Abstract
Evasion of apoptosis is considered to be one of the hallmarks of human cancers. This cell death modality is executed by caspases and several upstream regulatory factors, which direct their proteolytic activity, have been defined as either tumor suppressors or oncogenes. Often these regulatory factors, in addition to being potent apoptosis inducers, function in cell survival or repair signaling pathways in response to cellular stress. Thus, loss of function in a distinct regulatory mechanism does not necessarily mean that tumor formation is due to apoptosis malfunction resulting from insufficient caspase activation. Although each caspase has been assigned a distinct role in apoptosis, some redundancy with respect to their regulatory functions and substrate recognition is evident. Jointly, these proteases could be considered to possess solid tumor suppressor function, but what is the evidence that deregulation of specific caspases per se induces inappropriate cell survival, leading to enhanced tumorigenic potential? This question will be addressed in this review, which covers basic molecular mechanisms derived from in vitro analyses and emphasizes new insights that have emerged from in vivo and clinical studies.
Keywords: apoptosis, cancer, caspase, tumor suppressor
In organisms, excess cells during development, as well as potentially harmful cells resulting from pathophysiological conditions, are eliminated by various cell death modalities. Among them, apoptosis is the most investigated, and almost 40 years ago, it was described as a major defense strategy that prevents cells from acquiring tumorigenic potential.1 This suicide mechanism is controlled by multiple and interrelated pathways ensuring that caspases, the proteolytic initiators and executioners of apoptosis, are triggered only in cells requiring termination. There is overwhelming experimental evidence supporting the idea that disturbances in the regulation of caspase activation are central for the avoidance of cancer cell death, both in vitro and in vivo. However, what evidences suggest that caspases per se act as tumor suppressors? It was reported that cells do not necessarily undergo caspase-independent cell death in the absence of active caspases, but may instead survive insult and even promote clonogenic tumor growth.2 Yet, it was unclear which individual caspases could fulfill this function. Recently, certain initiator caspases have been suggested as putative tumor suppressor genes.3, 4 This review is focusing on caspases and their role in tumor suppression, and emphasizing new evidences that have emerged from in vivo observations and clinical material, taking into consideration supporting in vitro data that has enlightened basic molecular mechanisms.
Caspases
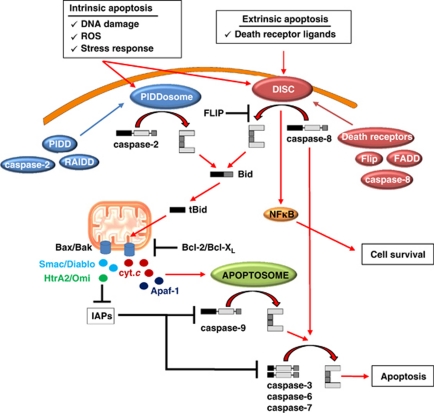
Classification of human caspases is based either on their function, the size of their pro-domain or cleavage specificity. Considering the first criteria, caspase-1, -4 and -5 belong to the group I (inflammatory) caspases that are involved in cytokine maturation. These proteases are primarily associated with the innate immunity against pathogens and, therefore, they fall outside the scope of this review. Regulation of apoptosis, on the other hand, is controlled by group II caspases, which are divided into two classes: initiator (apical) caspases (caspase-2, -8, -9 and -10) and effector (executioner) caspases (caspase-3, -6 and 7). Effector caspases are constitutively produced in cells as dimers and proteolytic processing by an initiator enzyme is required to trigger their activity (Figure 1). Being active, effector caspases target a broad spectrum of cellular proteins, ultimately leading to cell death. In contrast to effector caspases, initiator caspases are translated as monomeric zymogens. Formation of multi-component complexes triggers initiator caspase dimerization sufficient for their activation.5 The death-inducing signaling complex (DISC), the apoptosome and the p53-induced protein with a death domain (PIDD)osome are protein assembly platforms that can recruit caspase-8/-10, -9 and -2, respectively (Figure 1). The two main apoptosis pathways are the extrinsic receptor-mediated and intrinsic mitochondrial pathways. In the former, extracellular ligands stimulate receptor oligomerization and DISC assembly, whereas in the latter, proteins from the Bcl-2 family control the release of factors involved in apoptosome formation through preservation or disruption of mitochondrial integrity (Figure 1). The proteolytic activity of mature caspase-9 and -3 are inhibited by inhibitor of apoptosis proteins (IAPs). In turn, IAPs are inactivated and caspase activity restored by proteins, such as SMAC/Diablo or HtrA2/Omi, which are released from the mitochondria.
Figure 1.
Overview of caspase cascades and their inhibitors
Apoptosis as a Defense Strategy Against Tumorogenesis
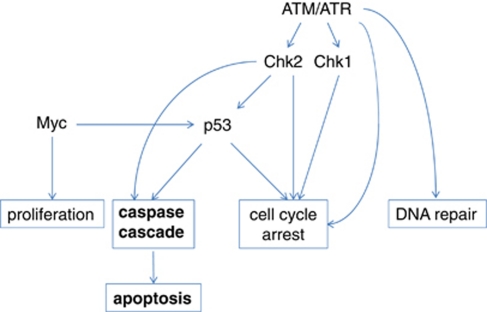
One of the pioneers of cell death research, Andrew Wyllie, suggested that pro-apoptotic genes might act as tumor suppressors, whereas oncogenes might fulfill antiapoptotic functions. Although this prediction turned out to be correct, most genes associated with tumorogenesis defined so far operate upstream of caspase activity regulating cell behavior during stress. For example, genomic damage initiates a delay in cell cycle progression and triggers the DNA repair response. Cell death through one of several modalities occurs only when a cell sustains severe damage that cannot be repaired. Several genes with oncogenic potential have been identified within the hierarchically ordered ataxia telangiectasia-mutated (ATM)/ataxia telangiectasia and Rad3-related (ATR)–checkpoint kinase 1/2 (Chk1/Chk2)–p53 signaling pathway, which mutually coordinates this process (Figure 2). DNA damage sensors transduce signals to ATM/ATR kinases, which in turn phosphorylate a large number of downstream substrates, including Chk1 and Chk2. Alterations in the tumor suppressor gene P53 are the most frequently observed genetic changes in clinical tumor samples, and epidemiological studies combined with molecular evidences have concluded that some ATM mutations correlate with breast cancer susceptibility.6 Neither Chk1+/− nor Chk2−/− mice show a predisposition to cancer, but it was recently revealed that Chk1/Chk2 double knockout mice have a progressive cancer-prone phenotype, indicating that these kinases have redundant functions and that either may be sufficient to mediate anti-tumor signaling.7
Figure 2.
Upstream regulatory pathways leading to apoptosis, cell cycle arrest, DNA repair and proliferation
Thus, apoptosis is only one of several possible outcomes modulated by the system maintaining genomic stability. Therefore, increased tumor susceptibility, due to loss of function in any of the regulatory proteins described above, may just as well be caused by inadequate cell cycle arrest, DNA synthesis or repair. A similar conclusion can be drawn with respect to other master regulatory signaling pathways, providing cells with multiple options during specific stress conditions. The Myc proto-oncoprotein synchronizes a number of physiological processes, including cell growth and division, metabolism and differentiation. Deregulation of MYC, as a consequence of carcinogenic events, forces cells into a hyperproliferative state. However, Myc activation also provokes intrinsic tumor suppressor mechanisms, including apoptosis, senescence and DNA damage responses that act as barriers to tumor development.8 Hence, although the MYC oncogene contributes to the progression of a variety of human cancers, this does not necessarily occur because of malfunctioned apoptotic signaling.
Tumor suppressor proteins and proto-oncogenes with a more direct effect on caspase activity can be found among p53-transregulated genes harboring apoptosis-specific functions. In human follicular lymphomas, the antiapoptotic BCL-2 gene is translocated to the immunoglobulin heavy-chain gene locus, resulting in constitutively high levels of functionally intact Bcl-2 protein, and enabling resistance of tumor cells to death.9 In addition, the soluble and non-signaling decoy receptor (DcR3) provides a mechanism for malignant cells to evade CD95 ligand-induced apoptosis in certain tumors and it was shown that elevated DcR3 expression can be used as a prognostic marker for survival of cancer patients.10, 11 The value of other caspase-regulating factors as prognostic markers has also been analyzed. In particular, downregulation of the mitochondrial IAP antagonist Smac/DIABLO is associated with renal cell carcinomas, and overexpression of survivin, another IAP, has been observed in most transformed cell lines and cancers tested to date, as compared with its absence in most normal adult tissues.12, 13 Although there is evidence suggesting an indirect caspase-9 processing interference by survivin,14 malfunctions of mitotic spindle checkpoint and aberrant progression of transformed cells may be due to non-apoptotic functions of this particular IAP.15 Association between X-linked inhibitor of apoptosis protein (XIAP) overexpression and poor clinical outcome has been reported for both renal cell carcinomas and acute myelogenous leukemia (AML). Yet, this correlation with AML was not confirmed by others.16 On the other hand, XAF1 (XIAP-associated factor 1), a ubiquitously expressed negative regulator of XIAP, is repressed in selected cancer cell lines,17 and XAF1 silencing by aberrant promoter methylation has been discovered in gastric tumors.18
The cellular FLICE inhibitory protein (c-FLIP) is a catalytically inactive homologue of caspase-8 and -10, which can prevent their activation by obstructing binding sites on the DISC. Multiple splice variants of c-FLIP have been reported, but three main isoforms, designated c-FLIPS, c-FLIPR and c-FLIPL, are detected on the protein level. Resistance to apoptosis induced by death receptors has been found to occur as a response to enhanced c-FLIPL expression in several human malignancies, and accordingly, it was shown that this variant can be used as a prognostic factor for certain cancer types.19 Interestingly, inhibition of procaspase-8 and -10 activations by c-FLIP may occur by more than one mechanism (for details see Shirley and Micheau19). Upon expression in non-small-cell lung cancer, c-FLIP evacuated DISC formation from lipid rafts to non-raft regions of the plasma membrane, thereby shifting initiation of caspase-8-mediated apoptosis towards cell survival through activation of NF-κB.20
Thus, several proteins involved in the complex signaling system controlling apoptosis have been reported as either oncogenes or tumor suppressors. Most of these are characterized by an ability to adjust the intrinsic or the extrinsic caspase cascade, or, in the case of certain IAPs, effector caspases. However, what do we know about the potential tumor suppressor functions of individual caspases?
CASP Polymorphism
The efficiency of apoptosis in an organism may be the consequence of polymorphisms in any gene regulating or executing apoptosis. Analysis of these polymorphisms may thus help to identify cancer patients with poor prognosis and tailor treatments accordingly. A few case-control studies have been performed, where the apoptotic capacity in cancer patients was evaluated and compared with healthy controls. Some data support the hypothesis that individuals with low apoptotic response are more prone to accumulate cells with potentially dangerous mutations, and thus have an increased risk of tumor manifestation.21 Evidences, which favor an alternative hypothesis, have also been presented, proposing that low apoptosis susceptibility may actually help to protect against cancer because of a more multi-faceted immune system.22 As it has been suggested that some gene polymorphisms may deregulate the expression or activity of caspases, an increasing number of reports have attempted to correlate caspase gene polymorphisms with cancer risk. A recent comprehensive review states that caspase gene polymorphisms may be involved in numerous tumor types.23 However, regardless of their importance as new prognostic biomarkers for tumor susceptibility, information regarding how these polymorphisms relate to protein function and expression is frequently missing. Moreover, most investigations are limited to one gene variation or multiple variations within the same gene. It is, therefore, possible that other, defined or undefined polymorphisms, which are not considered, contribute to the analysis outcome.
Because of the central role of caspase-8 in ligand-induced apoptosis of tumor cells triggered by cytotoxic T cells, more studies have been conducted investigating mutations and gene polymorphisms in CASP8 and their correlation with tumorogenesis than those focusing on other caspases. One such gene variation is a six-nucleotide deletion (−652 6N del) mapped to the CASP8 promoter. If present, a binding element for stimulatory protein 1 is eliminated and basal caspase-8 expression is reduced.24, 25 Even though conflicting evidences have been presented,26 this particular polymorphism has been associated with decreased risk of multiple cancers.25, 27 Similarly, a large study including 17 109 breast cancer patients (versus 16 423 controls) concluded that the CASP8 D302H polymorphism in some populations was associated with a reduced risk of breast cancer,28 a correlation that has been confirmed also for other tumor types.27 A CASP10 V410I substitution was also less frequently observed in patients suffering from breast cancer compared with control groups and the risk of carcinogenesis significantly decreased in individuals harboring a combination of these variant alleles, indicating a mutual protective effect.29 Although the CASP8 D302H polymorphism has not yet been functionally investigated, an in vitro expression study concluded that the V410I variant impairs caspase-10-mediated apoptosis.30 Although it may seem contradictory that some caspase mutations that decrease apoptotic potential can have tumor-protective effects, they may enable a more potent anti-tumor immune response owing to reduced apoptosis of tumor-infiltrating lymphocytes.25 Heterozygous mutations in CD95, CD95 ligand or caspase-10 trigger most cases of autoimmune lymphoproliferative syndrome (ALPS), a disorder that is characterized by impaired lymphocyte apoptosis, lymphadenopathy, splenomegaly and autoimmunity. Unlike individuals affected with ALPS, caspase-8 deficiency also leads to pleiotropic defects in lymphocyte activation, resulting in immunodeficiency.31 Thus, an individual may enhance their resistance to tumorogenesis from an alteration in caspase-8 activity as long as it is well balanced. More dramatic changes will cause unfavorable side effects.
It seems that although information regarding caspase polymorphism is important, it cannot be used as a main source for conclusion concerning the role of these enzymes in carcinogenesis. Mutational analyses of cell lines or in vivo, in combination with diverse apoptotic measurements, would provide useful tools to fully recapitulate the information provided by statistical gene variation studies.
Do Individual Caspases have Tumor Suppressor Functions?
Caspase-8
Initiator caspases are unique in their ability to be activated as a result of protein complex formation, with one specific caspase responding to one death stimulus. In fact, their sole obligatory role in apoptosis makes them good candidates for putative tumor suppressors. Several reports have described that CASP8 is silenced owing to gene deletion or promoter methylation, especially in pediatric tumors and their cell lines.32, 33 Loss of caspase-8 expression often correlates with amplification of the MYCN oncogene and elevated levels of the corresponding protein. However, it is not clear if these two genetic alterations are functionally linked or if they fit with a two-hit model providing a permissive environment for tumor growth. Gene methylation-independent reduction of caspase-8 expression has also been found in clinical glioma samples.34
The discovery of an antigen recognized by autologous cytolytic T lymphocytes in oral cavity human squamous cell carcinomas led to the identification of a CASP8 mutation that results in modification of the stop codon and adds an Alu repeat to the CASP8 coding region.35 CASP8 mutations have also been detected, albeit at relatively low frequency, in colorectal and gastric cancers.36, 37 An identical somatic frameshift mutation (1225–1226delTG) resulting in premature amino-acid synthesis termination in the p10 protease subunit was found in nine out of 69 patients suffering from hepatocellular carcinoma.38 Moreover, even though DISC-mediated apoptosis is considered to be an important regulator of the immune response by controlling the number of activated T- and B-cells, protection against this selection seems to ensure the maintenance of CD34+ hematopoietic progenitors.39 It appears that instead of the normal caspase-8a/b splice variants, hematopoietic stem cells express a smaller variant, caspase-8L, which acts as a dominant negative when recruited to DISC after CD95 triggering, thereby disrupting the link between CD95 and the caspase cascade. Because this caspase-8L isoform also has been found in samples obtained from AML and ALL patients, it has been hypothesized that caspase-8L participates in the development of stem cell-derived leukemias by providing protection from regulatory apoptosis.39 All caspase-8 alterations described result in reduced capacity to trigger apoptosis compared with normal caspase-8, strongly suggesting that caspase-8 deficiency might be a rationale for tumor outgrowth and that individual caspases indeed possess tumor suppressor functions (Table 1). Caspase-8 deficiency may have multiple effects on tumor development. Indeed, caspase-8 was shown to be involved in the suppression of oncogenic transformation, independent of its role in cell death induced by death receptors.4 Following continuous growth in culture, SV-40 T antigen immortalized caspase-8-null mouse embryonic fibroblasts (MEFs) became easily transformed, and in nude mice they have increased tumorogenic potential compared with MEFs derived from caspase-8 wild-type (wt) littermates. Because both wt and caspase-8-null cells underwent transformation, but the latter required fewer passages, it seems that loss of caspase-8 is not sufficient for transformations. Instead, the absence of caspase-8 might predispose cells to further oncogenic mutations, or allow spontaneous oncogenic changes to accumulate easier. The exact genetic mechanism(s) by which caspase-8 deficiency promotes transformation remain(s) to be investigated. Interestingly, the phosphorylation of caspase-8 on tyrosine 380 has been described as a switch trigger that inhibits proteolytic activation of the enzyme and enables functions that are uncoupled from apoptosis.40 Thus, caspase-8 has been found to associate with the p85 subunit of PI3-kinase and to mediate calpain-2 activity.41, 42 These results seem to be different faces of the same mechanism and together they strongly support a role for caspase-8 in cell adhesion and migration. Evidently, an imbalance between two caspase-8 modes of action (apoptotic versus migration) may lead to not only decreased apoptosis, but also to enhanced motility of affected cells. It is still not known to what extent the tyrosine 380 residue in caspase-8 is phosphorylated in tumors, but the PI3-kinase axis has been implicated in oncogenic progression, and considering the fact that selected tumors possess enhanced levels of caspase-8,43, 44 these findings are highly significant for the understanding of caspase-8 function.
Table 1. Mutations and imbalances of caspase expression reported in human tumors and cell lines.
| Caspase | Mutation | Protein expression | Cancer | Frequency | Reference |
|---|---|---|---|---|---|
| CASP8 | Promoter methylation | Low/absent | Childhood neuroblastomas | 11/42 | Teitz et al.32 |
| CASP8 | Nonsense | NA | Advanced gastric | 13/122 | Soung et al.37 |
| Missense | |||||
| Frameshift | |||||
| Deletion | |||||
| CASP8 | Nonsense | NA | Invasive colorectal | 5/98 | Kim et al.36 |
| Missense | |||||
| Frameshift | |||||
| CASP8 | Missense | NA | Head and neck | 1/1 | Mandruzzato et al.35 |
| CASP8 | Frameshift | NA | Hepatocellular | 9/69 | Soung et al.37 |
| Caspase-8 | NA | Low/absent | Lung and breast tumor cell lines | 6/55 | Kischkel et al.48 |
| Caspase-10 | NA | Low/absent | Lung and breast tumor cell lines | 31/55 | Kischkel et al.48 |
| Caspase-2 | NA | Low/absent | Gastric | 78/120 | Yoo et al.58 |
| Caspase-9 | NA | Low/absent | Colorectal | 12/26 | Palmerini82 |
| Caspase-6 | NA | Low/absent | Gastric | 57/120 | Yoo et al.58 |
| CASP7 | Nonsense | NA | Colorectal | 2/98 | Soung et al.81 |
| CASP7 | Missense | NA | Esophageal | 1/50 | Soung et al.81 |
| CASP7 | Missense | NA | Head and neck | 1/33 | Soung et al.81 |
| Caspase-7 | NA | Low/absent | Colorectal | 22/26 | Palmerini82 |
| Caspase-7 | NA | Low/absent | Gastric | 81/120 | Yoo et al.58 |
| Caspase-3 | NA | Low/absent | Breast | 23/31 | Devarajan et al.69 |
Abbreviation: NA, not analyzed
Caspase-10
Although both caspase-8 and -10 are involved in the extrinsic death receptor pathway, research to date has focused mainly on caspase-8, as shown by nearly 800 hits in a title-based PubMed search, compared with less than 40 publications for caspase-10. It has been shown that caspase-10 can function independently of caspase-8 in Fas- and TNF-related ligand-receptor-mediated apoptosis.45 Because overexpression of caspase-10 in caspase-8-deficient cells does not rescue these cells from apoptosis, it was proposed that caspase-10 is unable to fully substitute for caspase-8.46 In addition, to some extent caspase-8 and -10 appear to have distinct substrate cleavage specificities.47 These data indicate that DISC-mediated cell death involves not only caspase-8, but also caspase-10, and that each of these proteases may have unique roles in apoptosis initiation. In support of this, immunoblot analysis of 55 lung and breast carcinoma cell lines revealed that caspase-10 protein was more frequently absent than caspase-8 (Table 1), indicating possible selective pressure against caspase-10 expression in cancer cells.48 However, for a more general conclusion concerning the role of caspase-10 in tumorogenesis, this information is too limited and further in vivo observations are required.
Death effector domain (DED)-containing proteins, such as c-FLIP, FADD and caspase-8 and -10, in addition to regulating DISC-mediated apoptosis, may promote cell survival via their capacity to induce NF-κB activation.49, 50 Caspase-8 and -10 isoforms, which lack a protease domain, have been characterized and pro-domain-only polypeptides of caspase-10 have been reported to be pro-apoptotic in some experimental systems, but appeared to be antiapoptotic and capable of inducing NF-κB activity in others.51, 52 Because NF-κB is constitutively active in many tumors, it has been suggested that it might promote tumorogenesis by inducing transcription of antiapoptotic factors and proteins stimulating proliferation. Furthermore, NF-κB has been associated with angiogenesis and tumor metastasis.53 The correlation between DED-containing caspases, activation of the NF-κB pathway and tumorogenesis has not yet been examined; however, contribution from certain isoforms of caspase-8 or -10 to tumor growth cannot be excluded.
Caspase-2
Similarly to the observations described above for caspase-8-null mice, MEFs derived from caspase-2-deficient mice are resistant to cytotoxic drug- and γ-radiation-induced apoptosis and have a significantly faster growth rate than wt MEFs. Moreover, Eμ-Myc transgenic mice (which develop spontaneous lymphomas) crossed with caspase-2-deficient mice further accelerates tumor formation. Surprisingly, the loss of even one caspase-2 allele results in increased cell proliferation and accelerated tumorogenesis, suggesting a tumor suppression function for this enzyme.3
Like other genes involved in cell death regulation, caspase-2 generates two main splice variants (Casp-2L and Casp-2S), that fulfill either pro- or antiapoptotic functions, respectively.54 Transcriptional regulation by TAp73 and DNp73 might explain why Casp-2S accumulate as overexpression of both isoforms has been found in tumor cells concomitant with elevated Casp-2S levels.55 Silencing p73 expression in these cells led to reduced Casp-2S expression and increased cell death. These data raise the possibility that tumor suppression can be promoted by a caspase-2 splice variant. In addition, recent data indicate that the proteolytic activity of caspase-2 represses survivin transcription in tumor cells. Mechanistically, this pathway involves caspase-2 cleavage of the NF-κB activator, RIP1, which then shuts off transcription of NF-κB-responsive antiapoptotic genes, including survivin. In turn, acute loss of survivin causes mitotic defects, increased sensitivity to apoptosis and complete loss of tumorogenicity in vivo.56
Although primary MEFs from caspase-2 knockout mice show higher proliferation rates compared with those isolated from wt MEFs, transformation with oncogenic E1A/Ras further accelerated this process, suggesting that loss of caspase-2 leads to deregulation of the cell cycle, even before oncogenic transformation. The fact that both caspase-8 and -2 are involved in the same cell death pathway tempts us to speculate that the observed transformation might be a consequence of a deficiency in this death signaling pathway.57 Indeed, loss of caspase-2 expression has been observed in gastric cancer and the enzyme was proposed to be involved in pathogenesis via a deficient cell death program (Table 1).58
Notably, caspase-2-null MEFs are characterized by the loss of p53 function, leading to decreased p21 mRNA levels. Although no direct interaction between caspase-2 and p53 was found, p53-dependent activation of caspase-2 has been described and a possible reciprocal functional interaction between these proteins has been suggested. This assumption was based on several observations. First, decreased caspase-2 expression or activity was shown to affect both p53 stabilization and the expression of p53-inducible genes in response to DNA damage.59 Second, caspase-2 inhibitors affect silibinin-induced phosphorylation of Ser-139 in H2AX and Ser-15 in p53, without having any observable effect on ATM (Ser-1981) phosphorylation, suggesting a post-translational regulatory effect of caspase-2 on p53 and/or H2AX.60 Thus, these data imply the presence of a bi-directional functional connection between p53 and caspase-2. However, further investigation is required to clarify whether this link is responsible for the observed tumor suppressor function of caspase-2.
It is possible that the tumor suppressor function of caspase-2 is linked to its role in cell cycle regulation. Indeed, an enhanced caspase-2 activity within the DNA–PKcs–PIDDosome complex appears to delay the progression of G2 into the M phase in an ATM-dependent manner.61 Partial loss of the G2/M checkpoint has also been observed in caspase-2-deficient cells and in cells with downregulated DNA-PK, which might depend on its effect on p53, as caspase-2-null mice harbor reduced p21 expression owing to the loss of p53.
Caspase-2 localizes to the promyelocytic leukemia protein nuclear bodies (PML-NBs), which are unique as their tumor-suppressive functions may be attributed to both the protein and the conspicuous nuclear bodies that PML builds. Importantly, PML-NBs are involved in the regulation of apoptosis. Localization of caspase-2 in PML-NBs is suggested to require both the pro- and protease domains, but appears to be independent of adaptor proteins. It is unclear as to whether this interaction leads to caspase-2 activation or not. If not, then it is possible that formed caspase-2–PML-NBs complex preferentially operates as a tumor suppressor, rather than supports cell death. In agreement with this concept, high level of nuclear-inactive, non-processed caspase-2 together with caspase-3 was used as predictor of survival and complete remission in adults suffering from acute myeloblastic or lymphoblastic leukemias.62
Caspase-9
Although mutation or loss of CASP9 heterozygosity is not frequently observed in human cancers, it has been shown that abrogation of post-mitochondrial caspase activities facilitates oncogenic transformation and tumor progression (Table 1). MEFs retrovirally transduced with c-Myc from p53-, Apaf-1- and caspase-9-deficient mice were exposed to various death stimuli, chosen by their ability to mimic the conditions of tumor development. Using these settings, wtMEFs were sensitized to apoptosis by c-Myc, whereas inactivation of Apaf-1 or caspase-9 substituted for loss of p53 in promoting clonogenic survival and reduced the number of cells required for tumor formation in immune compromised mice.63 However, analyses of hematopoietic cell lines called into question the hypothesis that caspase-9 and Apaf-1 possess tumor suppressor functions.64, 65 In particular, in IL-3-dependent myeloid cell lines, Apaf-1, caspase-2 and caspase-9 are required for rapid, efficient apoptosis, but loss of these proteins does not enhance long-term survival of cells.65 Although no cell death protection was observed in mutant cells, several downstream apoptotic markers, including effector caspase processing and ICAD cleavage, were diminished. It was shown that the apoptosome is critical for tumor suppression in a cell-specific manner and that failure of apoptosome formation may be important for tumor cell survival only in cell types unable to switch their death program from apoptosis to caspase-independent cell death.2, 66 Mitochondrial outer membrane potential (MOMP)-dependent, caspase-independent cell death may be induced by damaged mitochondrial function or by the release of mitochondrial proteins, such as AIF, Omi/HtrA2 and endonuclease G. Cell survival in spite of MOMP, in contrast, requires recovery of mitochondrial function. Live cell imaging revealed that during apoptosis some mitochondria escape MOMP. This incomplete MOMP was observed in response to various stimuli and in different cell types, regardless of caspase activity. Interestingly, the presence of intact mitochondria associated with cellular recovery following MOMP requires silencing of caspase activity.67 These findings may have implications for oncogenesis in cells with reduced capacity to activate effector caspases.68 However, whether it happened in tissues prone to carcinogenesis is unclear.
Caspase-3
In a screen of primary breast tumor samples obtained from patients undergoing breast surgery, it was found that approximately 75% of the tumors as well as morphologically normal peritumoral tissue samples lacked caspase-3 transcripts and caspase-3 protein expression.69 In contrast, two reports describe upregulation of caspase-3 in clinical breast tumor samples.70, 71 The explanation for this discrepancy may be due to the fact that the CASP-3 gene can give rise to an alternative splicing variant known as caspase-3s, which has antiapoptotic function.72 Co-expression of caspase-3 and its splice variant has been found in diverse tumor cell lines, as well as in breast carcinomas, where the ratio of expression levels enabled identification of patients more likely to benefit from cyclophosphamide-containing chemotherapy.73 At present, the role of caspase-3 in tumor formation/progression and tumor sensitivity to treatment is still unclear.
Caspase-6
Caspase-6 is believed to be an effector caspase that is activated downstream of caspase-3 and -7 in apoptosome-mediated apoptosis.74 Apart from lamin A/C, few other specific substrates for caspase-6 have been described, calling into question to which extent this particular caspase actually participates in drug-induced apoptosis.75 However, it has been suggested that caspase-6 cleavage of tau protein leads to neurofibrillary tangle formation during Alzheimer's disease pathogenesis. Although caspase-3 seems to be the primary caspase in this case, colocalization with intracellular tau inclusions, in the form of neurophil threads and neurofibrillary tangles, and site-specific processing of tau protein also implicate caspase-6 in this process.76, 77 Moreover, accumulated caspase cleavage-mediated huntingtin fragments represent an early pathological change in the brains of Huntington's disease patients. Using a Huntington's disease mouse model that develops Huntington's-related characteristics with age, it was shown that individuals expressing a mutant huntingtin protein, which is resistant to caspase-6 cleavage, are protected from neuronal dysfunction.78 Taken together with the fact that very few reports have linked caspase-6 with tumorogenesis in vivo, these data argue for a specific role for caspase-6 in neurodegenerative diseases and aging, but not in cancer.
Caspase-7
Caspase-7 shares many similarities with caspase-3, both are effector caspases and are substrates for initiator caspases in extrinsic or intrinsic apoptotic pathways. Owing to their comparable specificity towards synthetic tetrapeptide substrates, these caspases are considered to be functionally redundant.79 However, these in vitro experimental results may be misleading, because they do not fully recapitulate caspase interactions with properly folded full-length substrates. Consequently, it is likely that caspase-3 and -7 do exhibit distinct activities in vivo, a hypothesis supported by immunodepletion experiments.80 The degree of involvement of CASP7 mutations in the development of human cancers has been analyzed by sequencing the entire coding region and all splice sites in solid cancers. CASP7 mutations were found in two out of 98 colon carcinomas, one out of 50 esophageal carcinomas and one out of 33 head/neck carcinomas. When these tumor-derived caspase-7 mutants were expressed in 293 T cells, apoptosis was reduced compared with controls.81 Moreover, immunohistochemistry analysis showed downregulation of caspase-7 and -9 in colon cancer samples compared with normal mucosa from the same tissue section (Table 1). No changes in the expression of caspase-8 were detected in these tumor samples.82 Even though effector caspases play significant roles in efficient apoptosis, malfunction of individual effector caspases are relatively infrequently reported as potential starting points for tumorogenesis. A plausible explanation for this discrepancy might be that effector caspases are able to perform redundant roles during the execution phase of apoptosis. Alternatively, because caspases seems to have rather distinct apoptotic roles, they might compensate for each other, but only in situations where inactivity of a certain caspase is caused by mutations or other deregulations.83 Caspase-7 knockout mice have a mild phenotype, and when MEFs originating from these animals were treated with inducers of apoptosis, they exhibited only a slight survival advantage compared with wtMEFs. This can be explained by a compensatory mechanism from caspase-3. In support of this, fibroblasts lacking both caspase-7 and -3 were highly resistant to extrinsic and intrinsic apoptosis.84 A third possibility is that deficiency in just one effector caspase fails to prevent apoptosis, which is supported by the fact that, although caspase-3 activity is absolutely required for DNA and nuclear fragmentation, caspase-3−/− hepatocytes undergo delayed CD95 receptor-mediated apoptosis, without nuclear events.83, 85
Concluding Remarks
Caspase activity is (in most cases) essential for apoptosis to occur and, accordingly, a multi-faceted regulatory system maintains their silence in normally dividing cells. In cells harboring defects in even a single caspase, compensatory mechanisms and alternative signaling pathways, including caspase-independent forms of apoptosis, may contribute to assure apoptotic termination. The observations discussed above underscore the notion that specific caspases can possess tumor suppressor functions, but, with the exception of caspase-8, in some tumor types mutations in other caspases are not commonly observed in human cancers (Table 1), and none of the caspase mouse knockouts analyzed under unperturbed conditions to date have been reported to be cancer prone. In fact, relatively little evidence supports a central role for individual caspases in tumorogenesis. Consequently, caspases might perform their tumor suppressor functions jointly, such that the combined apoptotic effect of all caspases, rather than any single members from this family of enzymes, is important for the prevention of tumorogenesis in an organism.
Acknowledgments
The work in the authors' laboratories was supported by grants from the Swedish Research Council, the Swedish and the Stockholm Cancer Societies, the Swedish Childhood Cancer Foundation, the EC FP-6 (Chemores) and the EC FP7 (Apo-Sys) programs. We apologize to authors whose primary references could not be cited owing to space limitation.
Glossary
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3 related
- Chk 1/2
checkpoint kinase 1/2
- DISC
death-inducing signaling complex
- IAP
inhibitor of apoptosis proteins
- MEFs
mouse embryonic fibroblasts
- PIDD
p53-induced protein with a death domain
- PML-NB
promyelocytic leukemia protein nuclear bodies
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Janssen K, Schulze-Osthoff K. Does caspase inhibition promote clonogenic tumor growth. Cell Cycle. 2007;6:3048–3053. doi: 10.4161/cc.6.24.5118. [DOI] [PubMed] [Google Scholar]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci USA. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krelin Y, Zhang L, Kang TB, Appel E, Kovalenko A, Wallach D. Caspase-8 deficiency facilitates cellular transformation in vitro. Cell Death Differ. 2008;15:1350–1355. doi: 10.1038/cdd.2008.88. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Ahmed M, Rahman N. ATM and breast cancer susceptibility. Oncogene. 2006;25:5906–5911. doi: 10.1038/sj.onc.1209873. [DOI] [PubMed] [Google Scholar]
- Niida H, Murata K, Shimada M, Ogawa K, Ohta K, Suzuki K, et al. Cooperative functions of Chk1 and Chk2 reduce tumour susceptibility in vivo. EMBO J. 2010;29:3558–3570. doi: 10.1038/emboj.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson LG, Henriksson MA. The Yin and Yang functions of the Myc oncoprotein in cancer development and as targets for therapy. Exp Cell Res. 2010;316:1429–1437. doi: 10.1016/j.yexcr.2010.03.025. [DOI] [PubMed] [Google Scholar]
- de Jong D. Molecular pathogenesis of follicular lymphoma: a cross talk of genetic and immunologic factors. J Clin Oncol. 2005;23:6358–6363. doi: 10.1200/JCO.2005.26.856. [DOI] [PubMed] [Google Scholar]
- Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P, et al. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396:699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- Takahama Y, Yamada Y, Emoto K, Fujimoto H, Takayama T, Ueno M, et al. The prognostic significance of overexpression of the decoy receptor for Fas ligand (DcR3) in patients with gastric carcinomas. Gastric Cancer. 2002;5:61–68. doi: 10.1007/s101200200011. [DOI] [PubMed] [Google Scholar]
- Mizutani Y, Nakanishi H, Yamamoto K, Li YN, Matsubara H, Mikami K, et al. Downregulation of Smac/DIABLO expression in renal cell carcinoma and its prognostic significance. J Clin Oncol. 2005;23:448–454. doi: 10.1200/JCO.2005.02.191. [DOI] [PubMed] [Google Scholar]
- Guha M, Altieri DC. Survivin as a global target of intrinsic tumor suppression networks. Cell Cycle. 2009;8:2708–2710. doi: 10.4161/cc.8.17.9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandele A, Prasad V, Jagtap JC, Shukla R, Shastry PR. Upregulation of survivin in G2/M cells and inhibition of caspase 9 activity enhances resistance in staurosporine-induced apoptosis. Neoplasia. 2004;6:29–40. doi: 10.1016/s1476-5586(04)80051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Welm A, Bishop JM. Cell division and cell survival in the absence of survivin. Proc Natl Acad Sci USA. 2004;101:15100–15105. doi: 10.1073/pnas.0406665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer AD, Dalili S, Batey RA, Riedl SJ. Targeting XIAP for the treatment of malignancy. Cell Death Differ. 2006;13:179–188. doi: 10.1038/sj.cdd.4401826. [DOI] [PubMed] [Google Scholar]
- Liston P, Fong WG, Kelly NL, Toji S, Miyazaki T, Conte D, et al. Identification of XAF1 as an antagonist of XIAP anti-caspase activity. Nat Cell Biol. 2001;3:128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- Byun DS, Cho K, Ryu BK, Lee MG, Kang MJ, Kim HR, et al. Hypermethylation of XIAP-associated factor 1, a putative tumor suppressor gene from the 17p13.2 locus, in human gastric adenocarcinomas. Cancer Res. 2003;63:7068–7075. [PubMed] [Google Scholar]
- Shirley S, Micheau O.Targeting c-FLIP in cancer Cancer Lett 2010. e-pub ahead of print 9 November 2010; PMID: 21071136. [DOI] [PubMed]
- Song JH, Tse MC, Bellail A, Phuphanich S, Khuri F, Kneteman NM, et al. Lipid rafts and nonrafts mediate tumor necrosis factor related apoptosis-inducing ligand induced apoptotic and nonapoptotic signals in non small cell lung carcinoma cells. Cancer Res. 2007;67:6946–6955. doi: 10.1158/0008-5472.CAN-06-3896. [DOI] [PubMed] [Google Scholar]
- Wang LE, Cheng L, Spitz MR, Wei Q. Fas A670G polymorphism, apoptotic capacity in lymphocyte cultures, and risk of lung cancer. Lung Cancer. 2003;42:1–8. doi: 10.1016/s0169-5002(03)00276-9. [DOI] [PubMed] [Google Scholar]
- Imyanitov EN. Gene polymorphisms, apoptotic capacity and cancer risk. Hum Genet. 2009;125:239–246. doi: 10.1007/s00439-009-0636-7. [DOI] [PubMed] [Google Scholar]
- Ghavami S, Hashemi M, Ande SR, Yeganeh B, Xiao W, Eshraghi M, et al. Apoptosis and cancer: mutations within caspase genes. J Med Genet. 2009;46:497–510. doi: 10.1136/jmg.2009.066944. [DOI] [PubMed] [Google Scholar]
- Liedtke C, Groger N, Manns MP, Trautwein C. The human caspase-8 promoter sustains basal activity through SP1 and ETS-like transcription factors and can be up-regulated by a p53-dependent mechanism. J Biol Chem. 2003;278:27593–27604. doi: 10.1074/jbc.M304077200. [DOI] [PubMed] [Google Scholar]
- Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–613. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- Haiman CA, Garcia RR, Kolonel LN, Henderson BE, Wu AH, Le Marchand L.A promoter polymorphism in the CASP8 gene is not associated with cancer risk Nat Genet 200840259–260.author reply 260–251. [DOI] [PubMed] [Google Scholar]
- Li C, Zhao H, Hu Z, Liu Z, Wang LE, Gershenwald JE, et al. Genetic variants and haplotypes of the caspase-8 and caspase-10 genes contribute to susceptibility to cutaneous melanoma. Hum Mutat. 2008;29:1443–1451. doi: 10.1002/humu.20803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- Frank B, Hemminki K, Wappenschmidt B, Meindl A, Klaes R, Schmutzler RK, et al. Association of the CASP10 V410I variant with reduced familial breast cancer risk and interaction with the CASP8 D302H variant. Carcinogenesis. 2006;27:606–609. doi: 10.1093/carcin/bgi248. [DOI] [PubMed] [Google Scholar]
- Park WS, Lee JH, Shin MS, Park JY, Kim HS, Kim YS, et al. Inactivating mutations of the caspase-10 gene in gastric cancer. Oncogene. 2002;21:2919–2925. doi: 10.1038/sj.onc.1205394. [DOI] [PubMed] [Google Scholar]
- Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- Harada K, Toyooka S, Shivapurkar N, Maitra A, Reddy JL, Matta H, et al. Deregulation of caspase 8 and 10 expression in pediatric tumors and cell lines. Cancer Res. 2002;62:5897–5901. [PubMed] [Google Scholar]
- Ashley DM, Riffkin CD, Muscat AM, Knight MJ, Kaye AH, Novak U, et al. Caspase 8 is absent or low in many ex vivo gliomas. Cancer. 2005;104:1487–1496. doi: 10.1002/cncr.21323. [DOI] [PubMed] [Google Scholar]
- Mandruzzato S, Brasseur F, Andry G, Boon T, van der Bruggen P. A CASP-8 mutation recognized by cytolytic T lymphocytes on a human head and neck carcinoma. J Exp Med. 1997;186:785–793. doi: 10.1084/jem.186.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Lee JW, Soung YH, Park WS, Kim SY, Lee JH, et al. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology. 2003;125:708–715. doi: 10.1016/s0016-5085(03)01059-x. [DOI] [PubMed] [Google Scholar]
- Soung YH, Lee JW, Kim SY, Jang J, Park YG, Park WS, et al. CASPASE-8 gene is inactivated by somatic mutations in gastric carcinomas. Cancer Res. 2005;65:815–821. [PubMed] [Google Scholar]
- Soung YH, Lee JW, Kim SY, Sung YJ, Park WS, Nam SW, et al. Caspase-8 gene is frequently inactivated by the frameshift somatic mutation 1225_1226delTG in hepatocellular carcinomas. Oncogene. 2005;24:141–147. doi: 10.1038/sj.onc.1208244. [DOI] [PubMed] [Google Scholar]
- Mohr A, Zwacka RM, Jarmy G, Buneker C, Schrezenmeier H, Dohner K, et al. Caspase-8L expression protects CD34+ hematopoietic progenitor cells and leukemic cells from CD95-mediated apoptosis. Oncogene. 2005;24:2421–2429. doi: 10.1038/sj.onc.1208432. [DOI] [PubMed] [Google Scholar]
- Cursi S, Rufini A, Stagni V, Condo I, Matafora V, Bachi A, et al. Src kinase phosphorylates Caspase-8 on Tyr380: a novel mechanism of apoptosis suppression. EMBO J. 2006;25:1895–1905. doi: 10.1038/sj.emboj.7601085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM. Caspase-8: fly or die. Cancer Res. 2008;68:4491–4493. doi: 10.1158/0008-5472.CAN-08-0952. [DOI] [PubMed] [Google Scholar]
- Finlay D, Howes A, Vuori K. Critical role for caspase-8 in epidermal growth factor signaling. Cancer Res. 2009;69:5023–5029. doi: 10.1158/0008-5472.CAN-08-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupack DG.Caspase-8 as a therapeutic target in cancer Cancer Lett 2010. e-pub ahead of print 1 September 2010; PMID: 20817393. [DOI] [PMC free article] [PubMed]
- Ishizawar RC, Tice DA, Karaoli T, Parsons SJ. The C terminus of c-Src inhibits breast tumor cell growth by a kinase-independent mechanism. J Biol Chem. 2004;279:23773–23781. doi: 10.1074/jbc.M312368200. [DOI] [PubMed] [Google Scholar]
- Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ. Caspase-10 is an initiator caspase in death receptor signaling. Proc Natl Acad Sci USA. 2001;98:13884–13888. doi: 10.1073/pnas.241358198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H. Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J. 2002;21:4520–4530. doi: 10.1093/emboj/cdf441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Stroh C, Schulze-Osthoff K. Unique and overlapping substrate specificities of caspase-8 and caspase-10. Oncogene. 2006;25:152–159. doi: 10.1038/sj.onc.1209015. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P, et al. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J Biol Chem. 2001;276:46639–46646. doi: 10.1074/jbc.M105102200. [DOI] [PubMed] [Google Scholar]
- Hu WH, Johnson H, Shu HB. Activation of NF-kappaB by FADD, Casper, and caspase-8. J Biol Chem. 2000;275:10838–10844. doi: 10.1074/jbc.275.15.10838. [DOI] [PubMed] [Google Scholar]
- Chaudhary PM, Eby MT, Jasmin A, Kumar A, Liu L, Hood L. Activation of the NF-kappaB pathway by caspase 8 and its homologs. Oncogene. 2000;19:4451–4460. doi: 10.1038/sj.onc.1203812. [DOI] [PubMed] [Google Scholar]
- Shikama Y, Yamada M, Miyashita T. Caspase-8 and caspase-10 activate NF-kappaB through RIP, NIK and IKKalpha kinases. Eur J Immunol. 2003;33:1998–2006. doi: 10.1002/eji.200324013. [DOI] [PubMed] [Google Scholar]
- Shikama Y, Shen L, Yonetani M, Miyauchi J, Miyashita T, Yamada M. Death effector domain-only polypeptides of caspase-8 and -10 specifically inhibit death receptor-induced cell death. Biochem Biophys Res Commun. 2002;291:484–493. doi: 10.1006/bbrc.2002.6482. [DOI] [PubMed] [Google Scholar]
- Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Mol Cell Biochem. 2010;336:25–37. doi: 10.1007/s11010-009-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Dupuis S, Jiang Z, Wu JY. Caspase-2 pre-mRNA alternative splicing: identification of an intronic element containing a decoy 3′ acceptor site. Proc Natl Acad Sci USA. 2001;98:938–943. doi: 10.1073/pnas.031564098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh WH, Logette E, Corcos L, Sabapathy K. TAp73beta and DNp73beta activate the expression of the pro-survival caspase-2S. Nucleic Acids Res. 2008;36:4498–4509. doi: 10.1093/nar/gkn414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M, Xia F, Raskett CM, Altieri DC. Caspase 2-mediated tumor suppression involves survivin gene silencing. Oncogene. 2010;29:1280–1292. doi: 10.1038/onc.2009.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Vakifahmetoglu H, Abruzzo PM, Hogstrand K, Grandien A, Zhivotovsky B. DISC-mediated activation of caspase-2 in DNA damage-induced apoptosis. Oncogene. 2009;28:1949–1959. doi: 10.1038/onc.2009.36. [DOI] [PubMed] [Google Scholar]
- Yoo NJ, Lee JW, Kim YJ, Soung YH, Kim SY, Nam SW, et al. Loss of caspase-2, -6 and -7 expression in gastric cancers. APMIS. 2004;112:330–335. doi: 10.1111/j.1600-0463.2004.apm1120602.x. [DOI] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M, Orrenius S, Zhivotovsky B. Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene. 2006;25:5683–5692. doi: 10.1038/sj.onc.1209569. [DOI] [PubMed] [Google Scholar]
- Tyagi A, Singh RP, Agarwal C, Agarwal R. Silibinin activates p53-caspase 2 pathway and causes caspase-mediated cleavage of Cip1/p21 in apoptosis induction in bladder transitional-cell papilloma RT4 cells: evidence for a regulatory loop between p53 and caspase 2. Carcinogenesis. 2006;27:2269–2280. doi: 10.1093/carcin/bgl098. [DOI] [PubMed] [Google Scholar]
- Shi M, Vivian CJ, Lee KJ, Ge C, Morotomi-Yano K, Manzl C, et al. DNA–PKcs–PIDDosome: a nuclear caspase-2-activating complex with role in G2/M checkpoint maintenance. Cell. 2009;136:508–520. doi: 10.1016/j.cell.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sanchez-Pulido L, Valencia A, Rojas AM. Are promyelocytic leukaemia protein nuclear bodies a scaffold for caspase-2 programmed cell death. Trends Biochem Sci. 2007;32:400–406. doi: 10.1016/j.tibs.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Soengas MS, Alarcon RM, Yoshida H, Giaccia AJ, Hakem R, Mak TW, et al. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- Marsden VS, O'Connor L, O'Reilly LA, Silke J, Metcalf D, Ekert PG, et al. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf-1/caspase-9 apoptosome. Nature. 2002;419:634–637. doi: 10.1038/nature01101. [DOI] [PubMed] [Google Scholar]
- Ekert PG, Read SH, Silke J, Marsden VS, Kaufmann H, Hawkins CJ, et al. Apaf-1 and caspase-9 accelerate apoptosis, but do not determine whether factor-deprived or drug-treated cells die. J Cell Biol. 2004;165:835–842. doi: 10.1083/jcb.200312031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen K, Pohlmann S, Janicke RU, Schulze-Osthoff K, Fischer U. Apaf-1 and caspase-9 deficiency prevents apoptosis in a Bax-controlled pathway and promotes clonogenic survival during paclitaxel treatment. Blood. 2007;110:3662–3672. doi: 10.1182/blood-2007-02-073213. [DOI] [PubMed] [Google Scholar]
- Tait SW, Parsons MJ, Llambi F, Bouchier-Hayes L, Connell S, Munoz-Pinedo C, et al. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev Cell. 2010;18:802–813. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- Devarajan E, Sahin AA, Chen JS, Krishnamurthy RR, Aggarwal N, Brun AM, et al. Down-regulation of caspase 3 in breast cancer: a possible mechanism for chemoresistance. Oncogene. 2002;21:8843–8851. doi: 10.1038/sj.onc.1206044. [DOI] [PubMed] [Google Scholar]
- O'Donovan N, Crown J, Stunell H, Hill AD, McDermott E, O'Higgins N, et al. Caspase 3 in breast cancer. Clin Cancer Res. 2003;9:738–742. [PubMed] [Google Scholar]
- Nakopoulou L, Alexandrou P, Stefanaki K, Panayotopoulou E, Lazaris AC, Davaris PS. Immunohistochemical expression of caspase-3 as an adverse indicator of the clinical outcome in human breast cancer. Pathobiology. 2001;69:266–273. doi: 10.1159/000064337. [DOI] [PubMed] [Google Scholar]
- Huang Y, Shin NH, Sun Y, Wang KK. Molecular cloning and characterization of a novel caspase-3 variant that attenuates apoptosis induced by proteasome inhibition. Biochem Biophys Res Commun. 2001;283:762–769. doi: 10.1006/bbrc.2001.4871. [DOI] [PubMed] [Google Scholar]
- Vegran F, Boidot R, Oudin C, Riedinger JM, Bonnetain F, Lizard-Nacol S. Overexpression of caspase-3s splice variant in locally advanced breast carcinoma is associated with poor response to neoadjuvant chemotherapy. Clin Cancer Res. 2006;12:5794–5800. doi: 10.1158/1078-0432.CCR-06-0725. [DOI] [PubMed] [Google Scholar]
- Inoue S, Browne G, Melino G, Cohen GM. Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ. 2009;16:1053–1061. doi: 10.1038/cdd.2009.29. [DOI] [PubMed] [Google Scholar]
- Loegering DA, Ruchaud S, Earnshaw WC, Kaufmann SH. Evaluation of the role of caspase-6 in anticancer drug-induced apoptosis. Cell Death Differ. 2006;13:346–347. doi: 10.1038/sj.cdd.4401791. [DOI] [PubMed] [Google Scholar]
- Horowitz PM, Patterson KR, Guillozet-Bongaarts AL, Reynolds MR, Carroll CA, Weintraub ST, et al. Early N-terminal changes and caspase-6 cleavage of tau in Alzheimer's disease. J Neurosci. 2004;24:7895–7902. doi: 10.1523/JNEUROSCI.1988-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC. Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am J Pathol. 2004;165:523–531. doi: 10.1016/S0002-9440(10)63317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276:7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- Soung YH, Lee JW, Kim HS, Park WS, Kim SY, Lee JH, et al. Inactivating mutations of CASPASE-7 gene in human cancers. Oncogene. 2003;22:8048–8052. doi: 10.1038/sj.onc.1206727. [DOI] [PubMed] [Google Scholar]
- Palmerini F, Devilard E, Jarry A, Birg F, Xerri L. Caspase 7 downregulation as an immunohistochemical marker of colonic carcinoma. Hum Pathol. 2001;32:461–467. doi: 10.1053/hupa.2001.24328. [DOI] [PubMed] [Google Scholar]
- Zheng TS, Hunot S, Kuida K, Momoi T, Srinivasan A, Nicholson DW, et al. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat Med. 2000;6:1241–1247. doi: 10.1038/81343. [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GA, Jr, Booth CJ, Mehal WZ, et al. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M, Hakem A, Elia AJ, Hakem R, Duncan GS, Patterson BJ, et al. In vivo evidence that caspase-3 is required for Fas-mediated apoptosis of hepatocytes. J Immunol. 1999;163:4909–4916. [PubMed] [Google Scholar]