest and apoptosis by positively regulating p53 transcriptional activity. Blood. 2006;108:4136–4145. doi: 10.1182/blood-2006-06-029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Vidal C, Wilhelm MT, Hellborg F, Qian W, Wiman KG. The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res. 2002;30:1991–1996. doi: 10.1093/nar/30.9.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez Vidal C, Prahl M, Wiman KG. The p53-induced Wig-1 protein binds double-stranded RNAs with structural characteristics of siRNAs and miRNAs. FEBS Lett. 2006;580:4401–4408. doi: 10.1016/j.febslet.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Hellborg F, Wiman KG. The p53-induced Wig-1 zinc finger protein is highly conserved from fish to man. Int J Oncol. 2004;24:1559–1564. [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Gillardon F, Spranger M, Tiesler C, Hossmann KA. Expression of cell death-associated phospho-c-Jun and p53-activated gene 608 in hippocampal CA1 neurons following global ischemia. Brain Res Mol Brain Res. 1999;73:138–143. doi: 10.1016/s0169-328x(99)00251-x. [DOI] [PubMed] [Google Scholar]
- Tomasevic G, Shamloo M, Israeli D, Wieloch T. Activation of p53 and its target genes p21(WAF1/Cip1) and PAG608/Wig-1 in ischemic preconditioning. Brain Res Mol Brain Res. 1999;70:304–313. doi: 10.1016/s0169-328x(99)00146-1. [DOI] [PubMed] [Google Scholar]
- Asanuma M, Miyazaki I, Higashi Y, Diaz-Corrales FJ, Shimizu M, Miyoshi K, et al. Suppression of p53-activated gene, PAG608, attenuates methamphetamine-induced neurotoxicity. Neurosci Lett. 2007;414:263–267. doi: 10.1016/j.neulet.2006.12.036. [DOI] [PubMed] [Google Scholar]
- Morimoto N, Nagai M, Miyazaki K, Ohta Y, Kurata T, Takehisa Y, et al. Induction of parkinsonism-related proteins in the spinal motor neurons of transgenic mouse carrying a mutant SOD1 gene. J Neurosci Res. 2010;88:1804–1811. doi: 10.1002/jnr.22341. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Miyazaki I, Higashi Y, Eslava-Alva MJ, Diaz-Corrales FJ, Asanuma M, et al. Specific induction of PAG608 in cranial and spinal motor neurons of L-DOPA-treated parkinsonian rats. Neurosci Res. 2008;60:355–363. doi: 10.1016/j.neures.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Higashi Y, Asanuma M, Miyazaki I, Haque ME, Fujita N, Tanaka K, et al. The p53-activated gene, PAG608, requires a zinc finger domain for nuclear localization and oxidative stress-induced apoptosis. J Biol Chem. 2002;277:42224–42232. doi: 10.1074/jbc.M203594200. [DOI] [PubMed] [Google Scholar]
- Enge M, Bao W, Hedstrom E, Jackson SP, Moumen A, Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15:171–183. doi: 10.1016/j.ccr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- Kriegebaum CB, Gutknecht L, Bartke L, Reif A, Buttenschon HN, Mors O, et al. The expression of the transcription factor FEV in adult human brain and its association with affective disorders. J Neural Transm. 2010;117:831–836. doi: 10.1007/s00702-010-0405-8. [DOI] [PubMed] [Google Scholar]
- Burda P, Laslo P, Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia. 2010;24:1249–1257. doi: 10.1038/leu.2010.104. [DOI] [PubMed] [Google Scholar]
- Hunger SP, Li S, Fall MZ, Naumovski L, Cleary ML. The proto-oncogene HLF and the related basic leucine zipper protein TEF display highly similar DNA-binding and transcriptional regulatory properties. Blood. 1996;87:4607–4617. [PubMed] [Google Scholar]
- Hsu SH, Hsieh-Li HM, Huang HY, Huang PH, Li H. bHLH-zip transcription factor Spz1 mediates mitogen-activated protein kinase cell proliferation, transformation, and tumorigenesis. Cancer Res. 2005;65:4041–4050. doi: 10.1158/0008-5472.CAN-04-3658. [DOI] [PubMed] [Google Scholar]
- Hsu SH, Shyu HW, Hsieh-Li HM, Li H. Spz1, a novel bHLH-Zip protein, is specifically expressed in testis. Mech Dev. 2001;100:177–187. doi: 10.1016/s0925-4773(00)00513-x. [DOI] [PubMed] [Google Scholar]
- Heng JC, Orlov YL, Ng HH.Transcription Factors for the Modulation of Pluripotency and Reprogramming Cold Spring Harb Symp Quant Biol 2010. e-pub ahead of print 3 November 2010. [DOI] [PubMed]
- Webber AL, Hodor P, Thut CJ, Vogt TF, Zhang T, Holder DJ, et al. Dual role of Nr2e3 in photoreceptor development and maintenance. Exp Eye Res. 2008;87:35–48. doi: 10.1016/j.exer.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Sugita M, Tanaka N, Davidson S, Sekiya S, Varella-Garcia M, West J, et al. Molecular definition of a small amplification domain within 3q26 in tumors of cervix, ovary, and lung. Cancer Genet Cytogenet. 2000;117:9–18. doi: 10.1016/s0165-4608(99)00135-1. [DOI] [PubMed] [Google Scholar]
- Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- Casas S, Aventin A, Fuentes F, Vallespi T, Granada I, Carrio A, et al. Genetic diagnosis by comparative genomic hybridization in adult de novo acute myelocytic leukemia. Cancer Genet Cytogenet. 2004;153:16–25. doi: 10.1016/j.cancergencyto.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Sakai N, Kajiyama Y, Iwanuma Y, Tomita N, Amano T, Isayama F, et al. Study of abnormal chromosome regions in esophageal squamous cell carcinoma by comparative genomic hybridization: relationship of lymph node metastasis and distant metastasis to selected abnormal regions. Dis Esophagus. 2010;23:415–421. doi: 10.1111/j.1442-2050.2009.01026.x. [DOI] [PubMed] [Google Scholar]
- Sattler HP, Lensch R, Rohde V, Zimmer E, Meese E, Bonkhoff H, et al. Novel amplification unit at chromosome 3q25-q27 in human prostate cancer. Prostate. 2000;45:207–215. doi: 10.1002/1097-0045(20001101)45:3<207::aid-pros2>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Singh B, Gogineni SK, Sacks PG, Shaha AR, Shah JP, Stoffel A, et al. Molecular cytogenetic characterization of head and neck squamous cell carcinoma and refinement of 3q amplification. Cancer Res. 2001;61:4506–4513. [PubMed] [Google Scholar]
- Or YY, Hui AB, Tam KY, Huang DP, Lo KW. Characterization of chromosome 3q and 12q amplicons in nasopharyngeal carcinoma cell lines. Int J Oncol. 2005;26:49–56. [PubMed] [Google Scholar]
- Soder AI, Hoare SF, Muir S, Going JJ, Parkinson EK, Keith WN. Amplification, increased dosage and in situ expression of the telomerase RNA gene in human cancer. Oncogene. 1997;14:1013–1021. doi: 10.1038/sj.onc.1201066. [DOI] [PubMed] [Google Scholar]
- Bea S, Ribas M, Hernandez JM, Bosch F, Pinyol M, Hernandez L, et al. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood. 1999;93:4365–4374. [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. Stemness': transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Keller MA, Addya S, Vadigepalli R, Banini B, Delgrosso K, Huang H, et al. Transcriptional regulatory network analysis of developing human erythroid progenitors reveals patterns of coregulation and potential transcriptional regulators. Physiol Genomics. 2006;28:114–128. doi: 10.1152/physiolgenomics.00055.2006. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci USA. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, et al. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process. Mol Endocrinol. 2006;20:1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Potireddy S, Vassena R, Patel BG, Latham KE. Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev Biol. 2006;298:155–166. doi: 10.1016/j.ydbio.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Prahl M, Vilborg A, Palmberg C, Jornvall H, Asker C, Wiman KG. The p53 target protein Wig-1 binds hnRNP A2/B1 and RNA Helicase A via RNA. FEBS Lett. 2008;582:2173–2177. doi: 10.1016/j.febslet.2008.04.065. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles. Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol Cell. 2004;96:479–498. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation. J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RA. mRNA stability control: a clandestine force in normal and malignant hematopoiesis. Leukemia. 2007;21:1158–1171. doi: 10.1038/sj.leu.2404656. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galban S, Martindale JL, Mazan-Mamczarz K, Lopez de Silanes I, Fan J, Wang W, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 2003;23:7083–7095. doi: 10.1128/MCB.23.20.7083-7095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Wilhelm MT, Wiman KG. Regulation of tumor suppressor p53 at the RNA level. J Mol Med. 2010;88:645–652. doi: 10.1007/s00109-010-0609-2. [DOI] [PubMed] [Google Scholar]
- Vilborg A, Glahder JA, Wilhelm MT, Bersani C, Corcoran M, Mahmoudi S, et al. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc Natl Acad Sci USA. 2009;106:15756–15761. doi: 10.1073/pnas.0900862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci USA. 2006;103:19842–19847. doi: 10.1073/pnas.0606343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Janicke RU, Sohn D, Schulze-Osthoff K. The dark side of a tumor suppressor: anti-apoptotic p53. Cell Death Differ. 2008;15:959–976. doi: 10.1038/cdd.2008.33. [DOI] [PubMed] [Google Scholar]
- Nindl I, Dang C, Forschner T, Kuban RJ, Meyer T, Sterry W, et al. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol Cancer. 2006;5:30. doi: 10.1186/1476-4598-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchstaller J, Sommer L, Bodmer M, Hoffmann R, Suter U, Mantei N. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci. 2004;24:2357–2365. doi: 10.1523/JNEUROSCI.4083-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]