Abstract
Ubiquitin modification of many cellular proteins targets them for proteasomal degradation, but in addition can also serve non-proteolytic functions. Over the last years, a significant progress has been made in our understanding of how modification of the substrates of the ubiquitin system is regulated. However, little is known on how the ubiquitin system that is comprised of ∼1500 components is regulated. Here, we discuss how the biggest subfamily within the system, that of the E3 ubiquitin ligases that endow the system with its high specificity towards the numerous substrates, is regulated and in particular via self-regulation mediated by ubiquitin modification. Ligases can be targeted for degradation in a self-catalyzed manner, or through modification mediated by an external ligase(s). In addition, non-proteolytic functions of self-ubiquitination, for example activation of the ligase, of E3s are discussed.
Keywords: ubiquitin, E3 ligase, self-ubiquitination, proteasomal degradation
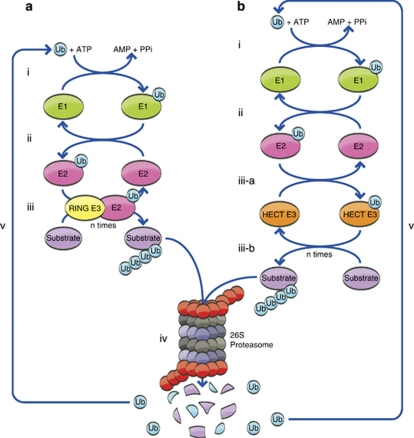
One of the major roles of the covalent modification of cellular proteins by ubiquitin is signaling them for proteasomal degradation (Figure 1). The first step of the modification is catalyzed by the ubiquitin-activating enzyme, E1, which generates a high-energy thiol ester intermediate that is subsequently transferred to the second enzyme, a ubiquitin-conjugating enzyme, E2. The third step ascertains substrate specificity, and is catalyzed by one of the numerous (∼650) ubiquitin ligases, E3s. Typically, it results in the formation of an isopeptide bond between the C-terminal Gly of ubiquitin and an ɛ-NH2 group of an internal Lys of the substrate. Less frequently, it can generate a linear peptide bond with the α-NH2 group, a thiol ester bond with an internal Cys, or an ester bond with a Thr or Ser. The three-step cascade of reactions is repeated, where additional ubiquitin moieties are attached sequentially to one another in an isopeptide bond involving one of the seven internal Lys residues in the ubiquitin moiety, thus generating a polyubiquitin chain. Lys48-based chains serve as a signal for proteasomal degradation, whereas chains based on other internal Lys residues, or modification by single moiety(ies) can serve non-proteolytic functions.
Figure 1.
The ubiquitin-proteasome system. Conjugation of ubiquitin catalyzed by RING (a) or HECT (b) domain-containing ligases. (ai, bi) ATP-dependent activation of ubiquitin catalyzed by the ubiquitin-activating enzyme, E1. (aii, bii) Transfer of the activated ubiquitin to a ubiquitin-carrier protein (ubiquitin-conjugating enzyme), E2. In the case of a RING ligase, the ubiquitin-charged E2 binds to the E3 and transfers the activated ubiquitin moiety directly to the substrate that is also bound to the E3 (aiii). In the case of a HECT domain ligase, ubiquitin is transferred from the E2 to a Cys residue in the E3 (biii-a) and then to the substrate (biii-b). (iv and v) The conjugated substrate is degraded to short peptides by the 26S proteasome (iv) with release of free and reusable ubiquitin mediated by DUB(s) (v). Some of the ubiquitin is degraded in this process along with the substrate (iv)
Ligases fall into two main families: RING (really interesting new gene) and HECT (homologous to the E6-AP carboxy terminus) domain-containing E3s. RING ligases serve as scaffolds that facilitate direct transfer of ubiquitin from the E2 to the target protein. HECT E3s contain an active Cys residue to which ubiquitin binds prior to its transfer to the substrate (Figure 1). There are ∼600 RING finger and ∼30 HECT ligases in humans. Smaller families of ligases (U-box, plant homology domain, and zinc finger) have also been described.
An important problem relates to regulation of the ubiquitin system components, and in particular to that of the ligases that are the specific substrate-recognizing elements.1, 2 Phosphorylation of an E3 can activate the protein, such as the case for CBL RING E3s,3 and for the anaphase-promoting complex/cyclosome (APC/C), which assembles with its substrate receptor in a phosphorylation-dependent manner.4 In contrast, phosphorylation of NEDD4-2 (neural precursor cell-expressed developmentally downregulated 4-2) inactivates it by preventing the binding to its substrate, ENaC (epithelial sodium channel).5 In addition, other mechanisms of regulation exist, such as intermolecular and intramolecular interactions. Examples include p19/Arf (alternative reading frame) that binds to mouse double minute (Mdm2) and inhibits p53 conjugation,6 and the polycomb group (PcG) protein BMI1 that stimulates the histone H2A ubiquitinating activity of RING1B.7, 8 Another way in which E3 activity can be modulated is via modification by ubiquitin (see below) and ubiquitin-like proteins, such as NEDD8. Conjugation of NEDD8 to the Cullin subunit of a Cullin-RING ligase (CRL) complex results in a conformational change that facilitates transfer of ubiquitin from the RING-bound E2 to the substrate.9, 10
A relatively unexplored area is how the degradation of the different ligases is regulated. E3s can be degraded by the proteasome via two main mechanisms – self-catalyzed ubiquitination and/or the activity of an exogenous ligase.
Degradation of Ligases via Self-catalyzed Ubiquitination
A typical feature of most ligases is the ability to catalyze their own ubiquitination.11, 12 Although this feature is widely used to follow E3s activity, the detailed molecular mechanisms and functional consequences have remained largely elusive. In particular, it is not clear whether the reaction is intermolecular or intramolecular. For several ligases, including E6-AP (E6-associated protein),13 and the RING ligase SIAH1 (seven in absentia 1),14 intermolecular transfer of ubiquitin has been observed. Consistently, some self-ubiquitinating RING ligases such as SIAH1 and TRAF (TNF receptor-associated factor) 6, have been detected as homodimers, and dimerization was found to be essential for the self-ubiquitination of TRAF6.14, 15 In other cases, self-ubiquitination could not be catalyzed in trans, implying that it is possibly an intramolecular event. This was described, for example, for the self-ubiquitination of the HECT ligase Rsp5,12 and the F-box protein Grr1p.16 Consistent with the concept of intramolecular modification, it appears that the self-ubiquitinating activity of ITCH and other HECT ligases like NEDD4-1, NEDD4-2, SMURF2, and WWP1, is regulated through intramolecular interactions that are modulated by modifications such as phosphorylation, and that involve the HECT domain.17, 18, 19
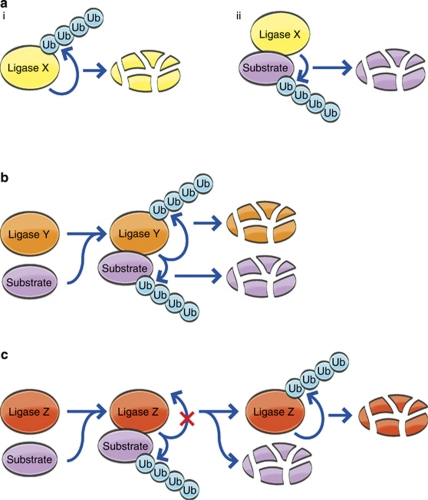
In an attempt to decipher the biological role of self-ubiquitination, it was proposed that it serves to target the ligase for degradation,11 which has been observed indeed as a means of negative feedback for Mdm2,6, 20 E6-AP,21 CBL ligases,22 and the substrate receptor subunits of CRL complexes.23 Self-ubiquitination can occur in substrate-independent24 (Figure 2a) and -dependent modes22 (Figure 2b). Also, binding to the substrate can protect the ligase from self-destruction23 (Figure 2c).
Figure 2.
Self-ubiquitination and degradation of E3 ligases occur in three substrate-related modes: (a) Self-ubiquitination and degradation of the ligase (i) are independent from ubiquitination of the substrate (ii). (b) Self-ubiquitination occurs concomitantly with ubiquitination of the substrate. (c) Inhibition of self-ubiquitination by the substrate
Substrate-independent self-ubiquitination of Mdm2
Due to its role as a ligase for the tumor suppressor protein p53, Mdm2 is probably one of the most studied ubiquitin ligases. RING-dependent self-ubiquitination of Mdm2 has been observed in vitro and upon ectopic expression in cells.6, 20 Self-ubiquitination of Mdm2 appears to be regulated in numerous ways that include DNA damage-induced phosphorylation, SUMOylation, and association with its partners, among them DAXX (death-associated protein 6), the Mdm2 homologue MdmX, and its inhibitor Arf. In addition, this modification can be reversed by deubiquitinating enzymes (DUBs) such as USP7.25 Interestingly, self-ubiquitination of Mdm2 appears to be independent of its activity towards p53 (Figure 2a). A chimeric mutant of Mdm2 in which the RING domain was replaced with that of PRAJA1, lost its ability to ubiquitinate p53, but retained its self-ubiquitinating activity.20 In vitro, binding to MdmX changes Mdm2 substrate preference by increasing its activity towards p53, while reducing its self-ubiquitination.24
It should be noted however that in embryonic fibroblasts derived from knock-in mice carrying a RING-inactivating mutation in Mdm2, no difference was observed in proteasome-dependent degradation rates of the mutant protein compared with that of the WT ligase.26 In the same cells, however, overexpressed HDM2 did undergo proteasomal degradation, which was dependent on its own RING activity, suggesting that the mechanisms of Mdm2 degradation in cells are dependent on its level. It appears therefore that under physiological levels, Mdm2 is targeted for degradation by an external ligase. Consistently, it was recently reported that the histone acetyl transferase p300-CBP-associated factor ubiquitinates Mdm2, resulting in its proteasomal degradation.27 At high levels of expression, however, Mdm2 directs its own ubiquitination and subsequent proteasomal degradation. One possible explanation for this concentration dependence is that self-ubiquitination of Mdm2 may occur in trans, which requires the generation of a large enough concentration of Mdm2 homodimers. It seems therefore that self-induced degradation of Mdm2 serves as a backup mechanism that occurs only when its level exceeds a certain threshold. It should be noted that these observations reveal an important caveat in the mechanistic analysis of self-ubiquitination of ligases, and should be taken into careful consideration when interpreting the functions and consequences of ‘suicide' of E3s based on overexpression or in vitro reconstitution experiments.
Degradation of CBL ligases along with their substrates
In some cases, ligases have been reported to be degraded along with their substrates (Figure 2b). CBL proteins regulate a wide array of signal transduction pathways, many of which involve targeting of receptor and non-receptor tyrosine kinases (TKs).22 Tyrosine phosphorylation of CBL proteins catalyzed by their TK substrates induces a conformational change, which results in activation of the ligase towards the substrate and itself.3, 22 Activation of the CBL substrate epidermal growth factor receptor (EGF-R) leads to a rapid decrease in the level of the EGF-R concomitant with a decrease in c-CBL, CBL-b, and CBL-c.22, 28 Similar decrease in CBL-b and c-CBL levels were observed after stimulation of the KIT receptor TK.29 In both cases, the degradation of the receptor and the CBL ligase was dependent on the RING domain of the E3. However, the exact mechanism of degradation of the two proteins remains elusive, as it appears sensitive to inhibitors of both the proteasome and lysosomes.28, 29, 30 Given the large number of substrates targeted by CBL proteins, a substrate-coupled activation and subsequent degradation of CBL proteins as observed for the EGF and KIT receptors, might serve to avoid undesired targeting of other substrates by the activated CBL, ensuring that the specificity of the signaling pathway is maintained. A similar relationship between the target substrate and self-ubiquitination has been observed for NEDD4 that undergoes more efficient ubiquitination in cells following co-expression of its substrate ENaC. Here, it was proposed that binding of ENaC to NEDD4 abolishes an autoinhibitory intramolecular interaction between the WW domains and a PY motif in the HECT domain of the ligase.18
Not in all cases, CBL is targeted for degradation along with its substrate. For example, whereas ligand engagement of the colony stimulating factor receptor results in ubiquitination of c-CBL, this ubiquitination does not target it for degradation, but rather serves as a membrane targeting signal.31 Thus, the susceptibility of CBL proteins to degradation along with their substrate appears to be substrate specific. Alternatively, the different fates of the CBL proteins can be determined by adaptor proteins that regulate CBL self-ubiquitination. ALIX associates with the EGF-R and the platelet-derived growth factor receptor (PDGF-R), and enhances the binding of CBL to the complex. In both cases, ALIX inhibits the ubiquitination of the receptor, and in the case of the EGF-R, also the ubiquitination of CBL.32, 33 However, in the case of the PDGF-R, ALIX stimulates the phosphorylation and proteasomal degradation of c-CBL.33 What causes these differential effects of ALIX is currently unknown, but it might be dependent on the composition of the complex or the nature of the stimulus.
Self-catalyzed ubiquitination of F-box proteins can be substrate inhibitable
Substrate specificity of CRL complexes is achieved through unique substrate receptor subunits (including F-box, SOCS/BC (suppressors of cytokine signaling/Elongin BC) box, DCAFs (DDB1- and Cul4-associated factors), or BTB (Bric-a-brac, Tramtrack, Broad complex) proteins), the large number of which allows these ligase complexes to recognize numerous substrates, and via their degradation, to have a function in the regulation of many basic cellular processes.2, 23 Whereas the other subunits of CRL ligases, including the RING finger-containing component, that are shared by all the complexes are generally stable, several studies indicate that a subset of the unique substrate receptors are short lived, and are targeted to ubiquitination and proteasomal degradation.23 Assembly of these substrate receptors to generate the intact complex is a prerequisite for their degradation. In the cases of the F-box proteins HOS (homologue of Slimb) and SKP2 (S-phase kinase-associated protein 2), and in the case of CDH1 – a substrate receptor for the APC/C complex, which is not an F-box protein – their ubiquitination could be reconstituted in vitro using the purified complex. This finding suggests that their degradation is mediated via an ‘autocatalytic' mechanism (mediated by the RING finger components).34, 35, 36, 37 Interestingly, it was observed that the degradation of several F-box proteins is attenuated by their respective substrates.16, 34, 36 It was recently uncovered that a WD40 repeat, which is present in a large subset of substrate receptors, can bind ubiquitin. This ubiquitin-binding property was required for self-ubiquitination and degradation of Cdc4 (cell division cycle 4). In addition, binding of a substrate or ubiquitin to Cdc4 are mutually exclusive, providing a possible mechanistic background for substrate-inhibited self-ubiquitination of F-box proteins.38 This type of regulation would ensure that sufficient levels of F-box proteins are maintained to target high level of substrates when they occur. However, after substrate concentration decreases, the F-box protein would become abundant, and should therefore be targeted for proteasomal destruction, while preserving the other components of the CRL complex. Such a mechanism would allow for a quick reassembly of the complex with different F-box proteins to adapt to changes in the desired specificity (Figure 2c). In agreement with this mechanism, it has recently been shown, though indirectly, that the architecture of CRL complexes is more likely to be dependent on the abundance of substrate receptors, rather than post-translational modifications such as NEDDylation.39 It should be noted, however, that such a mechanism for autocatalytic degradation does not exist for all substrate receptors of CRL complexes, as both the von Hippel Lindau (VHL) protein and the SOCS/BC protein SOCS-1 were shown to be stabilized following incorporation into the Elongin BC complex.40, 41 However, a chimera of VHL with part of Cdc4 (including the WD40 repeat) was self-ubiquitinated and degraded in the context of the Elongin BC complex, indicating that their susceptibility to self-ubiquitination is a property of the substrate receptor itself rather than of the complex it is associated with.40
Ligases Targeting Ligases: Exogenous Ubiquitination
Whereas self-ubiquitination has been thought for long to target ligases for degradation, it turns out that many of them, even those that catalyze their own ubiquitination, are targeted in trans by exogenous ligases. However, one should bear in mind that the landscape may be more complex than a simple division between self- and in trans-targeted ligases. At times, physiological adaptations may be needed in which abrogated self-induced degradation of ligase ‘X' is compensated by external ligase(s), or targeting by external ligases might occur only in response to specific stimuli (see for Mdm2 above). For other ligases it was described that self-ubiquitination does not lead to their degradation, but rather serves to regulate their activity (see below). It therefore appears that these ligases are obligatory and exclusively targeted for degradation in trans.
Degradation of E3 ligases mediated exclusively by external ligase(s) in trans
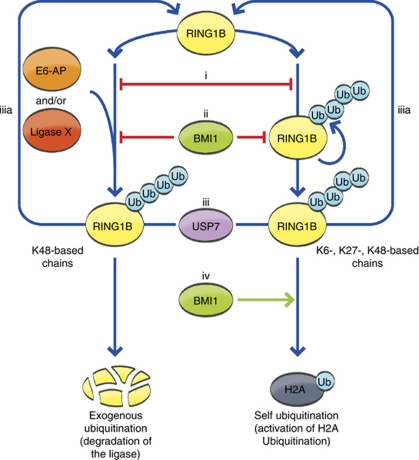
This group logically comprises of ligases incapable of self-destruction. One such ligase is the PcG protein RING1B. Employing cell free reconstituted systems, and expression experiments in cells, E6-AP was recently identified as a ligase that regulates the ubiquitination and proteasomal degradation of RING1B. Further corroborating these findings, it was shown that Ring1B levels were elevated in tissues of E6-AP-deficient mice.42 Notably, Ring1B levels appeared to be differentially regulated in the brain of these mice: elevated levels were observed in cerebellar Purkinje cells, but not in other brain cell types, suggesting that RING1B can be also targeted by other ligase(s). Importantly, RING1B catalyzes its own ubiquitination, but this modification generates Lys6-, Lys27-, and Lys48-based mixed and multiply branched chains that do not target the protein for degradation. Rather, they stimulate its activity as a monoubiquitinating ligase of histone H2A (Figure 3).
Figure 3.
Schematic representation of RING1B regulation by self-ubiquitination and ubiquitination by an exogenous ligase. RING1B is target to ubiquitination by E6-AP, and/or other E3 ligase(s), generating Lys48-based ubiquitin chains that target RING1B for proteasomal degradation. Self-ubiquitination of RING1B results in the formation of Lys6-, Lys27-, and Lys48-based mixed and multiply branched ubiquitin chains that activate it as a ligase for histone H2A. The balance between these two types of ubiquitination is regulated in several ways. (i) Since the activating and degrading forms of ubiquitination target the same lysine residues in RING1B, the two modifications are mutually exclusive. (ii) The PRC1 subunit BMI1 inhibits both types of ubiquitination. (iii) The DUB USP7 reverses both self- and E6-AP-mediated ubiquitination of RING1B, thereby returning it to its native state, potentially allowing the balance between the types of ubiquitination to be shifted. (iv) BMI1 stimulates ubiquitination of H2A by RING1B
In another case, the Drosophila melanogaster inhibitor of apoptosis 1 (Diap1) has been shown to undergo proteasomal degradation mediated by an external ligase, Diap2.43 This targeting of one IAP by another appears to be conserved in humans as well: XIAP was shown to be targeted for degradation by cIAP1.44 An additional mechanism of degradation of Diap1 is provided by caspase-catalyzed processing followed by the proteasomal degradation of the C-terminally released fragment by the N-end rule pathway.43, 45
Degradation of E3 ligases both through self-targeting and through external ligase(s)
For several ligases that mediate their own degradation, it has also been shown that they can be targeted by alternative mechanisms involving external ligases. These include the aforementioned Mdm2, and also GP78, a RING finger ligase implicated in ER-associated degradation of misfolded proteins. GP78 is capable of directing its own degradation,46 and in addition is also targeted for proteasomal degradation by HRD1.47, 48
CBL proteins also appear to be regulated by other ligases in trans. The HECT E3 ligases NEDD4 and ITCH bind to CBL proteins, and it was reported that ITCH stimulates EGF-R signaling.49, 50 It was shown that NEDD4 directly ubiquitinates CBL-b in vitro, and that both NEDD4 and ITCH stimulate the degradation of CBL-b and c-CBL.50 CBL-b degradation by NEDD4 appears to be induced by T-cell activation: co-stimulation of resting T cells by CD3 and CD28 results in a rapid degradation CBL-b, which is dependent on NEDD4.51 Loss of CBL-b following T-cell activation was shown to be mediated by CD28-induced phosphorylation of CBL-b by PKC-θ, providing a possible mechanism that targets CBL-b for ubiquitination by NEDD4.52
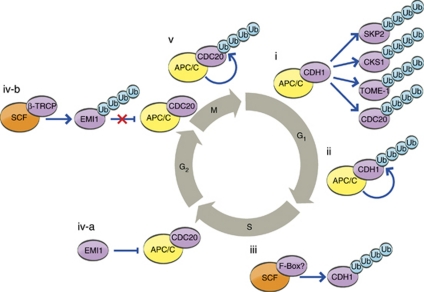
The CRL complexes SCF (SKP1–Cullin1–F-box) and APC/C have critical roles in cell-cycle control by providing timely degradation of key regulatory proteins. The activities of SCFs and APC/C are regulated in distinct manners, either by phosphorylation of the substrates that modulates their binding to the respective ligases, or by regulatory feedback between the SCF and APC/C ligases themselves (Figure 4). Two components of the SCFSKP2 complex, the F-box protein SKP2 and its essential cofactor CKS1 (cyclin-dependent kinases regulatory subunit 1), are required for entry and sustainment of the S phase. Both SKP2 and CKS1 are targeted by APC/CCDH1, preventing transition from G0/G1 to S phase.53, 54 In turn, it was shown that the APC/C substrate receptor CDH1 is degraded in an SCF-dependent manner during S phase, whereas, during G0/G1 phase, it mediates its own degradation catalyzed by the APC/C of which it is a component.37, 55 During the G1 phase, APC/C also mediates the degradation of TOME-1 (trigger of mitotic entry-1), an F-box protein required for mitotic entry through the activation of CDK1/Cyclin B.56 During the spindle-associated checkpoint in early M phase, the APC/C substrate receptor CDC20 undergoes degradation, which is thought to be self-catalyzed within the APC/C.57 CDC20 is also targeted to ubiquitination and degradation by APC/CCDH1, which most likely takes place during the G1 phase as that is when APC/CCDH1 is active.58 Mitotic progression is characterized by APC/C-dependent degradation of Cyclin B. During the S and G2 phases, the F-box protein EMI1 inhibits the activity of APC/C, preventing premature degradation of Cyclin B. Phosphorylation-induced degradation of EMI1 by SCFβ-TRCP results in the timely activation of APC/C, allowing the cell to progress though the M phase.59, 60 It is interesting to note that regulation of CRLs by external ligases aims exclusively at the substrate receptor in order to avoid unnecessary loss of the entire complex, and to allow rapid adaptation to the degradation of different substrates targeted by distinct receptors that share common basic complex components.
Figure 4.
Schematic representation of the interplay between the ubiquitinating activities of APC/C and SCF complexes during cell cycle. (i) During G1 phase, APC/CCDH1 targets several F-box substrate receptors of SCF complexes, including SKP2 and TOME-1, and the SKP2 cofactor CKS1. In addition, CDC20 is also targeted to ubiquitination and degradation by APC/CCDH1. While CDH1 is subject to self-ubiquitination during G0 and G1 phases within the context of the APC/C complex (ii), the degradation of the free released phosphorylated form is mediated by an SCF complex during S phase (iii). (iv) During S and G2 phases, APC/CCDC20 is inhibited by the F-box protein EMI1 (iv-a); however, in the M phase, EMI1 is targeted for destruction by SCFβ-TRCP alleviating the inhibition of APC/CCDC20 (iv-b). (v) CDC20 is targeted in a self-catalytic manner during the spindle-associated checkpoint
Hierarchical organization of the degradation of E3 ligases
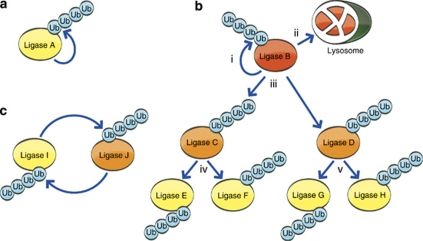
Based on these examples, several hierarchical organizations of degradation of E3s may exist (Figure 5). E3s can either induce their own degradation through self-ubiquitination or their degradation can be mediated by external ligases in trans. The latter can occur through a linear model, in which an E3 targets one, or several other ligases, and in turn is targeted itself by self-ubiquitination (or another proteolytic machinery, e.g., the lysosome). Alternatively, ligases can target one another in a circular manner, as exemplified by SCF and APC/C complexes that target each other in an oscillating manner parallel to the cell cycle (see above). Some E3s, such as Mdm2, RING1B, Diap1, and CBL proteins are regulated through multiple proteolytic pathways, highlighting an extensive regulation of the ubiquitin system itself (see above).
Figure 5.
Mechanisms for proteasomal targeting of E3 ligases. (a) Degradation mediated by self-ubiquitination. (b) Hierarchical linear control of ligases. Ligase B is controlled by autoubiquitination (i) or another proteolytic mechanism, e.g., lysosomal degradation; (ii). It modifies in trans ligases C and D (iii), which in turn can modify ligases E and F, and ligases G, H, respectively (iv and v), etc. (c) Two (or more) ligases target one another in a closed circular manner
Non-proteolytic Functions of Self-ubiquitination of E3s
Self-ubiquitination of E3 ligases has also been implicated in regulating the ligases' activity and the recruitment of substrates. RING1B-mediated monoubiquitination of histone H2A is a hallmark of PcG-mediated gene silencing, a process that is critical, for example, for the maintenance of stem cells. The activity of RING1B towards H2A is significantly increased once assembled into the polycomb repressive complex 1 (PRC1), and appears to be particularly enhanced by the RING domain-containing protein BMI1.8 A prerequisite for the ability of RING1B to ubiquitinate H2A is its self-ubiquitination, resulting in the generation of Lys6-, Lys27-, and Lys48-based, mixed, and multiply branched ubiquitin chains7 (Figure 3): addition of Lys0 ubiquitin (that cannot polymerize as it does not have any internal lysine residues), cannot support monoubiquitination of H2A, though this modification requires one single ubiquitin moiety. The exact mechanism through which RING1B self-ubiquitination activates it as a ligase for H2A has not been elucidated. The mechanism through which BMI1 stimulates H2A ubiquitination remains elusive as well, as it was also shown to attenuate self-ubiquitination of RING1B.7 A possible explanation for this apparent discrepancy can be that BMI1 functions to regulate the self-ubiquitination chain length or architecture, or the timing between self-ubiquitination and ubiquitination of H2A, as it appears that the two reactions must occur concomitantly.7 Both the activating self-ubiquitination and the degrading ubiquitination by E6-AP target the same lysine residues in RING1B, and are therefore mutually exclusive, suggesting that controlling the ubiquitination state of RING1B is critical in regulating polycomb-mediated gene silencing (Figure 3). Upstream of RING1B ubiquitination, BMI1 inhibits E6-AP-mediated ubiquitination and degradation of RING1B, while stimulating RING1B-mediated ubiquitination of H2A.7, 8, 42 In analogy to self-activation of RING1B, BRCA1 (breast cancer 1) is subject to self-ubiquitination generating Lys6-based chains, resulting in an increased potential to ubiquitinate histones in vitro.11, 61, 62 The activity of BRCA1 appears also to be regulated through binding to a RING domain-containing protein, BARD1 (BRCA1-associated RING domain 1) that, like BMI1, does not appear to have a ubiquitin ligating activity. It appears that BARD1 enhances the self-ubiquitinating activity of BRCA1.63
In contrast to the above-mentioned two examples, self-ubiquitination of Diap1 appears to negatively regulate its activity. Diap1 was shown to modify itself with Lys63-based polyubiquitin chains, which attenuate its ability to ubiquitinate its substrate Dronc in vitro. Strikingly, Diap1's activity was not inhibited when subjected to ubiquitination with Lys0 ubiquitin. It therefore seems that long polyubiquitin chains are required for this effect, suggesting that it may result from steric hindrance preventing binding to, or efficient conjugation of the substrate.43
Self-ubiquitination can also serve as a mechanism to recruit substrates with ubiquitin-binding properties, as has been shown for TRAF6 and NEDD4. TRAFs are RING domain-containing E3 ligases that have crucial roles in the initial activation of several signaling cascades including the NF-κB, JNK, and p38 kinase pathways. TRAF6 ubiquitinates itself following receptor stimulation by IL-1β, generating Lys63-based polyubiquitin chains.64 Dimerization of TRAF6 (through its C-terminus or N-terminal RING domain) is essential and sufficient to induce its own ubiquitination and subsequent activation of the JNK and NF-κB signaling pathways.15, 64, 65 The self-generated Lys63-based ubiquitin chains appear to function as recruitment adaptors to attract other substrates to TRAF6. TAB2 (TAK1-binding protein 2) binds specifically to Lys63-linked ubiquitin chains, which might serve to recruit TAB2 to the self-ubiquitinated TRAF6. In turn, TAB2 recruits the TAK1 (TGF-β activated kinase 1)/TAB1 (TAK1-binding protein 1) kinase complex, which is subsequently activated by a mechanism that could involve ubiquitination of TAK1 by TRAF6.66, 67, 68 Alternatively, TAK1 might be activated by autophosphorylation induced by binding of TAB2 to free Lys63-linked ubiquitin chains synthesized by TRAF6.69 NEMO (NF-κB essential modulator), through similar Lys63-linked ubiquitin chain binding properties, might also be recruited to TRAF6, resulting in its ubiquitination.70, 71
Another example for self-ubiquitination-dependent recruitment of substrates was reported for NEDD4: its self-catalyzed monoubiquitination serves to recruit EPS15, which is subsequently also monoubiquitinated by NEDD4.72 The function of monoubiquitination of EPS15 still remains an enigma, but it might have an inhibitory role in the function of EPS15 to facilitate clathrin-mediated endocytosis of transmembrane proteins.73
Regulation of E3s by Deubiquitination
The ubiquitin mark can be removed through the action of DUBs, a class of isopeptidases that specifically cleave ubiquitin linkages, allowing for fine-tuning, or reversal of the modification. Mdm2 can be stabilized through deubiquitination by USP7, a DUB that in addition also targets, and thereby protects, p53.74 The physiological significance of these, apparently opposing activities, is still not clear. USP2a, however, was shown to specifically deubiquitinate Mdm2, but not p53, thereby stimulating the degradation of p53.74 Other E3 ligases were shown to be targeted by USP7 including ICP0, MARCH7, and RING1B.74, 75, 76 USP7 removes both self- and E6-AP-generated ubiquitin chains from RING1B, thereby regulating its stability (Figure 3).75 Therefore, it is not surprising that USP7 was also shown to exert a regulatory effect on the expression of polycomb target genes.77 Activation of the NF-κB pathway trough ligand-induced oligomerization and self-ubiquitination of TRAF6 is regulated through the actions of ubiquitin linkage-specific DUBs. CYLD, a DUB mutated in familial cylindromatosis, was shown to specifically target Lys63-based ubiquitin chains on TRAF2 and TRAF6, and consequently has an inhibitory effect on the NF-κB pathway.78 In addition, TRAF6 might also be regulated by A20, another Lys63-linked ubiquitin chain-specific DUB that was shown to deubiquitinate TRAF6, among other substrates in the NF-κB pathway.79 Importantly, A20 was found to be a double function enzyme – a DUB and a ligase. Following deubiquitination of Lys63-based polyubiquitin chains, it generates on RIP (receptor interacting protein) Lys48-based chains, thus rendering it susceptible to degradation.80 Since the DUB activity of CYLD and A20 appears to be essential, though they both target the same type of chains in the NF-κB pathway, it is possible that they act in different cells and follow different stimuli.
Translational Implications of the Mechanisms that Underlie Degradation of Ligases
Although the versatility and complexity of the ubiquitin-proteasome system in health and disease is well established and our knowledge on its invovlement in pathogenetic mechanisms still grows exponentially, a detailed atlas of its broad landscape is still missing. First, we need to unravel the dynamic ubiquitome – the part of the proteome that undergoes ubiquitination for both proteolytic and non-proteolytic functions under different pathophysiological conditions and along time. We shall then need to identify the different ubiquitin ligases that target these proteins and unravel their recognition motifs. Related to the enigma of specific recognition, it will be necessary to investigate the regulation of modification of the different substrates – what renders them resistant or susceptible to ubiquitination at certain time points and under different conditions. Last, it will be necessary to decipher the mechanisms that underlie the regulation and turnover of the different components of the system itself, including the ligases. Currently, we have at hand only small and rather random pieces of this cumbersome chart, and for only a few of the ∼650 E3 ligases and the thousands substrates. However, even with this partial map, one can recognize a number of basic principals through which the degradation of ligases is regulated (Figures 2 and 5). Using such insights, one can design small molecule inhibitors that will prevent destructive autoubiquitination without affecting modification of exogenous substrates. Such inhibitors can increase the level of the respective ligases, consequently making them more effective towards their substrates. This can be useful, for example, in the case of pro-inflammatory (e.g., NF-κB) or oncogenic (e.g., Myc) substrates that are targeted by known components of the UPS. With respect to targeting of ligases by upperstream ligases, it is possible that in the future we shall discover regulating hubs, for example a ligase that controls several ligases with common general function, such as regulation of a signaling pathway. Inhibition of such a ligase will allow to control the entire pathway, which should be more efficient than targeting it at a single point. Once the entire map will be unraveled, the potential of this approach will grow dramatically.
Acknowledgments
Research in the laboratory of AC is supported by grants from the Dr. Miriam and Sheldon Adelson Foundation for Medical research (AMRF), the Israel Science Foundation (ISF), the German-Israeli Foundation for Research and Scientific Development (GIF), the Rubicon European Union (EU) Network of Excellence, an Israel Cancer Research Fund (ICRF) USA Professorship, and a grant from the Foundation for Promotion of Research in the Technion.
Glossary
- ARF
alternative reading frame
- APC/C
anaphase-promoting complex/cyclosome
- CRL
Cullin-RING ligase
- Diap1
D. melanogaster inhibitor of apoptosis 1
- DUB
deubiquitinating enzyme
- EGF-R
epidermal growth factor receptor
- ENaC
epithelial sodium channel
- HECT
homologous to the E6-AP carboxy terminus
- PcG
polycomb group
- PDGF-R
platelet-derived growth factor receptor
- PRC1
polycomb repressive complex 1
- RING
really interesting new gene
- RIP
receptor interacting protein
- SCF
Skip1–Cullin1–F-box
- TK
tyrosine kinase
- TRAF
TNF receptor-associated factor
- VHL
von Hippel Lindau
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun. 2007;354:329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-CBL/SLI-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Vodermaier HC. APC/C and SCF: controlling each other and the cell cycle. Curr Biol. 2004;14:R787–R796. doi: 10.1016/j.cub.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Debonneville C, Flores SY, Kamynina E, Plant PJ, Tauxe C, Thomas MA, et al. Phosphorylation of NEDD4-2 by SGK1 regulates epithelial Na(+) channel cell surface expression. EMBO J. 2001;20:7052–7059. doi: 10.1093/emboj/20.24.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Association of p19(Arf) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein RING1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24:701–711. doi: 10.1016/j.molcel.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of BMI-1 and RING1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of Cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by NEDD8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuber U, Schwarz SE, Scheffner M. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur J Biochem. 1998;254:643–649. doi: 10.1046/j.1432-1327.1998.2540643.x. [DOI] [PubMed] [Google Scholar]
- Hu G, Fearon ER. SIAH-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci USA. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce MC, Kanelis V, Fouladkou F, Debonneville A, Staub O, Rotin D. Regulation of NEDD4-2 self-ubiquitination and stability by a PY motif located within its HECT-domain. Biochem J. 2008;415:155–163. doi: 10.1042/BJ20071708. [DOI] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, et al. Autoinhibition of the HECT-type ubiquitin ligase SMURF2 through its C2 domain. Cell. 2007;130:651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- Kao WH, Beaudenon SL, Talis AL, Huibregtse JM, Howley PM. Human papillomavirus type 16 E6 induces self-ubiquitination of the E6AP ubiquitin-protein ligase. J Virol. 2000;74:6408–6417. doi: 10.1128/jvi.74.14.6408-6417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PE, Davies GC, Nau MM, Lipkowitz S. Regulating the regulator: negative regulation of CBL ubiquitin ligases. Trends Biochem Sci. 2006;31:79–88. doi: 10.1016/j.tibs.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of Cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583:2710–2714. doi: 10.1016/j.febslet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Coutts AS, Adams CJ, La Thangue NB. p53 ubiquitination by Mdm2: a never ending tail. DNA Repair (Amst) 2009;8:483–490. doi: 10.1016/j.dnarep.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, Latreille D, Cuvier O, et al. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein HDM2. Nat Cell Biol. 2007;9:331–338. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- Ettenberg SA, Magnifico A, Cuello M, Nau MM, Rubinstein YR, Yarden Y, et al. CBL-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J Biol Chem. 2001;276:27677–27684. doi: 10.1074/jbc.M102641200. [DOI] [PubMed] [Google Scholar]
- Zeng S, Xu Z, Lipkowitz S, Longley JB. Regulation of stem cell factor receptor signaling by CBL family proteins (CBL-b/c-CBL) Blood. 2005;105:226–232. doi: 10.1182/blood-2004-05-1768. [DOI] [PubMed] [Google Scholar]
- Alwan HA, van Zoelen EJ, van Leeuwen JE. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J Biol Chem. 2003;278:35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yeung YG, Langdon WY, Stanley ER. c-CBL is transiently tyrosine-phosphorylated, ubiquitinated, and membrane-targeted following CSF-1 stimulation of macrophages. J Biol Chem. 1996;271:17–20. doi: 10.1074/jbc.271.1.17. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Hoeller D, Yu J, Furnari FB, Cavenee WK, Dikic I, et al. ALIX/AIP1 antagonizes epidermal growth factor receptor downregulation by the CBL-SETA/CIN85 complex. Mol Cell Biol. 2004;24:8981–8993. doi: 10.1128/MCB.24.20.8981-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartsson J, Wardega P, Engstrom U, Hellman U, Heldin CH. ALIX facilitates the interaction between c-CBL and platelet-derived growth factor beta-receptor and thereby modulates receptor down-regulation. J Biol Chem. 2006;281:39152–39158. doi: 10.1074/jbc.M608489200. [DOI] [PubMed] [Google Scholar]
- Li Y, Gazdoiu S, Pan ZQ, Fuchs SY. Stability of homologue of Slimb F-box protein is regulated by availability of its substrate. J Biol Chem. 2004;279:11074–11080. doi: 10.1074/jbc.M312301200. [DOI] [PubMed] [Google Scholar]
- Wirbelauer C, Sutterluty H, Blondel M, Gstaiger M, Peter M, Reymond F, et al. The F-box protein SKP2 is a ubiquitylation target of a CUL1-based core ubiquitin ligase complex: evidence for a role of CUL1 in the suppression of SKP2 expression in quiescent fibroblasts. EMBO J. 2000;19:5362–5375. doi: 10.1093/emboj/19.20.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ungermannova D, Jin J, Harper JW, Liu X. Negative regulation of SCFSKP2 ubiquitin ligase by TGF-β signaling. Oncogene. 2004;23:1064–1075. doi: 10.1038/sj.onc.1207204. [DOI] [PubMed] [Google Scholar]
- Listovsky T, Oren YS, Yudkovsky Y, Mahbubani HM, Weiss AM, Lebendiker M, et al. Mammalian CDH1/FZR mediates its own degradation. EMBO J. 2004;23:1619–1626. doi: 10.1038/sj.emboj.7600149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol Cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of Cullin-RING ubiquitin ligase network revealed by systemic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Brower CS, Conaway RC, Conaway JW. A molecular basis for stabilization of the von Hippel-Lindau (VHL) tumor suppressor protein by components of the VHL ubiquitin ligase. J Biol Chem. 2002;277:30388–30393. doi: 10.1074/jbc.M203344200. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Jr, Conaway RC, et al. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaaroor-Regev D, de Bie P, Scheffner M, Noy T, Shemer R, Heled M, et al. Regulation of the polycomb protein RING1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of Angelman syndrome. Proc Natl Acad Sci USA. 2010;107:6788–6793. doi: 10.1073/pnas.1003108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ. 2007;14:861–871. doi: 10.1038/sj.cdd.4402079. [DOI] [PubMed] [Google Scholar]
- Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M, et al. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci USA. 2005;102:16182–16187. doi: 10.1073/pnas.0502828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, et al. Degradation of Diap1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, GP78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballar P, Ors AU, Yang H, Fang S. Differential regulation of CFTRDeltaF508 degradation by ubiquitin ligases GP78 and HRD1. Int J Biochem Cell Biol. 2010;42:167–173. doi: 10.1016/j.biocel.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Shmueli A, Tsai YC, Yang M, Braun MA, Weissman AM. Targeting of GP78 for ubiquitin-mediated proteasomal degradation by HRD1: cross-talk between E3s in the endoplasmic reticulum. Biochem Biophys Res Commun. 2009;390:758–762. doi: 10.1016/j.bbrc.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbard JR, Fiore F, Adelaide J, Borg JP, Birnbaum D, Ollendorff V. Interaction between two ubiquitin-protein isopeptide ligases of different classes, CBL-C and AIP4/ITCH. J Biol Chem. 2002;277:45267–45275. doi: 10.1074/jbc.M206460200. [DOI] [PubMed] [Google Scholar]
- Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, et al. WW domain HECT E3s target CBL RING finger E3s for proteasomal degradation. J Biol Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
- Yang B, Gay DL, MacLeod MK, Cao X, Hala T, Sweezer EM, et al. NEDD4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of CBL-b in activated T cells. Nat Immunol. 2008;9:1356–1363. doi: 10.1038/ni.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber T, Hermann-Kleiter N, Hinterleitner R, Fresser F, Schneider R, Gastl G, et al. PKC-θ modulates the strength of T cell responses by targeting CBL-b for ubiquitination and degradation. Sci Signal. 2009;2:ra30. doi: 10.1126/scisignal.2000046. [DOI] [PubMed] [Google Scholar]
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCF(SKP2-CKS1) ubiquitin ligase by the APC/C(CDH1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component SKP2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Benmaamar R, Pagano M. Involvement of the SCF complex in the control of CDH1 degradation in S-phase. Cell Cycle. 2005;4:1230–1232. doi: 10.4161/cc.4.9.2048. [DOI] [PubMed] [Google Scholar]
- Ayad NG, Rankin S, Murakami M, Jebanathirajah J, Gygi S, Kirschner MW. TOME-1, a trigger of mitotic entry, is degraded during G1 via the APC. Cell. 2003;113:101–113. doi: 10.1016/s0092-8674(03)00232-0. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by CDH1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, et al. Control of meiotic and mitotic progression by the F box protein β-TRCP1 in vivo. Dev Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- Margottin-Goguet F, Hsu JY, Loktev A, Hsieh HM, Reimann JD, Jackson PK. Prophase destruction of Emi1 by the SCF(β-TRCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Wu-Baer F, Lagrazon K, Yuan W, Baer R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem. 2003;278:34743–34746. doi: 10.1074/jbc.C300249200. [DOI] [PubMed] [Google Scholar]
- Mallery DL, Vandenberg CJ, Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002;21:6755–6762. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, et al. The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10:1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- Fan Y, Yu Y, Shi Y, Sun W, Xie M, Ge N, et al. Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor α- and interleukin-1β-induced IKK/NF-κB and JNK/AP-1 activation. J Biol Chem. 2010;285:5347–5360. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J Biol Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Woelk T, Oldrini B, Maspero E, Confalonieri S, Cavallaro E, Di Fiore PP, et al. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8:1246–1254. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- van Bergen En, Henegouwen PM. EPS15: a multifunctional adaptor protein regulating intracellular trafficking. Cell Commun Signal. 2009;7:24. doi: 10.1186/1478-811X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviet L, Colland F. Targeting ubiquitin specific proteases for drug discovery. Biochimie. 2008;90:270–283. doi: 10.1016/j.biochi.2007.09.013. [DOI] [PubMed] [Google Scholar]
- de Bie P, Zaaroor-Regev D, Ciechanover A. Regulation of the polycomb protein RING1B ubiquitination by USP7. Biochem Biophys Res Commun. 2010;400:389–395. doi: 10.1016/j.bbrc.2010.08.082. [DOI] [PubMed] [Google Scholar]
- Nathan JA, Sengupta S, Wood SA, Admon A, Markson G, Sanderson C, et al. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9:1130–1145. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 2010;29:2553–2565. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-κB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]