Abstract
Identification of the sex-determining genes of the Nile tilapia (Oreochromis niloticus) has important implications for commercial aquaculture. We previously identified an XX/XY sex-determining locus in this species within a 10cM interval between markers GM201 and UNH995 on linkage group one (LG1). In order to refine this region, we developed new AFLP markers using bulked segregant analysis of the mapping families. We identified three AFLP markers that showed a sex-specific pattern of segregation. All three mapped near, but just outside, the previously identified sex-determining region on LG1. Hybridization of BAC clones containing these markers to chromosome spreads confirmed that the XX/XY sex determining locus is on one of the small chromosomes in O. niloticus.
Keywords: AFLP, sex chromosomes, O. niloticus, tilapia, sex-linked markers
Introduction
The Nile tilapia (Oreochromis niloticus) is one of the most important species of farmed fish (FAO, 2007). A major challenge in commercial operations is to control reproduction, because tilapia often begin to breed before reaching market size. The usual method to control reproduction is to culture only phenotypic males produced by hormonal sex reversal (Hopkins et al., 1979; Rosenstein and Hulata, 1994; Mair et al., 1997). Monosex populations of males also can be produced by genetic manipulation, notably the use of YY male broodstock (Mair et al., 1997). Neither approach is perfectly efficient because temperature and minor genetic factors also can influence sex determination (Mair et al., 1991; Baroiller et al., 1995; Abucay et al., 1999; Karayucel et al., 2004). Further studies of the mechanisms of sex determination are needed to provide the technical basis for improvement of commercial production.
The genetic sex of Nile tilapia is thought to be an XX/XY male heterogametic system controlled by a major gene (Mair et al., 1991). There are no gross morphological differences between the sex chromosomes (Majumdar and McAndrew, 1986). However, several studies have aimed to identify the sex chromosomes by observing the synaptomenal complex in meiotic chromosomes (Carrasco et al., 1999; Campos-Ramos et al., 2001; Harvey et al., 2002; Ocalewicz et al., 2009). These studies identified a region of incomplete or delayed chromosome pairing that suggested that the longest chromosome (chromosome 1) is the sex chromosome in this species.
We have used genetic markers to identify the sex-determining regions of several tilapia species (Lee et al., 2003; Lee et al., 2004; Cnaani et al., 2008). In O. niloticus we identified a sex-determining region in a 10cM interval between microsatellite markers UNH995 and GM201 on linkage group 1 (Lee et al., 2003). Physical mapping of BAC clones containing these genetic markers using fluorescent in situ hybridization (FISH) to chromosome spreads showed that these sex-linked markers are on one of the smaller chromosomes (Cnaani et al., 2008).
Here we report the development of new AFLP markers closely linked to the sex-determining locus in O. niloticus. To resolve the controversy over whether these sex-linked markers map to a large or small chromosome in O. niloticus, we prepared an integrated genetic and physical map of the region, using FISH probes derived from BAC clones containing these new markers.
Material and methods
AFLP/BSA Analysis
We developed sex-linked AFLP markers based on bulk segregant analysis (BSA) (Michelmore et al., 1991; Vos et al., 1995). From each of two informative families of O. niloticus used in the previous study to identify the sex-determining regions (Lee et al., 2003), we constructed pools of 10 male or 10 female fish based on their genotypes at markers flanking the sex locus. The final concentration of DNA from each individual in the pool was 10 ng/ul. DNAs were simultaneously digested with EcoR1 and MseI and ligated to adaptor pairs (EcoRI F: 5’ CTCGTAGACTGCGTACC 3’ and R: 5’ AATTGGTACGCAGTCTAC 3’; MseI F: 5’ GACGATGAGTCCTGAG 3’ and R: 5’ TACTCAGGACTCAT 3’) in a single reaction that contained 1X T4 DNA ligase buffer (30 mM Tris-HCl, pH=7.8; 10 mM MgCl2; 10 mM DTT; 1 mM ATP), 50 mM NaCl, 50 ng/ul BSA, 1 unit MseI, 5 units EcoR1, 1 unit T4 DNA ligase, 50 pmol MseI adaptor pair and 5 pmol EcoRI adaptor pair. This reaction was incubated at 37°C for 2 hours and then was diluted 1:20 with TE/10. The diluted restriction-ligation reaction then was PCR-amplified using pre-selective primers to create a quantity of partially selected DNA fragments. Pre-selective primers contained one additional selective base beyond the adaptor sequence (EcoR I-5’ GACTGCGTACCAATTC[A] 3’, MseI 5’ GATGAGTCCTGAGTAA[C] 3’), which reduced the fragment complexity 16-fold. The cycling conditions were: 72°C for 2 min; 20 cycles of 94°C for 20 sec, 56°C for 30 sec, and 72°C for 2 min; and finally a hold at 60°C for 30 min. Pre-selective reactions were diluted 1: 20 with TE/10 and used for selective PCR with selective primers (1 pmol EcoRI primer and 5 pmol MseI primer). The selective primers contained two additional selective bases beyond that of the pre-selective primers, which further reduced the fragment complexity by a factor of 256. The cycling conditions for the selective amplification were: 94°C for 2 min; 10 cycles of 94°C for 2 min, 66°C 30 sec, 72°C for 2 min decreasing 1°C after each cycle; 20 cycles of 94°C for 20 sec, 56°C for 30 sec, 72°C for 2 min. All selective reaction products were run on an ABI377 automated sequencer and fragments on gels were analyzed with GeneScan (ver. 3.1.2). (Applied Biosystems, Foster City CA).
Genotyping and linkage analysis
AFLP markers that were specific to one phenotypic pool were tested on each individual in both family 5 (n=39) and family 7 (n=43) as described above. The genotypes were coded as dominant markers and linkage analysis was performed using JoinMap version3.0 (Stam, 1993) with a LOD-score threshold of 3.0.
Isolation of BAC clones
Primer sequences for the sex-linked AFLP markers (OniY382, OniY227, OniY425, and OniX420) developed by Ezaz et al. (2004) were retrieved from the NCBI database (accession #’s BV012734, BV012735, BV012736, and BV012737) and used to test whether these markers could be amplified from tilapia genomic DNA. PCR was performed in 50 ul reaction containing 1X PCR buffer (50ml KCl; 10mM Tris-HCl, pH9.0;, 0.1% Triton-X; 2mM MgCl2), 0.8 mM dNTPs, 0.2uM each forward and reverse primer, and 1 U of Taq DNA polymerase in the cycling condition: 94C for 3 min; 35 cycles of 94C for 20sec, 55C for 30sec, 72C for 1 min; 72C for 5 min. The primers that successfully amplified gDNA then were used for PCR screening of BAC libraries (Katagiri et al. 2001). DNA was extracted from the positive BAC clones using the BIG BAC DNA kit (Princeton Separations, Freehold, NJ).
Fluorescence In Situ Hybridization (FISH)
Chromosome preparations were obtained as previously described (Fischer et al., 2000). BAC clones from the tilapia libraries were used as probes and labeled with biotin- or digoxygenin (DIG)-coupled nucleotide by random priming using ‘high prime’ kits (Roche), according to the supplier's protocol. For double-FISH, precipitated labeled probes were dissolved at 7 ng/ul in the hybridization buffer (50% deionized formamide, 2X SSC, 10% dextran sulfate, 50 mM sodium phosphate, pH 7) with a 1000-fold excess of sheared bovine carrier DNA and a 200-fold excess of sheared specific (tilapia) competitor DNA. They were denatured 5 min. at 75°C, briefly chilled and pre-hybridized separately for 1 h at 37°C in order to eliminate nonspecific sequences generated from the BAC vector and abundant repeats present in the insert. Probes were pooled prior to hybridization. Chromosome preparations were thawed and air-dried a few minutes before use, denatured in 70% formamide, 2X SSC, pH 7 for 10 sec. and dehydrated in an ice-cold ethanol series. Fifteen µl of pre-annealed probe mixture were hybridized under a cover slip in a moist chamber for 48 h. Slides were washed 15 min. in 50% formamide, 2X SSC, pH 7 at 43°C with intermittent agitation for 15 min, then 15 min in 0.1X SSC, pH 7 at 60°C with continuous agitation. Hybridization signals were detected with avidin-FITC and rhodamine-anti-DIG (Roche Applied Sciences, Indianapolis IN) according to the supplier's protocol. After three washes of 2 min in phosphate buffer detergent (4X SSC, 1% Tween 20), slides were mounted with DAPI/antifade. Results were recorded with a Zeiss Axiophot microscope equipped with a CoolSNAP CCD camera (Photometrics, Tuscon AZ) and Genus software for animal karyotyping and FISH-imaging (Genetix Europe Ltd., New Milton, UK).
Results
Identification of sex-specific AFLP
Among the thousands of fragments amplified by 128 selective AFLP primer pairs, we found three sex-specific markers. MCTT/EACG-382 and MCAC/EAAC-391 showed sex-specific patterns in male and female BSA pools of both Family 5 and 7 (Figure 1). MCTA/EAAG-425 was informative in Family 7, but not in Family 5.
Figure 1.
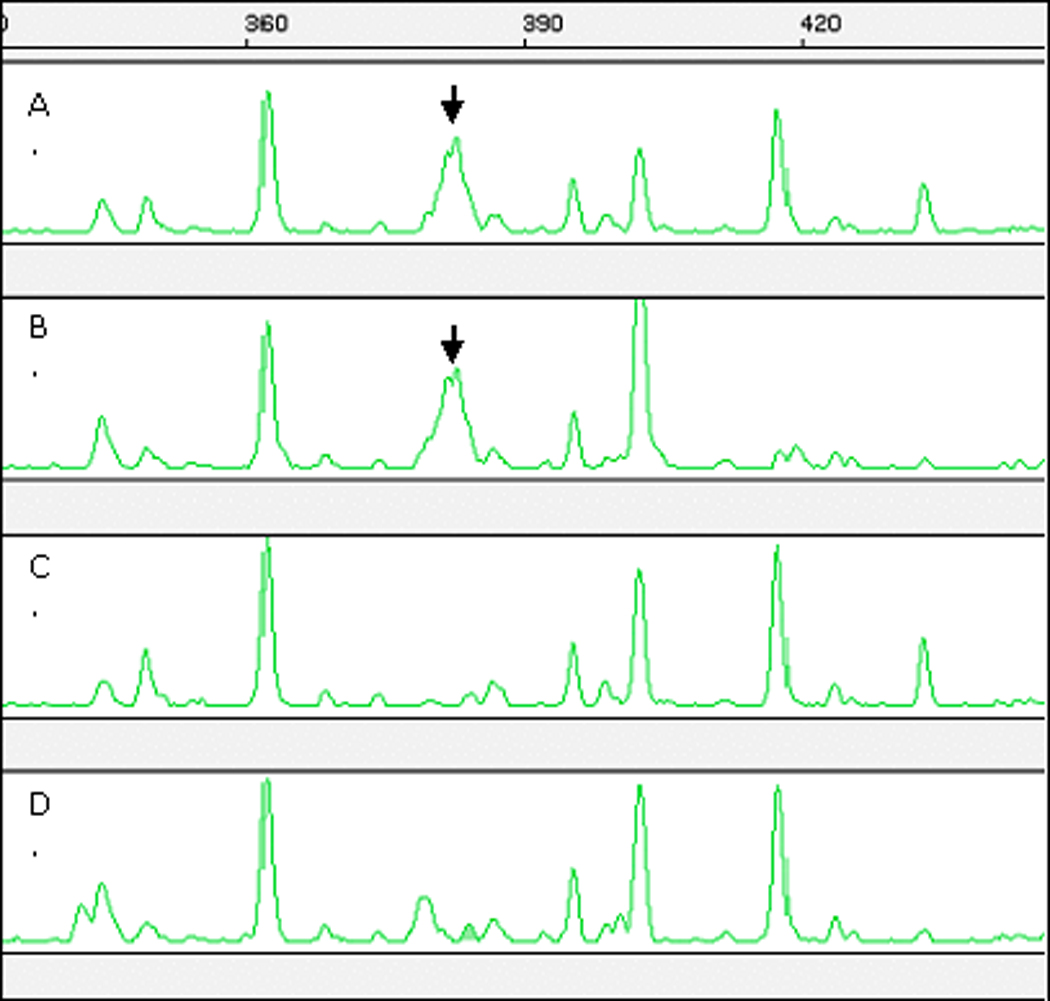
Identification of a sex-specific AFLP marker (MCTT/EACG-382) in the O. niloticus sex-determining region using bulked segregant analysis. A. Family 5 male pool; B. Family 7 male pool; C. Family 5 female pool; D. Family 7 female pool. Sex-specific peaks are indicated by the arrows. Only male pools show the 382 bp peak.
Genetic Mapping
The three AFLP markers showing sex-specific segregation mapped to LG1, near the sex-determining locus mapped to 35cM in our earlier study (Lee et al., 2003). MCTT/EACG-382 and MCAC/EAAC-391 mapped at 31cM and 30cM in LG1. Linkage analysis put MCTA/EAAG-425 coincident with UNH104 and UNH995 at 39cM. While these new markers help to confirm the position of the sex locus on this chromosome, they do not contribute to refining the sex-determining region because they map outside the critical interval identified by microsatellite markers in our previous work (Figure 2).
Figure 2.
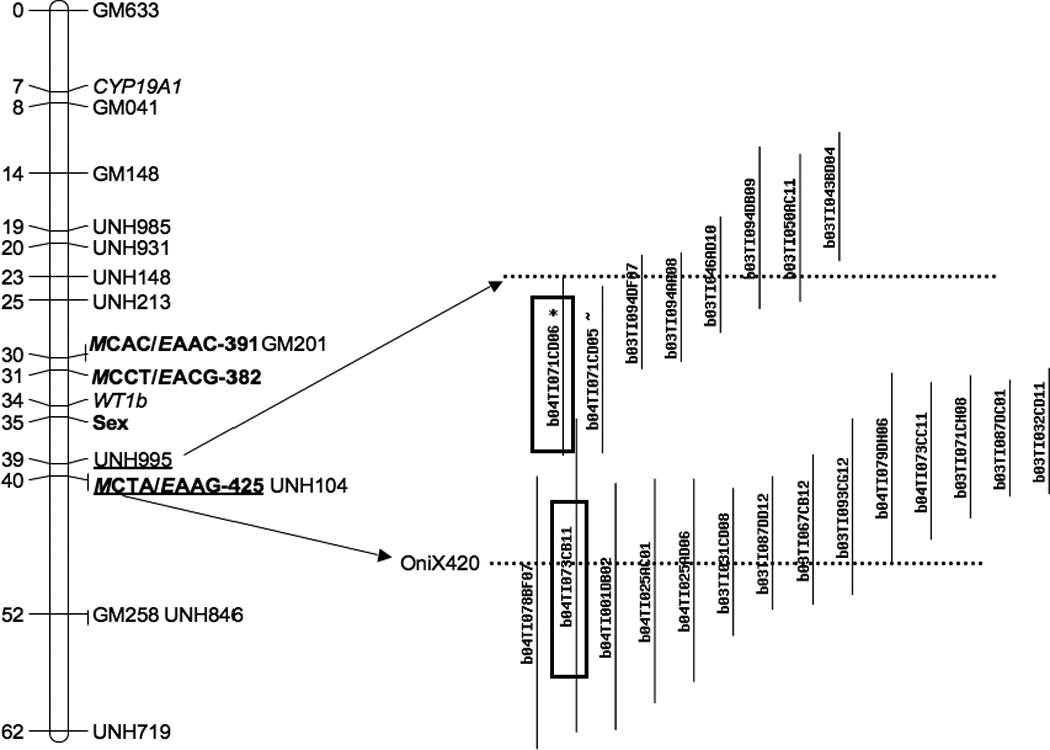
Genetic mapping of sex-linked AFLP markers (boldic) and BAC contig assembled from restriction fingerprints containing AFLP marker OniX420 and microsatellite marker UNH995. Two BACs (boxes) were used as FISH probes. Dotted lines indicate where the markers were located in the contig.
Physical mapping
Ezaz et al. (2004) reported four sex-linked AFLP markers from O. niloticus. The primers amplified OniY227, OniY382 and OniX420, but we were unable to amplify OniY425 from tilapia genomic DNA. We successfully isolated clones containing OniY227, OniY382 and OniX420 from tilapia BAC libraries that have been fingerprinted and assembled into contigs by analysis of restriction fingerprints (Katagiri et al., 2001; Katagiri et al., 2005). Based on the analysis of contig assembly software called FPC (Soderlund et al., 1997) , BAC clones containing OniY227 belonged to a contig consisting of 17 clones at a tolerance of 5 and e-value of e−8. OniY382 was found in a contig containing sequences similar to a forkhead box gene of Haplochromis species (GenBank acc#BJ690985). Comparative mapping suggests this sequence is homologous to a forkhead gene on Tetraodon chr 5 (4912k) and lies adjacent to the sex-determining interval on tilapia LG1 (Lee and Kocher, 2007b). OniX420 was found in a contig which also contains the microsatellite marker UNH995 (Figure 3). Two BAC clones (b04TI071CD06 and b04TI073CB11) from this contig were selected as probes for FISH mapping. The two probes hybridized close together on one of the small tilapia chromosomes (Figure 2).
Figure 3.
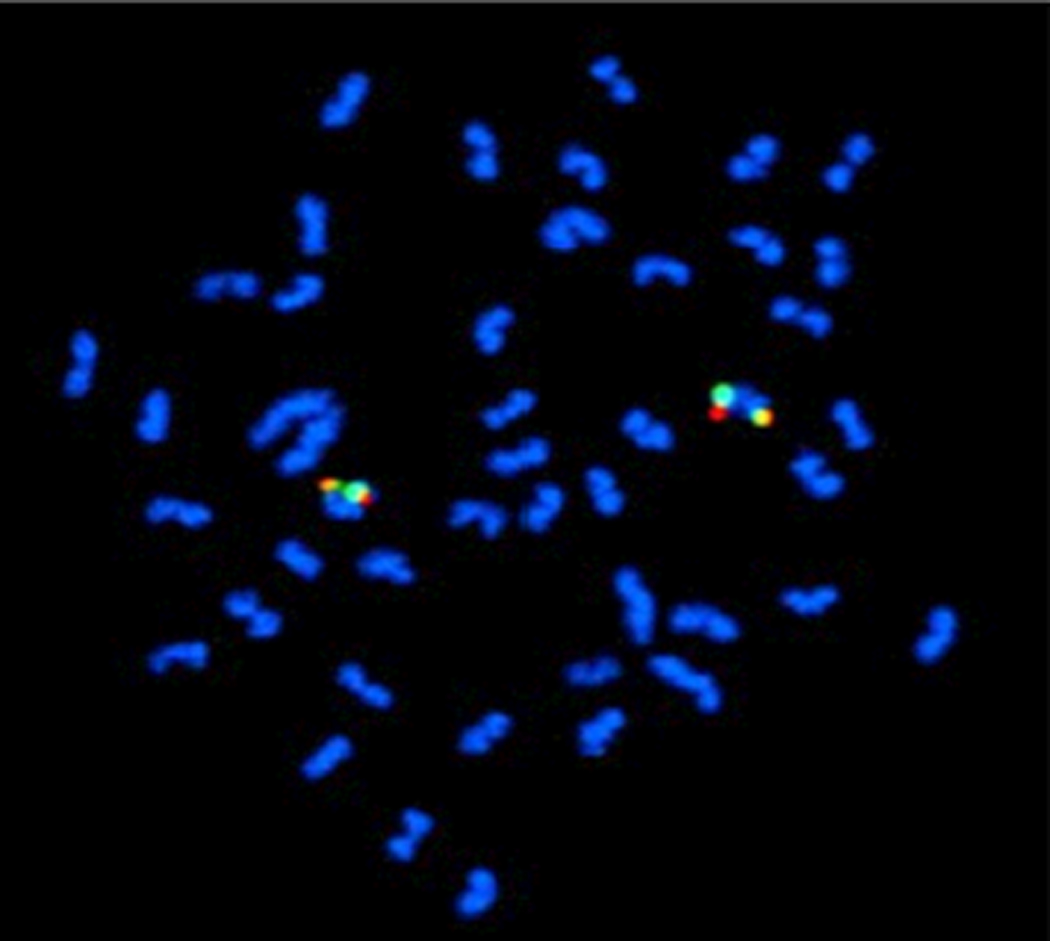
Hybridization of BAC clones to metaphase chromosomes by FISH. Clone b04TI071CD06 containing UNH995 was labeled with DIG (red) and clone b04TI073CB11 containing Onix420 was labeled with biotin (green). Both hybridized to one of the smaller chromosomes in the karyotype.
Discussion
We previously located an XX/XY sex-determining locus in O. niloticus to a region spanning about 10cM on LG1 between the microsatellite markers UNH995 and GM201(Lee et al., 2003). In an attempt to develop additional markers within this interval, we screened several thousand AFLP markers by bulked segregant analysis of DNA from males and females. We found three sex-linked markers, but unfortunately they mapped outside the critical interval. Afterward a newly developed SNP-based marker, Wt1b narrowed the interval by 6 cM (Lee and Kocher, 2007a).
The markers we developed appear to correspond with markers developed by Ezaz et al. (2004) because they include the same combination of selective bases in the AFLP protocol, and produce DNA fragments of the same size. OniX420 and OniY425 are alleles of the same locus (Ezaz et al. 2004). We found that OniX420 and UNH995 are within 100kb of each other on a single contig of BACs assembled by restriction fingerprint analysis. We also found that MCTA/EAAG-425 is very tightly linked to UNH995 by genetic linkage analysis. These results suggest that MCTA/EAAG-425 and OniY425/OniX420 are in fact the same locus. We genetically mapped MCTT/EACG382 coincident with GM201. OniY382 was amplified in the BACs of the contig that contains a forkhead box gene. Comparative mapping suggests this gene is about 100 kb away from GM201 in the Tetraodon genome, and a BAC end sequence of b03TI096BA12 shares the same scaffold with GM201 in the Fugu genome (Lee, 2004; Lee and Kocher, 2007b). These results suggest that MCTT/EACG-382 and OniY382 might be the same locus, but without sequence information for MCTT/EACG-382, we cannot reach a definitive conclusion.
Two previous studies have reported FISH mapping of sex-linked markers in O. niloticus (Ezaz et al., 2004; Cnaani et al., 2008). Ezaz et al. (2004) reported that BACs containing markers OniY227 and OniY382 hybridized to the largest chromosome (chromosome 1). A BAC containing marker OniY420 showed strong hybridization to a small chromosome (chromosome 2) with a weak signal on the large chromosome (chromosome 1). They concluded that either the strong signal on the small chromosome came from a repeat sequence shared with chromosome 1, or the BAC clone used as the probe was a chimera consisting of sequences from chromosome 2 attached to a small piece of chromosome 1 containing OniX420/OniY425. Cnaani et al. (2008) reported that a BAC contig containing microsatellite marker UNH995 mapped to a small tilapia chromosome. The same study showed that markers from tilapia LG3, which contains a ZZ/ZW sex-determining system in some species of tilapia, hybridized to the large chromosome (chromosome 1).
In this study, we have shown that a BAC clone containing OniX420 is part of a fingerprinted contig of BACs that also contains UNH995. BACs from this contig hybridize to the same region of a small chromosome, with no detectable hybridization on the largest chromosome pair (Figure 3). Further, we identified BAC contigs containing each of the three markers reported by Ezaz et al. (2004) and show that these all map to a small region of the Tetraodon genome. We cannot explain the hybridization signal reported by Ezaz et al. (2004) on the large chromosome, or the incomplete meiotic synapsis which first suggested the large pair might be the sex chromosome. Nonetheless, our new data clearly indicate that the XX/XY sex-determining locus identified in previous reports maps to one of the smaller chromosomes in the Nile tilapia karyotype.
Acknowledgements
This project was supported by National Research Initiative competitive grant no. 2006-04830 from the USDA National Institute of Food and Agriculture Animal Genome Program and grant no. #IS-3995-07 US-Israel Binational Agricultural Research and Development Fund.
References
- Abucay JS, Mair GC, Skibinski DOF, Beardmore JA. Environmental sex determination: the effect of temperature and salinity on sex ratio in Oreochromis niloticus L. Aquaculture. 1999;173:219–234. [Google Scholar]
- Baroiller JF, Chourrout D, Fostier A, Jalabert B. Temperature and sexchromosomes govern sex-ratios of the mouthbrooding Cichlid fish Oreochromisniloticus. J Exp Zool. 1995;273:216–223. [Google Scholar]
- Campos-Ramos R, Harvey SC, Masabanda JS, Carrasco LAP, Griffin DK, Mcandrew BJ, Bromage NR, Penman DJ. Identification of putative sex chromosomes in the blue tilapia, Oreochromis aureus, through synaptonemal complex and FISH analysis. Genetica. 2001;111:143–153. doi: 10.1023/a:1013707818534. [DOI] [PubMed] [Google Scholar]
- Carrasco LAP, Penman DJ, Bromage N. Evidence for the presence of sex chromosomes in the Nile tilapia (Oreochromis niloticus) from synaptonemal complex analysis of XX, XY and YY genotypes. Aquaculture. 1999;173:207–218. [Google Scholar]
- Cnaani A, Lee BY, Zilberman N, Ozouf-Costaz C, Hulata G, Ron M, D'hont A, Baroiller JF, D'cotta H, Penman DJ, Tomasino E, Coutanceau JP, Pepey E, Shirak A, Kocher TD. Genetics of sex determination in tilapiine species. Sex Dev. 2008;2:43–54. doi: 10.1159/000117718. [DOI] [PubMed] [Google Scholar]
- Ezaz MT, Harvey SC, Boonphakdee C, Teale AJ, Mcandrew BJ, Penman DJ. Isolation and physical mapping of sex-linked AFLP markers in nile tilapia (Oreochromis niloticus L.) Mar Biotechnol (NY) 2004;6:435–445. doi: 10.1007/s10126-004-3004-6. [DOI] [PubMed] [Google Scholar]
- Fao . The State of World Fisheries and Aquacultrue 2006. Rome: Food and Agriculture Organization of the United Nations; 2007. [Google Scholar]
- Fischer C, Ozouf-Costaz C, Roest Crollius H, Dasilva C, Jaillon O, Bouneau L, Bonillo C, Weissenbach J, Bernot A. Karyotype and chromosome location of characteristic tandem repeats in the pufferfish Tetraodon nigroviridis. Cytogenet Cell Genet. 2000;88:50–55. doi: 10.1159/000015484. [DOI] [PubMed] [Google Scholar]
- Harvey SC, Masabanda J, Carrasco LA, Bromage NR, Penman DJ, Griffin DK. Molecular-cytogenetic analysis reveals sequence differences between the sex chromosomes of Oreochromis niloticus: evidence for an early stage of sex-chromosome differentiation. Cytogenet Genome Res. 2002;97:76–80. doi: 10.1159/000064036. [DOI] [PubMed] [Google Scholar]
- Hopkins KD, Shelton WL, Engle CR. Estrogen sex-reversal of Tilapia aurea. Aquaculture. 1979;18:263–268. [Google Scholar]
- Karayucel I, Ezaz T, Karayucel S, Mcandrew BJ, Penman DJ. Evidence for two unlinked "sex reversal" loci in the Nile tilapia, Oreochromis niloticus, and for linkage of one of these to the red body colour gene. Aquaculture. 2004;234:51–63. [Google Scholar]
- Katagiri T, Asakawa S, Minagawa S, Shimizu N, Hirono I, Aoki Construction and characterization of BAC libraries for three fish species; rainbow trout, carp and tilapia. Anim Genet. 2001;32:200–204. doi: 10.1046/j.1365-2052.2001.00764.x. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Kidd C, Tomasino E, Davis JT, Wishon C, Stern JE, Carleton KL, Howe AE, Kocher TD. A BAC-based physical map of the Nile tilapia genome. Bmc Genomics. 2005;6:89. doi: 10.1186/1471-2164-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY. Program in Genetics. Durham: University of New Hampshire; 2004. Approach to the identification of sex-determining genes in the tilapia genome by genetic mapping and comparative positional cloning. [Google Scholar]
- Lee BY, Hulata G, Kocher TD. Two unlinked loci controlling the sex of blue tilapia (Oreochromis aureus) Heredity. 2004;92:543–549. doi: 10.1038/sj.hdy.6800453. [DOI] [PubMed] [Google Scholar]
- Lee BY, Kocher TD. Exclusion of Wilms tumour (WT1b) and ovarian cytochrome P450 aromatase (CYP19A1) as candidates for sex determination genes in Nile tilapia (Oreochromis niloticus) Anim Genet. 2007a;38:85–86. doi: 10.1111/j.1365-2052.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Lee BY, Kocher TD. Chapter 19. Comparative Genomics and Positional Cloning. In: Liu Z, editor. Aquaculture Gnome Technologies. Ames, IA: Wiley-Blackwell; 2007b. pp. 323–335. [Google Scholar]
- Lee BY, Penman DJ, Kocher TD. Identification of a sex-determining region in Nile tilapia (Oreochromis niloticus) using bulked segregant analysis. Anim Genet. 2003;34:379–383. doi: 10.1046/j.1365-2052.2003.01035.x. [DOI] [PubMed] [Google Scholar]
- Mair GC, Abucay JS, Skibinski DOF, Abella TA, Beardmore JA. Genetic manipulation of sex ratio for the large-scale production of all-male tilapia, Oreochromis niloticus. Can J Fish Aquat Sci. 1997;54:396–404. [Google Scholar]
- Mair GC, Scott AG, Penman DJ, Beardmore JA, Skibinski DOF. Sex determination in the genus Oreochromis .1. Sex reversal, gynogenesis andtriploidy in Oreochromis niloticus (L) Theor Appl Genet. 1991;82:144–152. doi: 10.1007/BF00226205. [DOI] [PubMed] [Google Scholar]
- Majumdar KC, Mcandrew BJ. Relative DNA content of somatic nuclei and chromosomal studies in 3 genera, Tilapia, Sarotherodon, and Oreochromis of the Tribe Tilapiini (Pisces, Cichlidae) Genetica. 1986;68:175–188. [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis - a rapid method to detect markers in specific genomic regions by using segregating populations. P Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocalewicz K, Mota-Velasco JC, Campos-Ramos R, Penman DJ. FISH and DAPI staining of the synaptonemal complex of the Nile tilapia (Oreochromis niloticus) allow orientation of the unpaired region of bivalent 1 observed during early pachytene. Chromosome Res. 2009;17:773–782. doi: 10.1007/s10577-009-9071-9. [DOI] [PubMed] [Google Scholar]
- Rosenstein S, Hulata G. Sex reversal in the genus Oreochromis: optimization of feminization protocol. Aquac Res. 1994;25:329–339. [Google Scholar]
- Soderlund C, Longden I, Mott R. FPC: a system for building contigs from restriction fingerprinted clones. Comput Appl Biosci. 1997;13:523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- Stam P. Construction of integrated genetic linkage maps by means of a new computer package: Join Map. The Plant Journal. 1993;3:739–744. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Van De Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]


