Abstract
Mutational activation of KRAS is a common event in human tumors. Identification of the key signaling pathways downstream of mutant KRAS is essential for our understanding of how to pharmacologically target these cancers in patients. We show that PD0325901, a small molecule MEK inhibitor, decreases MEK/ERK pathway signaling, and destabilizes Cyclin D1, resulting in significant anti-cancer activity in a subset of KRAS mutant tumors in vitro and in vivo. Mutational activation of PIK3CA, which commonly co-occurs with KRAS mutation, provides resistance to MEK inhibition through reactivation of AKT signaling. Genetic ablation of the mutant PIK3CA allele in MEK inhibitor-resistant cells restores MEK pathway sensitivity, and re-expression of mutant PIK3CA reinstates the resistance, highlighting the importance of this mutation in resistance to therapy in human cancers. In KRAS mutant tumors, PIK3CA mutation restores Cyclin D1 expression and G1/S cell cycle progression so that they are no longer dependent on KRAS and MEK/ERK signaling. Furthermore, the growth of KRAS mutant tumors with coexistent PIK3CA mutations in vivo is profoundly inhibited with combined pharmacologic inhibition of MEK and AKT. These data suggest that tumors with both KRAS and PI3K mutations are unlikely to respond to inhibition of the MEK pathway alone but will require effective inhibition of both MEK and PI3K/AKT pathway signaling.
Introduction
The three members of the Ras family of small GTPase proteins, KRAS, HRAS, and NRAS, play central roles in the transduction of growth factor receptor-induced signals (1). Mutations of RAS that impair its GTPase function are oncogenic and occur at high frequency in many human malignancies (2, 3). Activation of RAS has been implicated in mediating many aspects of the transformed phenotype, including deregulated proliferation, survival, invasion and metastasis.
The mechanisms through which RAS supports these processes are not completely understood. In the activated, GTP-bound state, RAS binds to and activates multiple effector proteins of which more than ten have been identified (4). Of these, the most studied are the RAF kinase and phosphoinositide 3-kinase (PI3K) protein families and Ral-GDS (Ral guanine dissociation stimulator)(5–8). Early work focused on the RAF family of serine/threonine protein kinases. RAS-GTP binds to and activates the three RAF kinase family members which phosphorylate their main substrates, the serine/threonine kinases MEK1 and MEK2, which in turn activate the two ERK kinases (5, 9, 10). ERK phosphorylates multiple substrates and has pleiotrophic cellular effects, including induction of proliferation and invasion. This observation, the finding that ERK activation was required for the transformation of NIH3T3 cells by mutant RAS (11), the isolation of gag-RAF as a retroviral oncogene (12), and the recent discovery that BRAF is mutated and oncogenic in a significant fraction of human tumors (13) led to the idea that the RAF kinases and subsequent activation of MEK/ERK signaling are key effectors of mutant KRAS-induced transformation. Therefore, pharmaceutic efforts have focused on developing therapeutic agents that inhibit the components of this KRAS effector pathway (14, 15). However, the ability to pharmacologically and genetically block key KRAS effector pathways unveiled unexpected complexities of KRAS signaling in human cancer. Unlike the findings reported for BRAF mutant tumors (16, 17), many KRAS mutant tumor cells are resistant to MEK/ERK pathway inhibition. In fact, recent studies in vitro and in vivo suggest that KRAS mutant tumors require dual inhibition of both the MEK and PIK3CA pathways in order to achieve inhibition of tumor growth (18–20).
Here, we use a selective, allosteric MEK inhibitor to determine the MEK-dependence of tumors with mutational activation of the pathway. These studies indicate that many KRAS mutant tumor cell lines are, contrary to the prevailing view, sensitive to the MEK inhibitor PD0325901 and hence dependent on the RAF/MEK/ERK signaling arm. Resistance to MEK inhibitors in the relevant cell lines is not an intrinsic feature of KRAS oncogenic function but instead mutational activation of PIK3CA is present in most, but not all, MEK-resistant KRAS mutant cancers. In such resistant lines, sensitivity is restored by functional ablation of mutant PI3K activity. Furthermore, our data show that combined inhibition of both MEK/ERK and PI3K/AKT pathways in tumors with both KRAS and PIK3CA mutations is effective in profoundly inhibiting their growth in vivo. Overall, our data define genotypes that suggest which KRAS tumors may be effectively treated with MEK inhibition and support the possibility that MEK/ERK inhibition, alone or in combination with inhibition of PI3K/AKT signaling may be effective therapeutic strategies for mutant KRAS-dependent malignancy.
Materials and Methods
Cell culture
Human colon, pancreas and lung cancer cell lines were obtained from the ATCC (Manassas, VA) and maintained as described in the Supplemental Methods. The MEK inhibitor, PD0325901 was synthesized as described (21). The dual AKT1/AKT2 inhibitor (AKTi-1/2) was obtained from Merck and Co. Inc (Whitehouse Station, NJ) (22–25). The PIK3CA isogenic HCT116 and DLD-1 cell lines have been previously described (26), and were kindly provided by Drs. Bert Vogelstein and Victor Velculescu (The Johns Hopkins University, Baltimore, MD). Cell proliferation assay and Western blot analysis were performed as previously described (17). Antibodies used for immunoblotting are listed in the Supplemental Methods.
Colony formation in soft agar
To assess anchorage-independent growth, triplicate samples of 1×104 cells were mixed in complete growth medium containing 0.3% low-melting agarose and the indicated concentration of PD0325901. In a single well of a six-well plate, 2ml of cell mixture was plated on top of a 2ml of solidified layer of 0.6% agarose containing growth medium. The agarose was overlaid with 200µl of complete medium. Cells were stained with crystal violet (Sigma-Aldrich) and photographed after 21 days using a dissecting microscope. Assays were done in triplicates and in all cases independently, at least twice.
siRNA experiments
ON-TARGETplus KRAS smartpool consisting of four siRNA duplexes (L-005069, Dharmacon) was used for KRAS knockdown. 3×105 cells were plated in six-well plates and the next day transfected with 20nM KRAS siRNA or a control Non-Targeting siRNA#2 using DharmaFECT-1 transfection reagent and accompanying protocol (Dharmacon).
Cell cycle analysis
Cells were seeded in 6-well plates (3×105 cells/well) in normal growth medium, and the following day, the cells were treated with vehicle (DMSO), 50nM PD0325901, or siRNA duplexes in the siRNA experiments. Both adherent and floating cells were harvested. Nuclei were isolated by the Nusse method, stained with ethidium bromide and analyzed for DNA content by flow cytometry (27).
Generation of cells stably expressing mutant PIK3CA
HCT116 wild-type PIK3CA cell line stably expressing mutant PI3K (H1047R) was generated by infection with retroviruses carrying mutant PIK3CA encoding gene (pBabe-puro-HA-PIK3CA-H1047R, Addgene plasmid 12524) (28). Vector-only plasmid (pBabe-puro, Addgene plasmid 1764) was used as a control. The plasmids were transfected into the GPG293 amphotropic packaging cell line using Lipofectamine transfection reagent to produce retrovirus. GPG293 cells were provided by Dr. Joan Massague and maintained in DMEM with 10% FBS, doxycycline (20ng/ml), puromycin (2ug/ml), and G418 (0.3mg/ml). Virus was collected 72 hours post transfection and filtered to remove cell contamination. Filtered retrovirus was used to infect cells in the presence of 8mg/ml polybrene. After infection, successfully transduced polyclonal cell populations were obtained by selection with puromycin (2µg/ml) as a pool of stable clones.
Animal studies
Animal experiments were carried out as previously described (16), under an IACUC-approved protocol, and institutional guidelines for the proper, humane use of animals in research were followed. PD0325901 was formulated in 0.5% hydroxypropyl methylcellulose plus 0.2% Tween 80 and administered by oral gavage × 5 consecutive days each week for 3–4 weeks. AKTi-1/2 was formulated in 25% hydroxypropyl β-cyclodextrin (p H 4–5), and administered subcutaneously. Additional details are provided in the supplemental methods.
Results
A subset of KRAS mutant cells depends on MEK/ERK signaling
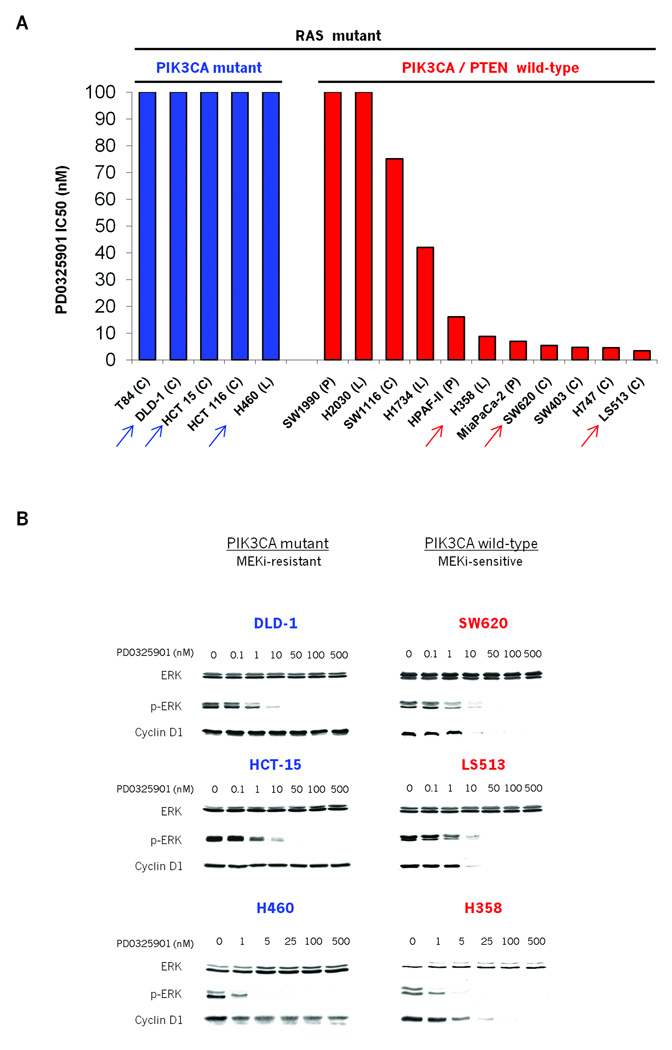
We first exposed a panel of sixteen KRAS mutant colorectal, lung, and pancreatic cancer cell lines to PD0325901, a potent and selective MEK inhibitor, in order to ascertain their dependence on MEK signaling for proliferation. At a concentration of 10nM, PD0325901 effectively inhibited MEK kinase signaling in all cells in the panel, as measured by loss of ERK phosphorylation (p-ERK) (Fig. 1B). In this panel however, MEK inhibition led to anti-proliferative activity in seven of these lines with IC50 values of less than 20nM PD0325901 (Fig. 1A). Seven cell lines showed no anti-proliferative response to PD0325901 at concentrations greater than 100nM (Fig. 1A) and two cell lines had an intermediate phenotype with IC50 values between 20 and 100nM. MEK dependence was not correlated with tumor lineage or the specific site of KRAS mutation. On the other hand, the majority of MEK-independent cell lines harbored PIK3CA mutations, as previously reported (Fig. 1A and S1) (29, 30). Importantly all the cell lines sensitive to MEK inhibition were wild-type for PIK3CA (Fig. 1A and S1).
Figure 1. MEK/ERK dependence of tumor cell lines with mutant KRAS.
A: Colon (C), pancreatic (P) and lung cancer (L) cell lines harboring KRAS mutation were grown in the presence of the MEK inhibitor PD0325901. Day 5 IC50 values were determined using Alamar Blue. Arrows indicate the representative cell lines illustrated in Fig 1B.
B: Effects of PD0325901 on ERK, p-ERK and Cyclin D1 expression in MEK-inhibitor sensitive (red) and resistant (blue) cell lines. Cells were treated with PD0325901, harvested 24hrs post treatment and cell lysates immunoblotted with the indicated antibodies.
In cells sensitive to MEK inhibition, expression of Cyclin D1 was MEK-dependent. In MEK inhibitor-resistant cells, Cyclin D1 expression was unaffected even at concentrations in vast excess of those required for inhibition of MEK activity (Fig. 1B). Thus, in a subset of KRAS mutant tumor cells, MEK/ERK signaling is absolutely required for Cyclin D1 expression and anchorage-dependent proliferation.
Coexistent KRAS and PIK3CA mutations prevent Cyclin D degradation and sensitivity to MEK inhibition
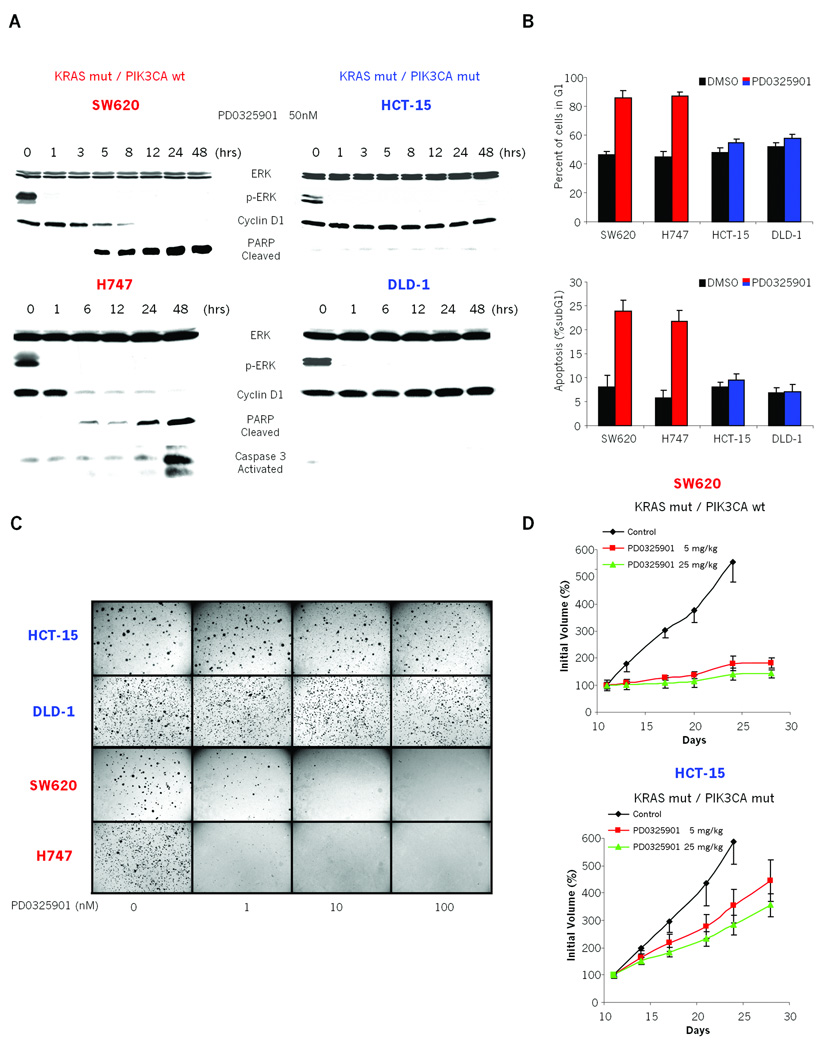
We further characterized the biological outcome of MEK antagonism in the above tumor cell lines. For example, in KRAS mutant/PIK3CA wild-type cell lines (SW620 and H747), MEK inhibition led to a rapid and persistent downregulation of Cyclin D1 expression and arrest of the cells in the G1 phase of the cell cycle (Fig. 1B, 2A and 2B). In these cells MEK inhibition also resulted in an increase in apoptosis as shown by an increase in cleaved PARP (Poly ADP-ribose polymerase) expression and a four-fold increase in the fraction of the cells with sub-G1 DNA content (Fig. 2A and 2B). As a result, MEK inhibition with PD325901 prevented the anchorage-independent growth of KRAS mutant/PIK3CA wild-type cells in soft agar (Fig. 2C). Notably, as little as 1nM PD0325901 was sufficient to inhibit colony formation by more than 50% in these lines (Fig. 2C).
Figure 2. Coexistent PIK3CA mutation is associated with decreased dependency on MEK signaling for tumor growth in vitro and in vivo.
A: Immunoblots showing the effects of MEK inhibition in PD0325901 sensitive (SW620, H747) and resistant (HCT-15, DLD-1) cell lines. Cells were treated with 50nM PD0325901 and lysates immunoblotted with the specified antibodies.
B: MEK-dependent (SW620, H747) and MEK-independent (HCT-15, DLD-1) cells were treated with 50nM PD0325901 or DMSO control and harvested 48 hours later. Graphs show: (Top) the percent of cells with G1 DNA content and (Bottom) the apoptotic fraction reported as the percent of cells with sub-G1 DNA content. The results represent the mean ± SE of two independent experiments done in triplicate.
C: MEK-dependent (SW620, H747) and MEK-independent (HCT-15, DLD-1) cells were treated with PD0325901 in a soft agar colony formation assay. Representative images at 21 days post plating are shown.
D: Mice with established SW620 and HCT15 xenografts were treated with PD0325901 × 5 days/week, for 3 weeks, or with vehicle only. The results represent the mean percent increase in tumor volume ± SE (n = 5 mice/group).
In contrast, upon MEK inhibition KRAS mutant/PIK3CA mutant cell lines (HCT-15 and DLD-1) exhibited neither Cyclin D1 degradation, cell cycle arrest, nor induction of apoptotsis (Fig. 2A and 2B). Again, these differences did not arise due to a lack of ERK inhibition (Fig. 1B and 2A). Consistent with this, anchorage-independent growth of these cells was unaffected by up to 100nM PD0325901 in soft agar assays (Fig. 2C).
We next determined whether MEK/ERK signaling was required for the growth of KRAS mutant colorectal tumors in vivo. To do this, we focused on two MEK-sensitive (SW620, H747) and two MEK-insensitive (HCT-15 and DLD-1) cell lines. 5 or 25 mg/kg of PD0325901 potently suppressed the growth of KRAS mutant/PIK3CA wild-type, SW620 and H747 xenografts (Fig. 2D and S2B). As seen in vitro, inhibition of tumor growth was associated with downregulation of p-ERK, Cyclin D1, and Cyclin D2 expression and induction of cleaved PARP (Fig. S2A). In contrast, KRAS/PIK3CA-double mutant HCT-15 and DLD-1 xenograft tumors grew in the presence of MEK inhibition, albeit at a somewhat slower rate than the control (Fig. 2D and S2B). In these tumors, no change in Cyclin D expression or induction of PARP cleavage was observed following PD0325901 treatment (Fig. S2A). Thus, Cyclin D expression, PARP cleavage, anchorage-independent in vitro growth, and in vivo growth of mutant KRAS colon tumor cells are not dependent on MEK/ERK signaling when PIK3CA mutations are present. Together these data help to reconcile previously observed complexity regarding MEK inhibitor sensitivity in RAS mutant cancers (16, 18) and support the notion that RAS mutation can predict sensitivity to MEK inhibition, but that primary resistance can be observed when RAS and PIK3CA mutations co-occur.
Selective knockout of mutant PIK3CA allele confers MEK/ERK dependence
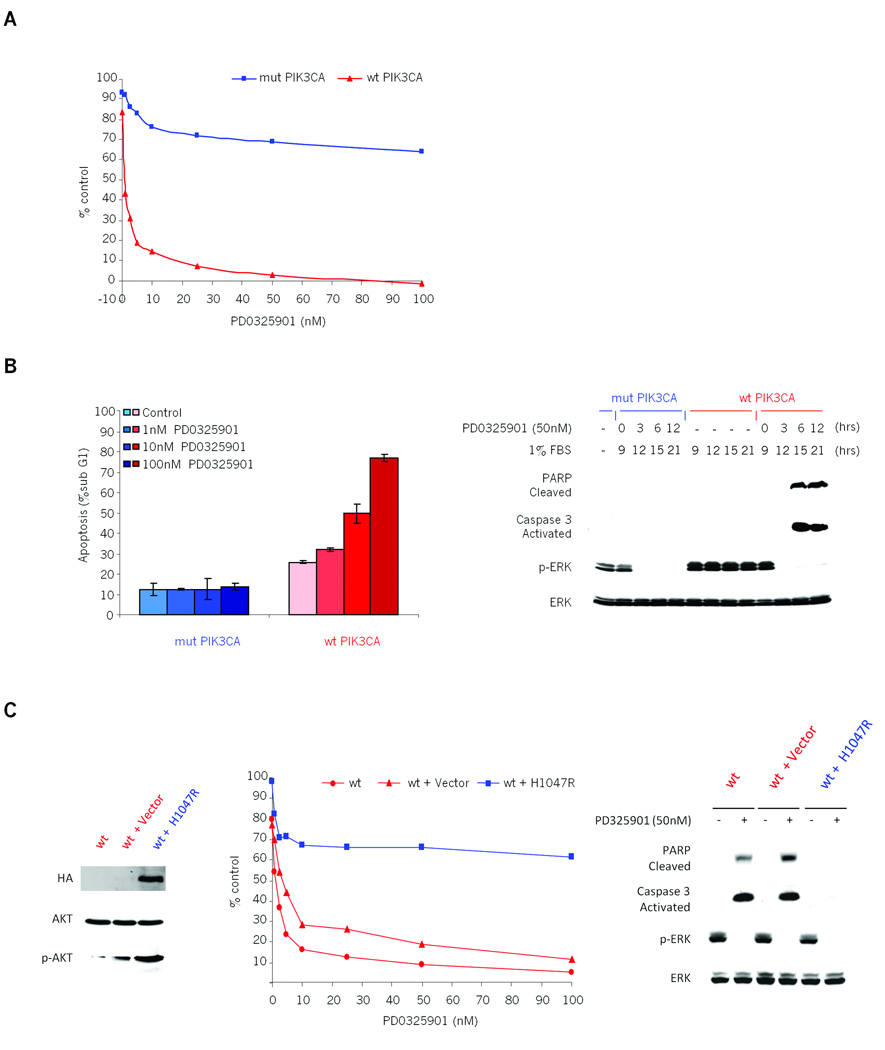
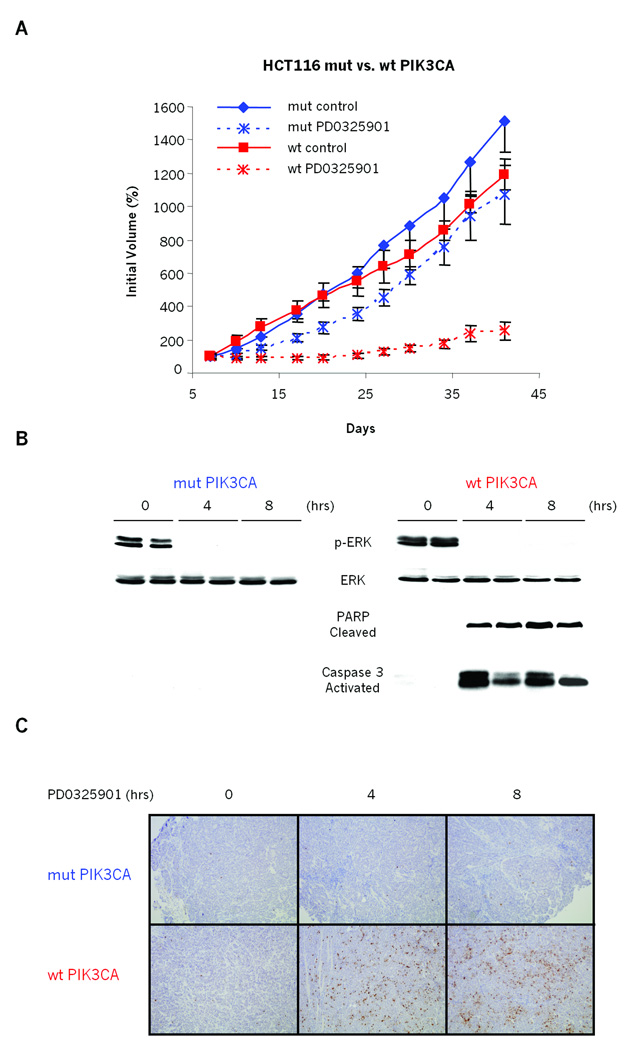
PIK3CA mutation could either be causal of MEK inhibitor resistance or simply associated with the phenomenon. To determine whether mutant PIK3CA was sufficient to cause resistance to MEK inhibition, we utilized KRAS mutant colorectal cancer cell lines that are isogenic with respect to PIK3CA mutational status (26). Both HCT116 and DLD-1 harbor heterozygous G13D KRAS mutations and are also heterozygous for H1047R or E545K PIK3CA mutations, respectively. Homologous recombination was used by Samuels and colleagues to knock out either the mutant or wild-type allele of PIK3CA in these cell lines, thus creating pairs of KRAS-mutant cell lines that are isogenic except with respect to their endogenous PIK3CA mutational status (26). Both HCT116 and DLD-1 cell lines with mutant PIK3CA were insensitive to PD0325901, as expected from results above (Fig. 3A and S3). In contrast, knock-out of mutant PIK3CA in the paired cell lines conferred sensitivity to MEK inhibition. This sensitivity correlated with induction of apoptotic markers such as an increase in the sub-G1 fraction of cells and induction of PARP cleavage and Caspase-3 activation (Fig. 3B and S3B). As expected from the original description of these lines, the effects of the MEK inhibition were more pronounced in 1% FBS (Figure 3A and S3). Importantly, in an in vivo setting, we further demonstrated that loss of the mutant PIK3CA allele was sufficient to confer sensitivity to PD0325901 when HCT116 cells were grown as xenografts (Fig. 4). Specifically, the growth of PIK3CA mutant tumors was only marginally affected by treatment with the MEK inhibitor. The growth of the tumor xenografts with wild-type PIK3CA tumors, however, was dramatically suppressed by MEK inhibition (Fig 4A). Similarly to in vitro data, in these PIK3CA wild-type xenografts, MEK inhibition caused a significant induction of cleaved PARP expression and Caspase 3 activation (Fig 4B and 4C). Formally, it is possible that selection for PIK3CA wild-type isogenic clones had co-selected for other background mutations that could confound these findings. To confirm that restored MEK dependence in these lines was specifically due to genetic knock-out of mutant PIK3CA, we exogenously re-expressed the mutant PIK3CA gene into the HCT116 wild-type PIK3CA cells and tested their dependence on MER/ERK signaling. Stable transfection of H1074R mutant PIK3CA into these cells restored resistance to MEK inhibition, and prevented PARP cleavage and Caspase-3 activation (Fig. 3C). Together these results strongly suggest that mutant PIK3CA is necessary for the MEK-independent growth of mutant KRAS colon cancers in tissue culture and in vivo and is consistent with studies using shRNA and PIK3CA inhibitors (18, 19).
Figure 3. Coexistent PIK3CA mutation causes KRAS mutant colorectal tumor cells to grow and survive in a MEK/ERK independent manner.
A: Isogenic HCT116 KRAS mutant/PIK3CA wild-type (wt PIK3CA, red) and HCT116 KRAS mutant/PIK3CA mutant (mut PIK3CA, blue) cells were grown in the presence of various concentrations of PD0325901 in 1% FBS growth medium and proliferation was measured using Alamar Blue. Results are represented as percent of untreated control on day 3 plotted as a function of the drug concentration.
B: Left panel: Apoptotic fraction of PD0325901 treated PIK3CA isogenic HCT116 cells, reported as the percent of cells with sub-G1 DNA content 48 hr post treatment. The results represent the mean ± SE of two independent experiments performed in triplicate. Right panel: PIK3CA isogenic HCT116 cells were grown in 1% FBS growth media for 9 hours and then treated with 50nM PD0325901. The cells were harvested at the indicated times post treatment, lysed and immunoblotted with the specified antibodies.
C: Left panel: Immunoblots of HCT116 KRAS mutant/PIK3CA wild-type cells (wt, red) transduced with H1047R-mutant PIK3CA cDNA (wt+H1047R, blue) or vector-only control (wt+Vector, red). The stable expression was confirmed by immunoprecipitation and western blot with an anti-HA tag antibody. Middle panel: HCT116 KRAS mutant/PIK3CA wild-type cells (wt, red) and cells stably expressing mutant PIK3CA (wt + H1047R, blue) or vector-only (wt + vector, red) were grown in the presence of various concentrations of PD0325901 in 1% FBS growth medium and proliferation was measured using Alamar Blue. Results are represented as the percentage of untreated control on day 3, plotted as a function of the drug concentration. Right panel: Western blot of apoptotic markers and p-ERK in HCT116 wild-type PIK3CA cells (wt) and these cells with either stably transduced mutant PIK3CA (H1047R) or control vector in the presence of PD0325901. The cells were grown in 1% FBS growth media for 9 hours and then treated with 50nM PD0325901. The cells were harvested 12 hours post treatment, lysed and immunoblotted with the specified antibodies.
Figure 4. PIK3CA mutation causes mutant KRAS colorectal tumor HCT116 to grow in vivo in a MEK-independent manner.
A: Mice with established HCT116 KRAS mutant/PIK3CA wild-type (wt PIK3CA, red) and HCT116 KRAS mutant/PIK3CA mutant (mut PIK3CA, blue) xenografts were treated with 5mg/kg of PD0325901 or vehicle only, 5 days/week for the indicated number of days. The results are represented as in Figure 2D.
B and C: Immunoblots of homogenized xenograft tissue after a single 5mg/kg dose of PD0325901. Tumors were excised pretreatment and at indicated times and split in half; (B) one half was flash frozen for immunoblot analysis with indicated antibodies; (C) the other half was formalin fixed, paraffin embedded and used for tissue-section preparation. 8 µm tissue sections were cut and stained with human specific antibody for cleaved PARP.
Sustained Cyclin D expression and bypass of MEK inhibitor-induced G1 arrest correlates with MEK antagonist efficacy
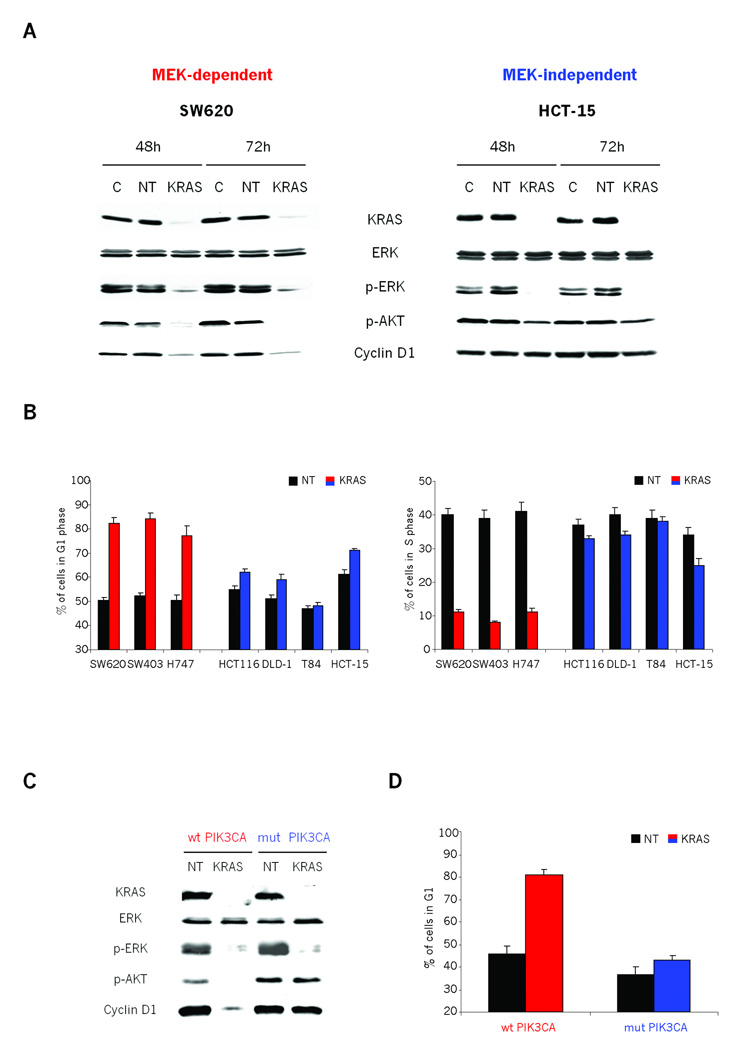
It is important to note that PI3K mutation, while inducing proliferative and apoptotic resistance to MEK inhibition, did not alter the activity of ERK. Thus, there might be PI3K pathway intersecting effector molecules further downstream of ERK. Our observation that Cyclin D1 regulation correlates with MEK sensitivity, further supported by the work of Smalley et al. (31) which shows that increased Cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF mutant melanomas, led us to further explore whether Cyclin D1 might be a candidate for one such downstream effector molecule. First, we transfected multiple cell lines with KRAS siRNA in order to determine whether MEK-dependent Cyclin D1 expression was also dependent on expression of KRAS. KRAS siRNA effectively reduced KRAS expression, resulting in marked inhibition of p-ERK (Fig. 5A and S4A). In MEK-dependent cell lines, KRAS knockdown also resulted in a more than 90% decrease in AKT phosphorylation on S473 (p-AKT) (Fig. 5A and S4A). In these cells, KRAS knockdown resulted in a greater than 90% decrease in Cyclin D1 expression, an average increase in the proportion of cells in G1 from 50% to 82% and average decrease in proportion of cells in S phase from 40% to 10% (Fig. 5A, 5B and S4B). KRAS knockdown caused a similar decrease in p-ERK levels in MEK-independent PIK3CA mutant cell lines (Fig. 5A and S4A). In contrast, KRAS knockdown in these cells had no effect on either AKT phosphorylation or Cyclin D1 expression (Fig. 5A and S4A). Consistent with the absence of Cyclin D1 degradation, KRAS knockdown in these cells only slightly altered the fraction of cells in G1 (an average increase from 55% to 60%) or S phase (an average decrease from 38% to 33%) (Fig. 5B). Together, these data suggest that while Cyclin D1 expression and G1/S cell cycle progression are dependent on KRAS and MEK/ERK signaling, mutation of PIK3CA relieves this dependence.
Figure 5. MEK-dependent Cyclin D1 expression is also dependent on expression of KRAS in cells with wild-type PIK3CA.
A: Immunoblots showing the effects of KRAS siRNA in both MEK-dependent (SW620) and MEK-independent (HCT15) cells. Cells were transfected with lipid carrier control (C), non-targeting control (NT) or KRAS siRNA and harvested at 48 and 72 hours post-transfection. Cell lysates were immunoblotted with the indicated antibodies.
B: MEK-dependent (SW620, SW403, H747) and MEK-independent (HCT116, DLD-1, T-84, HCT-15) cells were transfected with either non-targeting (NT) or KRAS siRNA, and collected 48hr later. Nuclei were isolated, stained with ethidium bromide, and cell cycle distribution analyzed by flow cytometry. Graphs show: 1) the percent of cells with G1 DNA content and 2) the percent of cells with S phase DNA content. The results represent the mean ± SE of two independent experiments performed in triplicate.
C and D: PIK3CA wild-type and mutant isogenic HCT116 cells were transfected with non-targeting control (NT) or KRAS siRNA, harvested 48 hr post transfection and (C) cell lysates immunoblotted with the indicated antibodies or (D) analyzed by flow cytometry. The results are represented as in Figure 5B.
To confirm that Cyclin D1 expression and cell cycle progression is dependent upon mutant PIK3CA in MEK-independent cell lines, we used the PIK3CA isogenic cell pairs in HCT116 to further explore this relationship. As expected, KRAS knockdown in cells lacking the mutant PIK3CA allele caused, in addition to downregulation of ERK phosphorylation, a marked downregulation of AKT phosphorylation and Cyclin D1 expression (Fig 5C). Consistent with the loss of Cyclin D1 expression, these cells accumulated in G1 (45% to 81%) (Fig. 5D) with a concomitant loss of cells in S phase (from 36% to 9%) (Fig S4C). In contrast, potent KRAS knockdown with siRNA did not alter AKT phosphorylation or the expression of Cyclin D1 and had only a marginal effect on cell cycle distribution in HCT116 cells bearing mutant PIK3CA (Fig. 5C, 5D and S4C). This strongly suggests that resistance to MEK inhibition and diminished requirement for KRAS in KRAS mutant cells directly correlates with persistent activation of a PI3K/AKT signaling axis.
Combined inhibition of both MEK/ERK and PI3K/AKT pathways suppresses the growth of tumors with coexisting KRAS and PIK3CA mutations
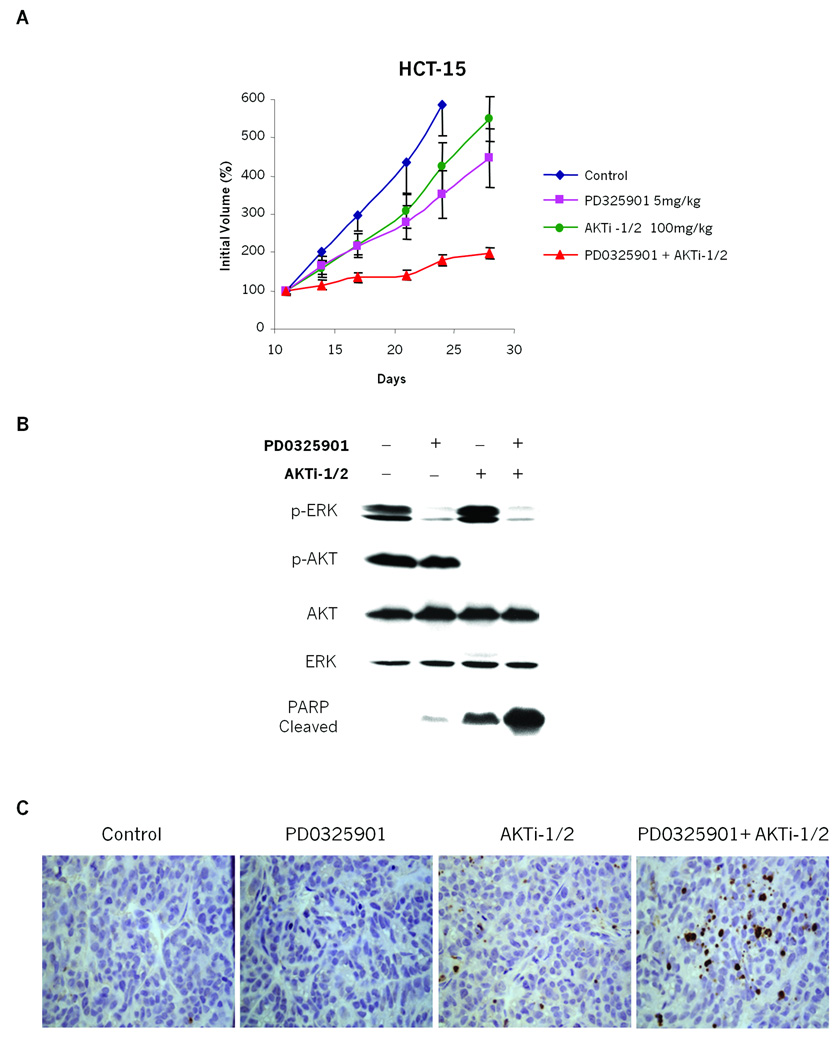
Genetic ablation of PIK3CA mutation is sufficient to restore sensitivity to MEK inhibitors in KRAS/PIK3CA mutant cell lines. This implies that pharmacological inhibition of the PI3K/AKT signaling axis might have similar effects. To explore this further, a potent and selective inhibitor of AKT1 and AKT2 (AKTi-1/2) (22–25) was utilized to block signaling downstream of mutant PIK3CA. Mice bearing KRAS/PIK3CA-mutant HCT15 tumor xenografts were treated with the MEK inhibitor PD0325901 and AKTi-1/2 alone or in combination. Daily treatments with either 5mg/kg of PD0325901 or 100mg/kg of AKTi-1/2 as single agents showed no significant effects on tumor growth. However, when both inhibitors were administered in combination, growth of tumor xenografts was profoundly abrogated (Fig 6A). Chronic administration of both drugs together was well tolerated (Fig S5). In mice treated with either inhibitor alone, the expected pathway inhibitory effect was achieved, as evident by downregulation of p-ERK with PD0325901 and p-AKT with AKTi-1/2 (Fig 6B). Neither agent alone resulted in elevated apoptosis, as judged by cleaved PARP. However, the dual inhibition of MEK and AKT signaling with both inhibitors cooperated to induce a profound accumulation of cleaved PARP, suggesting that the combination therapy induced apoptosis (Fig.6B). Using TUNEL staining (32) as an alternate measure of apoptosis, significant staining of xenograft tissue was only seen in the presence of both inhibitors (Fig 6C). Consistent with experiments using genetic ablation of mutant PIK3CA signaling, these data indicate that the concurrent pharmacological inhibition of both the PI3K/AKT and MEK/ERK pathways is an effective strategy to overcome the resistance to inhibiting either pathway alone in cancer cells bearing both KRAS and PIK3CA mutant alleles.
Figure 6. Combined inhibition of both MEK/ERK and PI3K/AKT pathways shows benefits in suppressing growth of tumors with coexisting KRAS and PIK3CA mutations.
A: Mice with established HCT15 (KRAS mutant/PIK3CA mutant) xenografts were dosed with 5mg/kg of PD0325901 and 100mg/kg of AKTi-1/2 alone or in combination. The results are represented as in Figure 2D.
B and C:Immunoblots of homogenized xenograft tissue. Tumors were excised 8 hours post treatment and split in half; (B) one half was flash frozen for immunoblot analysis with indicated antibodies; (C) the other half was formalin fixed, paraffin embedded and used for analysis by TUNEL assay (described in Gavrieli et al. (32)).
Discussion
Oncogenic activation of RAS isoforms by mutation is a highly recurrent phenomenon in human tumors. Although mutant RAS is an attractive target for therapy, efforts to develop pharmacologic inhibitors have to date been unsuccessful. This has led to efforts focused on inhibiting KRAS-dependent transformation by targeting downstream effector pathways. While more than ten families of RAS effector proteins have been identified (4), there is strong evidence that at least three of these, the RAF and PI3 kinases and the RAL exchange factors, play predominant roles in mediating its transforming effects (5–8). This suggests multiple effector pathways downstream of RAS must be blocked to achieve a therapeutic effect. Indeed, recent research has identified that dual inhibition of both the MEK (downstream of RAF) and PI3K kinase effector pathways is essential to cause regression of genetically engineered KRAS mutant tumors in mice (18). However, in human cancer cell lines, our results demonstrate that at least a subset of KRAS mutant tumors, particularly those wild-type for PIK3CA, retain sensitivity to MEK inhibition as a monotherapy, consistent with another recent report (19). We observe that KRAS mutant and PIK3CA wild-type cancer cells show a decrease in both p-ERK and p-AKT signaling as well as growth inhibition through Cyclin D1 degradation and G1 arrest in vitro and in vivo. In human cancers, the situation will undoubtedly be more complex, and the question remains; how can MEK inhibitor sensitive and resistant tumors be identified in patient populations? Our results indicate that p-ERK is a poor biomarker on its own. Because both mutant KRAS and PIK3CA signaling may converge on Cyclin D1 stability and G1/S cell cycle transition, the rapid and persistent loss of Cyclin D1 could be an important biomarker for the efficacy of MEK inhibition in cancers, a notion consistent with a recent publication which shows that increased Cyclin D1 expression can mediate BRAF inhibitor resistance (31).
It is important to note that while all identified MEK inhibitor-sensitive cancer cells were KRAS mutant/PIK3CA wild-type, not all KRAS mutant/PIK3CA wild-type cancer cells were MEK inhibitor sensitive. Interestingly, the downstream signaling features present in PIK3CA mutant tumors (e.g. stabilized Cyclin D1 expression) are retained in these inhibitor-resistant cells (Fig. 1B), suggesting that as-yet-unidentified genetic lesions that generate MEK resistance in KRAS mutant tumors remain to be discovered. For example, SW1990 and H2030 cells, despite having wild-type PIK3CA and PTEN, have augmented PIK3CA/AKT pathway activation, as evident by higher expression of phosphorylated AKT in these cells (Fig S1B). This upregulation of AKT pathway signaling, could possibly be due to mutations in other elements of PIK3CA signaling (33–35) and might be the basis for resistance in MEK inhibitor-insensitive PIK3CA/PTEN wild-type cell lines like SW1990 and H2030. However, because p-AKT levels do not appear to be predictive of resistance across all the cell lines, PIK3CA mutation remains the strongest predictor. Alternatively, it is also plausible that common downstream effectors of KRAS and PIK3CA that serve as integrators of signaling could be also mutated. Identification of these mutations and downstream signaling pathways will undoubtedly aid in our understanding of the common signaling pathways upon which KRAS and PIK3CA converge.
There is currently no therapeutic agent that directly inhibits KRAS function. The results reported here have important implications for the development of treatments for tumors with mutant RAS. Inhibition of MEK/ERK signaling may be useful in some of these patients as a single modality. However, only tumors with a wild-type PIK3CA pathway would be likely to respond to MEK/ERK inhibition alone. In early trials of MEK inhibitors, clinical responses were noted in a patient with pancreatic cancer and in a patient with mutant NRAS melanoma, suggesting that in some cases MEK inhibition can indeed have single-agent efficacy in humans (36–38). However, our results suggest that patients with coexistent PIK3CA pathway mutations should be excluded from such trials. Instead, our data with in vivo xenograft models suggest that patients with such tumors would require combined inhibition of both the MEK/ERK and PI3K/AKT pathways to observe efficacy. Since the RAF/MEK/ERK and PI3K/AKT pathways are two key RAS effector pathways, the combined inhibition of MEK and AKT may constitute a general “anti-RAS” therapeutic strategy. Regardless of PIK3CA mutational status, this strategy could be of utility in cancers with mutated RAS (pancreatic, colon, lung carcinoma) for which there are few and only marginally effective therapies. The tolerability of the combined inhibition of AKT and MEK and its marked effects on reducing tumor growth in mouse models suggest such a strategy could be useful in a variety of advanced tumors bearing both RAS and PIK3CA mutations.
Why do tumors mutate multiple oncogenes that appear to converge on similar downstream signaling events? We speculate that part of the selection for the second mutation is to reduce a cell’s dependency on any one pathway and provide redundancy to reduce oncogene addiction, perhaps in the face of environmental cues such as hypoxia or low nutrient conditions that attenuate tumor growth. The downstream convergence of PI3K/AKT and ERK signaling may account for the significant frequency of coexistent mutations in these pathways in cancer. In fact, the requirement for combined inhibition suggests that the two pathways activate converging targets that integrate their function. These integrators, for example, may include components of the networks that regulate cellular survival and apoptosis (39), protein translation (40) or cell cycle (such as Cyclin D1).
Supplementary Material
Acknowledgments
We would like to kindly thank Bert Vogelstein and Victor Velculescu for providing the HCT116 and DLD-1 isogenic cell lines; Ayana Sawai, Elisa De Stanchina, Manickam Janakiraman and Jose Lobo for technical assistance and Sarat Chandarlapaty for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (RO1-CA127240) and (PO1-CA129243), the Samuel Waxman Foundation, the William H. Goodwin and Alice Goodwin Foundation and the MSKCC Experimental Therapeutics Program.
References
- 1.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: 'it ain't over 'til it's over'. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 2.Campbell PM, Der CJ. Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin Cancer Biol. 2004;14:105–114. doi: 10.1016/j.semcancer.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Bowen DT, Frew ME, Hills R, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106:2113–2119. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 4.Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004;14:639–647. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Moodie SA, Willumsen BM, Weber MJ, Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 6.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Viciana P, Warne PH, Dhand R, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 8.Fuller SJ, Finn SG, Downward J, Sugden PH. Stimulation of gene expression in neonatal rat ventricular myocytes by Ras is mediated by Ral guanine nucleotide dissociation stimulator (Ral.GDS) and phosphatidylinositol 3-kinase in addition to Raf. Biochem J. 1998;335(Pt 2):241–246. doi: 10.1042/bj3350241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catling AD, Schaeffer HJ, Reuter CW, Reddy GR, Weber MJ. A proline-rich sequence unique to MEK1 and MEK2 is required for raf binding and regulates MEK function. Mol Cell Biol. 1995;15:5214–5225. doi: 10.1128/mcb.15.10.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crews CM, Alessandrini A, Erikson RL. The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science. 1992;258:478–480. doi: 10.1126/science.1411546. [DOI] [PubMed] [Google Scholar]
- 11.Gire V, Marshall CJ, Wynford-Thomas D. Activation of mitogen-activated protein kinase is necessary but not sufficient for proliferation of human thyroid epithelial cells induced by mutant Ras. Oncogene. 1999;18:4819–4832. doi: 10.1038/sj.onc.1202857. [DOI] [PubMed] [Google Scholar]
- 12.Rapp UR, Goldsborough MD, Mark GE, et al. Structure and biological activity of v-raf, a unique oncogene transduced by a retrovirus. Proc Natl Acad Sci U S A. 1983;80:4218–4222. doi: 10.1073/pnas.80.14.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 14.Sebolt-Leopold JS. Advances in the development of cancer therapeutics directed against the RAS-mitogen-activated protein kinase pathway. Clin Cancer Res. 2008;14:3651–3656. doi: 10.1158/1078-0432.CCR-08-0333. [DOI] [PubMed] [Google Scholar]
- 15.Saxena N, Lahiri SS, Hambarde S, Tripathi RP. RAS: target for cancer therapy. Cancer Invest. 2008;26:948–955. doi: 10.1080/07357900802087275. [DOI] [PubMed] [Google Scholar]
- 16.Solit DB, Garraway LA, Pratilas CA, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pratilas CA, Hanrahan AJ, Halilovic E, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68:9375–9383. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wee S, Jagani Z, Xiang KX, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009;69:4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 20.Yu K, Toral-Barza L, Shi C, Zhang WG, Zask A. Response and determinants of cancer cell susceptibility to PI3K inhibitors: combined targeting of PI3K and Mek1 as an effective anticancer strategy. Cancer Biol Ther. 2008;7:307–315. doi: 10.4161/cbt.7.2.5334. [DOI] [PubMed] [Google Scholar]
- 21.Barrett SD, Bridges AJ, Dudley DT, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorganic & Medicinal Chemistry Letters. 2008;18:6501–6504. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 22.DeFeo-Jones D, Barnett SF, Fu S, et al. Tumor cell sensitization to apoptotic stimuli by selective inhibition of specific Akt/PKB family members. Mol Cancer Ther. 2005;4:271–279. [PubMed] [Google Scholar]
- 23.Barnett SF, Defeo-Jones D, Fu S, et al. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem J. 2005;385:399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilodeau MT, Balitza AE, Hoffman JM, et al. Allosteric inhibitors of Akt1 and Akt2: a naphthyridinone with efficacy in an A2780 tumor xenograft model. Bioorg Med Chem Lett. 2008;18:3178–3182. doi: 10.1016/j.bmcl.2008.04.074. [DOI] [PubMed] [Google Scholar]
- 25.She QB, Chandarlapaty S, Ye Q, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS ONE. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Nusse M, Beisker W, Hoffmann C, Tarnok A. Flow cytometric analysis of G1- and G2/M-phase subpopulations in mammalian cell nuclei using side scatter and DNA content measurements. Cytometry. 1990;11:813–821. doi: 10.1002/cyto.990110707. [DOI] [PubMed] [Google Scholar]
- 28.Zhao JJ, Liu Z, Wang L, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikediobi ON, Davies H, Bignell G, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 31.Smalley KS, Lioni M, Dalla Palma M, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther. 2008;7:2876–2883. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasudevan KM, Barbie DA, Davies MA, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaiswal BS, Janakiraman V, Kljavin NM, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 36.Wang D, Boerner SA, Winkler JD, LoRusso PM. Clinical experience of MEK inhibitors in cancer therapy. Biochim Biophys Acta. 2007;1773:1248–1255. doi: 10.1016/j.bbamcr.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Lorusso PM, Adjei AA, Varterasian M, et al. Phase I and pharmacodynamic study of the oral MEK inhibitor CI-1040 in patients with advanced malignancies. J Clin Oncol. 2005;23:5281–5293. doi: 10.1200/JCO.2005.14.415. [DOI] [PubMed] [Google Scholar]
- 38.Adjei AA, Cohen RB, Franklin W, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.She QB, Solit DB, Ye Q, et al. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell. 2005;8:287–297. doi: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.She QB, Halilovic E, Ye Q, et al. 4E-BP1 Is a Key Effector of the Oncogenic Activation of the AKT and ERK Signaling Pathways That Integrates Their Function in Tumors. Cancer Cell. 2010 doi: 10.1016/j.ccr.2010.05.023. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.