Abstract
The large-scale BAC end-sequencing project of Nile tilapia (Oreochromis niloticus) has generated extensive sequence data that allowed the examination of the repeat content in this fish genome and building of a repeat library specific for this species. This library was established based on Tilapiini repeat sequences from GenBank; sequences orthologous to the repeat library of zebrafish in Repbase; and novel repeats detected by genome analysis using MIRA assembler. We estimate that repeats constitute about 14% of the tilapia genome and also give estimates for the occurrence of the different repeats based on the BLAST searches within the database of known tilapia sequences. The frequent occurrence of novel repeats in the tilapia genome indicates the importance of using the species-specific repeat masker prior to sequence analyses. A web tool based on the RepeatMasker software was designed to assist tilapia genomics.
Keywords: transposable-element, sequence-assembly, repeat-masking
Introduction
Repeat masking is a crucial step in many sequence analyses including assembly of genomic and EST sequences (Tang, 2007, Malde and Jonassen, 2008); sequence searches as well as gene identification and annotation (Smith et al., 2007); and the design of PCR primers and hybridization probes (Andreson et al., 2006). However, repeat libraries are not available for most fish species and it is a common practice to mask against known repeats from other model organisms such as zebrafish (Danio rerio) and pufferfish (Takifugu rubripes), which is less effective than masking with repeat libraries that are species- specific (Malde and Jonassen, 2008). Several classes of repeats have been described in cichlid fish mostly in Oreochromis niloticus (O. niloticus). They include satellite DNAs (Oliveira and Wright, 1998); LINES (Oliveira et al., 1999); telomeric (TTAGGG)n repeats (Chew et al., 2002); rDNA repeats (Martins et al., 2002); SINES (Terai et al., 2003); and heterochromatic repetitive sequences (Ferreira and Martins, 2008, Mazzuchelli and Martins, 2009). This work has annotated a repeat library for tilapia by combining the previously annotated repeats from Tilapiini, sequences from O. niloticus BAC-end project that were orthologous to the zebrafish repeat library, and novel repeats of tilapia classified by genome sequence analysis.
Materials and Methods
Preparation of tilapia repeat library
GenBank records (209) of annotated repetitive sequence in Tilapiini were located (http://www.ncbi.nlm.nih.gov/) within 231 non-mRNA sequences that corresponded to the following limits ((Tilapiini[Organism]) AND ((repeat[All Fields]) OR (transposon*[All Fields]) OR (repetitive[All Fields]) OR (SINE*[All Fields]) OR (LINE*[All Fields]) OR (satellite*[All Fields]) OR (“ribosomal RNA”[All Fields]))) NOT (mitochondri*[All Fields]). We frequently encountered failure to identify and remove all of the vector sequence in the finished BAC-end sequences (Fig. 1A). In order to detect such sequence contamination common to the current genomic projects of this fish tribe, the BAC cloning vector (FJ160466) was added to our repeat library records. BAC-end sequences (153,216) of O. niloticus were downloaded from Trace Archive (CENTER_PROJECT=”G1447”, http://www.ncbi.nlm.nih.gov/Traces/) and combined with 7,855 Tilapiini records available from GenBank to form a local database that was searchable by BLAST (blastall 2.2.17) on an Ubuntu (8.04 Hardy) 64-bit Q6600 Linux machine. Danio rerio (DR) records for high complexity repeats in Repbase13 (Jurka et al., 2005) were used as the queries in BLAST searches against this local database to reveal BAC-end sequences with significant (EXPECT threshold <1.0e−3) similarity matches. Characterization of tilapia sequence orthologues was semi-automated using PERL scripts. Sequences with 20% similarity or greater were assembled using GAP4 software (Staden 1.7.0) (Staden et al., 2000) in 169 databases, one for each of the Repbase matching entries. The resulting 327 contigs, which had no known annotation in tilapia, were mostly annotated as follows: [repeat record number]_[frequency in the database]_[name of DR repeat]_[matching location in this repeat]_[SRA accession number of the contig most 5′ BAC-end]. The annotated entries were constantly used to update the tilapia repeat library which was searchable using a CGI web tool based on RepeatMasker (version Open 3.2.6 A.F.A. Smit, R. Hubley & P. Green RepeatMasker at http://repeatmasker.org) that we created. During contig assembly 15 entries for repetitive sequences, which had no significant orthology to known repeats were annotated as “unknown”. To further characterize such repeats, we used MIRA software (V2.9.43) (Chevreux et al., 2004) under the –highlyrepetitive switch to assemble all BAC-ends. Output contained 27 Mb of consensus sequence in 23,722 contigs and the 528,828 filtered repeats in the file with the suffix _int_skimmarknastyrepeats_nastyseq_preassembly2.0.lst. Using Linux commands (grep, awk, sort) 332,244 unique repeats were detected. Following masking and resorting, 21,814 repeats longer than 36 bp were left. Of these the 2,003 sequences that were longer than 200 bp were GAP4 assembled into 38 contigs that were mostly annotated as “unknown” with the indication “_MIRA” in the field used for the accession number. The rest of the repeats that were not masked by the updated library (7,452 records) were GAP4 assembled into 1,871 contigs. The 16 main contigs (repeat frequency above 60, length ~200 bp) were then added to the repeat library and the rest 1574 unique contigs with average length of 51 bp were unified using ‘NNNNN’ spacers into one record under the entry annotation “Misc_short_repeats_generated_by_MIRA”. The complete repeat library is searchable and downloadable from http://cowry.agri.huji.ac.il/cgi-bin/TilapiaRM.cgi.
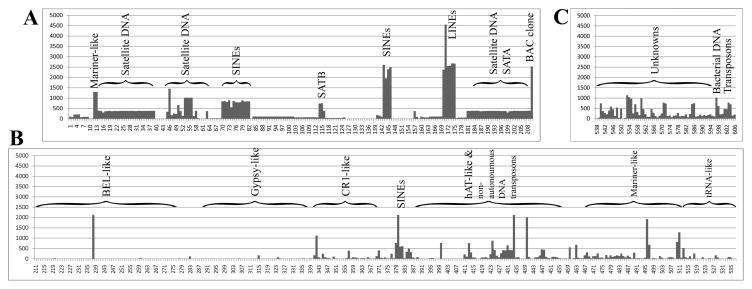
Fig. 1. Frequencies of repetitive elements in the tilapia genome.
A map of the tilapia repeat library that was established from three sequence sources is shown: A. Tilapiini repeat-sequences from GenBank; B. O. niloticus sequences orthologous to the repeat library of zebrafish in Repbase; C. novel O. niloticus repeats mostly detected by genome analysis using MIRA assembler. Under the horizontal axis the repeat number within this library is indicated. The vertical axis denotes the number of significant hits obtained by BLAST search against our local database, which consist mostly of O. niloticus BAC-end sequences. Types of repeats that form major landmarks in this map are indicated above major frequency peaks and above regions of repeat superfamilies delineated with brackets.
Results and Discussion
A total of 607 records were gathered to form the tilapia specific repeat library (Fig. 1). About a third of them (Fig. 1A) consist of entries imported directly from GenBank. To the original annotation of these records we added: a serial repeat number (RN) for the repeat in our library followed by an indication that may help to estimate the repeat frequency in the genome. This indication is the number of sequences that produced a significant alignment (EXPECT score better than 1.0e−3) in a local BLAST search against the known tilapia sequences (161,071 records, ~120 Mbp), mostly obtained from the BAC-end sequencing project. As the RNs also reflect batch submissions and creation dates in the original databank, the distribution of peaks (Fig. 1A) is not random and it is associated with the repeat types indicated above major landmarks on the map of the repeat library (Fig. 1). The most frequent repeat detected (Fig. 1A, RN171, frequency 4546) was annotated as CiLINE2 repeat sequence (Oliveira et al., 1999). Indeed L2 class of LINE-like retrotransposons from the CR1 superfamily are the most numerous repeat in Fugu (>6500 copies) (Jurka et al., 2005; Poulter et al., 1999), however the RN171 frequency is too high to be explained by CiLINE2 alone as it was obtained from a fraction equivalent to about 11% of the genome, assuming genome size of 1100 Mbp (Lee et al., 2005). A careful search of RN171 using Censor web tool (Kohany et al., 2006) showed that while the 5′ end contains the LINE2-like element, its 3′ end was similar to hAT-N3_FR element of the ancient and common hAT superfamily of transposons (Rubin et al., 2001). Hence, the RN171 annotation is problematic and this chimaeric element brought together the two major classes of repeats: a Class I element (retrotransposon) that moves via an RNA intermediate, and a Class II element (transposon) that migrates via a DNA intermediate. This combination was the reason for the particularly numerous BLAST hits encountered. Transposable elements nested within one another are a common situation and a known problem in repeat annotation (Kronmiller and Wise, 2008). The rest of the CiLINE2 repeats (Fig. 1A, RN170–175, frequency ~2500) with an estimated copy number of about 5500 for the haploid genome of O. niloticus (Oliveira et al., 1999) suggest that in order to estimate the number of occurrences in the genome the frequencies reported in this work should be at least doubled. Similar conclusion can be drawn from analysis of the frequency of 1900 bp SATB (Fig. 1A, RN115, frequency 745), which is one of the two main satellite DNA sequences in O. niloticus. SATB (1,000–10,000 copies per genome) is restricted to the centromeric region of a single chromosome (Oliveira and Wright, 1998). Unexpectedly, the other main satellite, the 237 bp SATA, which is distributed in the centromeric regions of all chromosomes, with tenfold higher copy number (Oliveira and Wright, 1998), had fewer BLAST hits (Fig 1A, RN206, frequency 384) than SATB.
Another notable class I repeats (Fig 1A, RN70–82, frequency 850; RN143–146, frequency ~2500) are the AFC SINEs (Terai et al., 2003), which were suggested as useful probes for the analysis of speciation of African cichlids. The most frequent group of class II repeats contained the recently identified MMTS transposons (Ahn et al., 2008) of the Mariner/Tc1-superfamily (Fig 1A, RN12–13, frequency 1296). The identification of numerous (2525) BLAST hits against the BAC vector prompted us to include the BAC vector sequence (RN210) at the end of the GenBank derived records (Fig 1A, most right peak).
Most (~54%) of the entries of our tilapia repeat library were derived from orthologous repeats that were present in the zebrafish repeat library in Repbase (Fig.1B). These were used to BLAST search the BAC-end database and to form orthologous entries in our library. A few fossilized copies of the large ancient Class I repeat superfamilies with direct-orientation flanking long-terminal repeats (LTRs) of BEL (Frame et al., 2001) and of Gypsy (Britten et al., 1995) were detected (Fig. 1B, RN211–274 and RN288–337, respectively). It should be noted that the exceptional frequency of the BEL13-like element (Fig. 1B, RN238, frequency 2132) results from chimerism of this element with a Mariner-like sequence.
The non-LTR retrotransposon of Class I repeats were more abundant (Fig. 1B, CR1-like, frequencies up to 1129 in RN342) and have been also previously reported as widespread LINEs in teleosts (Mazzuchelli and Martins, 2009). SINEs were an even more frequent non-LTR retrotransposons. Noteworthy was SINE_TE-like element (Fig. 1B, RN379–382, frequencies 588–2131), which is a member of the V-SINE superfamily (Ogiwara et al., 2002).
A substantial portion of the high-frequency repetitive elements detected using the zebrafish Repbase library was Class II transposons (Fig. 1B). These include repeats similar to hAT and Mariner superfamilies and non-autonomous DNA transposons which rely on other active intact elements of Class II type to move them. It should be noted that in this category chimaeric repeats produced numerous BLAST hits because they contained a nested SINE within (RN434, frequency 2125; RN496 frequency 1929) or a combination of hAT- and Mariner-like elements (RN440, frequency 2017). Moderate frequencies (1–174) for occurrences of repeats orthologous to tRNA pseudogenes were observed (Fig. 1B, to the right).
The third stage in assembling our tilapia repeat library was the addition of repeats that were not previously annotated or could not be detected by similarity search against GenBank or the zebrafish Repbase records (Fig. 1C). A whole genome sequence analysis to systematically detect such repeats has been recommended (Malde and Jonassen, 2008). As the MIRA genome assembler (Chevreux et al., 2004) is a specialized assembler for sequencing projects with a high number of similar repeats, we took advantage of the sophisticated algorithms implemented in this assembler for disentangling repeats. These take into account the number of sequence occurrences relative to the expected coverage as well as the number of nucleotide variations within the repeated region. While assembling the available O. niloticus BAC-end sequences, 14% of the input sequences were annotated as repeats with an average of 3.5 repetitive sequences per read. Repetitive sequences constitute about 50% of the human genome (Tang, 2007) and consequently its size is larger than the O. niloticus genome. Assuming that vertebrates have similar number of genes (Aparicio et al., 2002), it is indeed expected that the repeat content in O. niloticus genome would be of smaller proportion and similar to that of chicken (~11% repeat content in 1200 Mbp genome (Tang, 2007)). It should be noted that although most of the genome size differences can ultimately be attributed to repeats, the precise annotation of the repeat content and the estimation of its size are complicated as there are ancient repeats that are degenerated beyond recognition.
Repeat sequences that we detected using the MIRA assembler and that were not masked by the repeat library created in the first two stages, were assembled and the consensus sequences were added to this library. A total of 59 entries that belonged to 43 groups with no significant similarity to known repeats were annotated as “unknowns”. The importance of characterizing these repeats and creating the species-specific repeat library is evident from the relative abundance of these repeats (e.g. RN554, frequency 1044, Fig. 1C). The MIRA assembler also pointed out frequent bacterial and vector DNA contaminations that were not masked by the RepeatMasker defaults. These were added to repeat library as RN597-8 (Fig. 1C). The MIRA assembler also detected repeats that were orthologous to previously annotated transposons and escaped our analysis as they were not present in the zebrafish Repbase library. Repbase libraries for fugu, and even invertebrates such as planaria and hydra, seem to be valuable for identifying repetitive sequences that have escaped our analysis. This work was aimed at producing a practical web-tool in the form of RepeatMasker that would assist tilapia genomics. Based on O. niloticus repeats that our tilapia repeat-masker failed to mask and that we encountered while practically using this tool, we estimate that this library represents about 80% of the repeats that would be present following similar analysis using the complete genome data.
Acknowledgments
This is a contribution from the Agricultural Research Organization, Institute of Animal Science, Bet Dagan, Israel, No. 549/09. The research was supported by research grant IS-3995-07 from the United States-Israel Bi-national Agricultural Research and Development (BARD) Fund.
Contributor Information
A. Shirak, Agricultural Research Organization, Institute of Animal Science, Bet-Dagan 50250, Israel
M. Grabherr, Broad Institute of Harvard and MIT, 320 Charles Street, Cambridge, Massachusetts 02141, USA
F. Di Palma, Broad Institute of Harvard and MIT, 320 Charles Street, Cambridge, Massachusetts 02141, USA
K. Lindblad-Toh, Broad Institute of Harvard and MIT, 320 Charles Street, Cambridge, Massachusetts 02141, USA
G. Hulata, Agricultural Research Organization, Institute of Animal Science, Bet-Dagan 50250, Israel
M. Ron, Agricultural Research Organization, Institute of Animal Science, Bet-Dagan 50250, Israel
TD. Kocher, Department of Biology, University of Maryland, College Park, MD, 20742 USA
E. Seroussi, Email: Seroussi@agri.huji.ac.il, Agricultural Research Organization, Institute of Animal Science, Bet-Dagan 50250, Israel
References
- Ahn SJ, Kim MS, Jang JH, Lim SU, Lee HH. MMTS, a New Subfamily of Tc1-like Transposons. Molecules and Cells. 2008;26:387–395. [PubMed] [Google Scholar]
- Andreson R, Reppo E, Kaplinski L, Remm M. GENOMEMASKER package for designing unique genomic PCR primers. Bmc Bioinf. 2006:7. doi: 10.1186/1471-2105-7-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia J, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MDS, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJK, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Mccormack TJ, Mears TL, Davidson EH. Gypsy/Ty3-Class Retrotransposons Integrated in the DNA of Herring, Tunicate, and Echinoderms. J Mol Evol. 1995;40:13–24. doi: 10.1007/BF00166592. [DOI] [PubMed] [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WE, Wetter T, Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew JSK, Oliveira C, Wright JM, Dobson MJ. Molecular and cytogenetic analysis of the telomeric (TTAGGG)(n) repetitive sequences in the Nile tilapia, Oreochromis niloticus (Teleostei: Cichlidae) Chromosoma. 2002;111:45–52. doi: 10.1007/s00412-002-0187-3. [DOI] [PubMed] [Google Scholar]
- Ferreira IA, Martins C. Physical chromosome mapping of repetitive DNA sequences in Nile tilapia Oreochromis niloticus: Evidences for a differential distribution of repetitive elements in the sex chromosomes. Micron. 2008;39:411–418. doi: 10.1016/j.micron.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Frame IG, Cutfield JF, Poulter RTM. New BEL-like LTR-retrotransposons in Fugu rubripes, Caenorhabditis elegans, and Drosophila melanogaster. Gene. 2001;263:219–230. doi: 10.1016/s0378-1119(00)00567-9. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. Bmc Bioinf. 2006:7. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronmiller BA, Wise RP. TEnest: Automated chronological annotation and visualization of nested plant transposable elements. Plant Physiol. 2008;146:45–59. doi: 10.1104/pp.107.110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY, Lee WJ, Streelman JT, Carleton KL, Howe AE, Hulata G, Slettan A, Stern JE, Terai Y, Kocher TD. A second-generation genetic linkage map of tilapia (Oreochromis spp.) Genetics. 2005;170:237–244. doi: 10.1534/genetics.104.035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malde K, Jonassen I. Repeats and EST analysis for new organisms. Bmc Genomics. 2008:9. doi: 10.1186/1471-2164-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C, Wasko AP, Oliveira C, Porto-Foresti F, Parise-Maltempi PP, Wright JM, Foresti F. Dynamics of 5S rDNA in the tilapia (Oreochromis niloticus) genome: repeat units, inverted sequences, pseudogenes and chromosome loci. Cytogenetic and Genome Res. 2002;98:78–85. doi: 10.1159/000068542. [DOI] [PubMed] [Google Scholar]
- Mazzuchelli J, Martins C. Genomic organization of repetitive DNAs in the cichlid fish Astronotus ocellatus. Genetica. 2009;136:461–469. doi: 10.1007/s10709-008-9346-7. [DOI] [PubMed] [Google Scholar]
- Ogiwara I, Miya M, Ohshima K, Okada N. V-SINEs: A new superfamily of vertebrate SINEs that are widespread in vertebrate Genomes and retain a strongly conserved segment within each repetitive unit. Genome Res. 2002;12:316–324. doi: 10.1101/gr.212302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C, Chew JSK, Porto-Foresti F, Dobson MJ, Wright JM. A LINE2 repetitive DNA sequence from the cichlid fish, Oreochromis niloticus: sequence analysis and chromosomal distribution. Chromosoma. 1999;108:457–468. doi: 10.1007/s004120050397. [DOI] [PubMed] [Google Scholar]
- Oliveira C, Wright JM. Molecular cytogenetic analysis of heterochromatin in the chromosomes of tilapia, Oreochromis niloticus (Teleostei: Cichlidae) Chromosome Res. 1998;6:205–211. doi: 10.1023/a:1009211701829. [DOI] [PubMed] [Google Scholar]
- Poulter R, Butler M, Ormandy J. A LINE element from the pufferfish (fugu) Fugu rubripes which shows similarity to the CR1 family of non-LTR retrotransposons. Gene. 1999;227:169–179. doi: 10.1016/s0378-1119(98)00600-3. [DOI] [PubMed] [Google Scholar]
- Rubin E, Lithwick G, Levy AA. Structure and evolution of the hAT transposon superfamily. Genetics. 2001;158:949–957. doi: 10.1093/genetics/158.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Edgar RC, Yandell MD, Smith DR, Celniker SE, Myers EW, Karpen GH. Improved repeat identification and masking in Dipterans. Gene. 2007;389:1–9. doi: 10.1016/j.gene.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- Tang H. Genome assembly, rearrangement, and repeats. Chemical Reviews. 2007;107:3391–3406. doi: 10.1021/cr0683008. [DOI] [PubMed] [Google Scholar]
- Terai Y, Takahashi K, Nishida M, Sato T, Okada N. Using SINEs to probe ancient explosive speciation: “Hidden” radiation of African cichlids? Mol Biol and Evol. 2003;20:924–930. doi: 10.1093/molbev/msg104. [DOI] [PubMed] [Google Scholar]