Abstract
Mutations in SIMPLE cause an autosomal dominant, demyelinating form of peripheral neuropathy termed Charcot–Marie–Tooth disease type 1C (CMT1C), but the pathogenic mechanisms of these mutations remain unknown. Here, we report that SIMPLE is an early endosomal membrane protein that is highly expressed in the peripheral nerves and Schwann cells. Our analysis has identified a transmembrane domain (TMD) embedded within the cysteine-rich (C-rich) region that anchors SIMPLE to the membrane, and suggests that SIMPLE is a post-translationally inserted, C-tail-anchored membrane protein. We found that CMT1C-linked pathogenic mutations are clustered within or around the TMD of SIMPLE and that these mutations cause mislocalization of SIMPLE from the early endosome membrane to the cytosol. The CMT1C-associated SIMPLE mutant proteins are unstable and prone to aggregation, and they are selectively degraded by both the proteasome and aggresome–autophagy pathways. Our findings suggest that SIMPLE mutations cause CMT1C peripheral neuropathy by a combination of loss-of-function and toxic gain-of-function mechanisms, and highlight the importance of both the proteasome and autophagy pathways in the clearance of CMT1C-associated mutant SIMPLE proteins.
Key words: Charcot–Marie–Tooth disease, LITAF, SIMPLE, Aggresome, Misfolded protein
Introduction
Charcot–Marie–Tooth disease (CMT) is the most prevalent inherited peripheral neuropathy, characterized by progressive motor weakness, sensory loss and muscle wasting (Patzko and Shy, 2011). CMT is divided into two types, the demyelinating type, which includes the majority of CMT cases (80%) and the axonal degeneration type, which is less common (20%). The primary defect of demyelinating CMT is the inability for Schwann cells to properly myelinate peripheral axons, which manifests as slowed conduction velocities at the peripheral nerves and secondary degeneration of axons (Patzko and Shy, 2011). Examination of the peripheral nerves from patients and rodent models implicate abnormal targeting and accumulation of myelin proteins as potential contributing factors to demyelinating CMT (Nishimura et al., 1996; Naef and Suter, 1999; Notterpek et al., 1999; Tobler et al., 1999; Fortun et al., 2003). However, the pathogenic mechanisms underlying demyelinating CMT remain largely unknown.
Genetic studies have identified eight missense mutations in small integral membrane protein of lysosome/late endosome (SIMPLE) that cause autosomal dominant, demyelinating CMT type 1C (CMT1C) (Street et al., 2003; Bennett et al., 2004; Saifi et al., 2005; Latour et al., 2006; Gerding et al., 2009). SIMPLE, also known as lipopolysaccharide-induced TNF-α factor (LITAF), is a 161 amino acid protein that has been implicated in cytokine signaling (Moriwaki et al., 2001; Bolcato-Bellemin et al., 2004) and tumor suppression (Mestre-Escorihuela et al., 2007; Wang et al., 2009), but its precise cellular function remains elusive. Northern blot analyses showed a ubiquitous expression pattern of SIMPLE mRNA in multiple tissues (Moriwaki et al., 2001; Street et al., 2003), therefore it is puzzling as to how mutations in SIMPLE can cause a demyelinating neuropathy phenotype that specifically affects the peripheral nervous system. The subcellular distribution of endogenous SIMPLE is unknown, although a subpopulation was suggested to associate with the late endosome and lysosome (Moriwaki et al., 2001). Sequence analysis revealed a cysteine-rich (C-rich) domain at the C-terminus of SIMPLE (Moriwaki et al., 2001). Although this C-rich domain was proposed to be a putative RING finger domain that may have E3 ubiquitin-protein ligase activity (Moriwaki et al., 2001; Saifi et al., 2005), the function of this domain remains to be defined. Interestingly, CMT1C-associated SIMPLE mutations are all clustered within the C-rich domain. The pathogenic effects of the disease-linked SIMPLE mutations have not yet been examined.
In this study, we undertook the characterization of the tissue distribution, subcellular localization, and membrane association of endogenous SIMPLE protein and investigated the effects of CMT1C-associated mutations on SIMPLE protein stability, localization, aggregation and degradation. Our results reveal that SIMPLE is an early endosome membrane protein enriched in the peripheral nerves and Schwann cells, and indicate that CMT1C-associated mutations not only disrupt the endosome membrane association of SIMPLE, but also promote SIMPLE protein aggregation and degradation by both the proteasome and aggresome–autophagy pathways. Our findings provide new insights into the pathogenic mechanisms of CMT1C-associated SIMPLE mutations and have important implications for understanding and treating peripheral neuropathy.
Results
SIMPLE protein is highly expressed in peripheral nerves and Schwann cells
To study endogenous SIMPLE protein, we generated and characterized a rabbit polyclonal anti-SIMPLE antibody (supplementary material Fig. S1). Immunoblot analysis showed that our anti-SIMPLE antibody specifically recognized endogenous SIMPLE protein in HeLa and HEK293 cells at the expected size of 18 kDa (supplementary material Fig. S1A), as well as a recombinant SIMPLE protein (supplementary material Fig. S1B). The specificity of our anti-SIMPLE antibody was confirmed by selective loss of the SIMPLE-immunoreactive band upon depletion of endogenous SIMPLE protein in HeLa cells with a SIMPLE-specific short hairpin RNA (shRNA) (supplementary material Fig. S1C). Moreover, we showed that the anti-SIMPLE antibody is able to recognize both recombinant and endogenous SIMPLE proteins by immunostaining (supplementary material Fig. S1D,E) and confirmed its specificity by using the SIMPLE shRNAs in immunostaining experiments (supplementary material Fig. S1E).
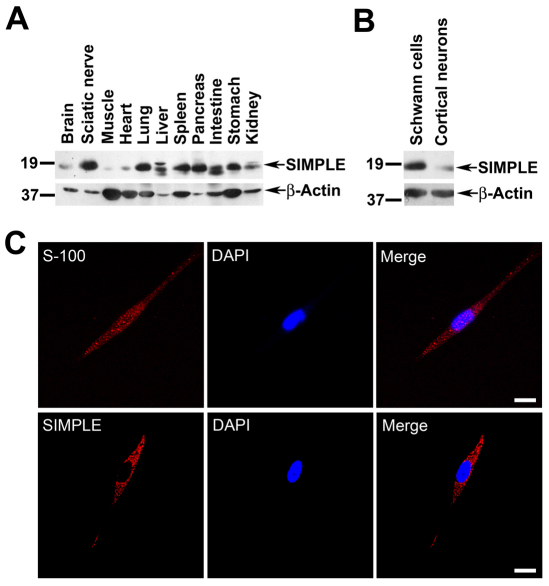
We then used the anti-SIMPLE antibody to examine the expression of SIMPLE protein in multiple mouse tissues and in sciatic nerves by immunoblot analysis. The result showed that the 18 kDa SIMPLE protein is widely expressed in many tissues, although at different abundance (Fig. 1A). We observed a second SIMPLE protein band at ~19 kDa in liver and kidney (Fig. 1A). Although its identity remains to be determined, this upper band might represent a phosphorylated form of SIMPLE protein because there are several predicted phosphorylation sites in the SIMPLE sequence (Moriwaki et al., 2001). In liver and intestine, there was an additional SIMPLE protein band at ~17 kDa (Fig. 1A), which might represent a degradation product because its relative intensity compared with the 18 kDa band varied from preparation to preparation. We found that SIMPLE protein was highly enriched in the sciatic nerves compared with the brain and muscle (Fig. 1A). Furthermore, our immunoblot analysis showed that SIMPLE protein expression in Schwann cells was substantially higher than its expression in primary cortical neurons (Fig. 1B). Immunostaining analysis revealed that SIMPLE was highly expressed in a punctate pattern throughout the cytoplasm of primary Schwann cells that are positive for the Schwann-cell-specific marker S-100 (Scarpini et al., 1986) (Fig. 1C). By contrast, SIMPLE expression was much lower in S-100-negative primary fibroblasts (data not shown). Together, our results indicate that SIMPLE protein is highly expressed in peripheral nerves and Schwann cells.
Fig. 1.
SIMPLE is widely expressed in several tissues and is highly expressed in the peripheral nerves and Schwann cells. (A,B) Equal amounts of total proteins (100 μg protein/lane) from the indicated mouse tissues (A) or primary Schwann cells and cortical neurons (B) were subjected to immunoblot analysis with anti-SIMPLE and anti-β-actin antibodies. (C) Primary Schwann cells immunostained with antibodies against S-100 or SIMPLE. Nuclei are visualized using DAPI stain. Scale bars: 10 μm.
SIMPLE is a C-tail-anchored integral membrane protein
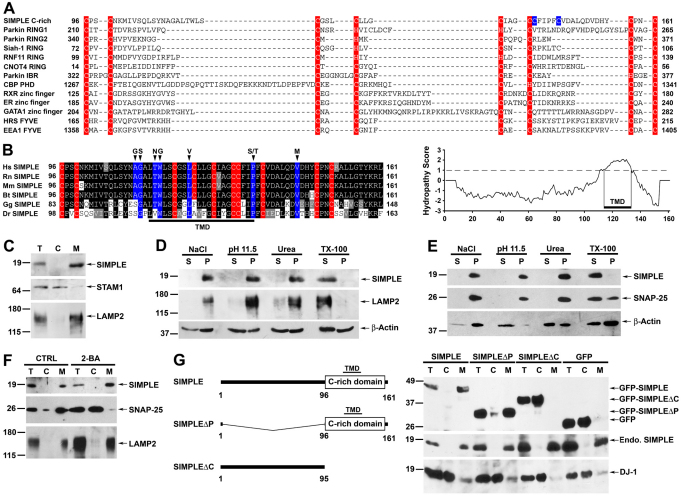
SIMPLE contains an evolutionarily conserved C-rich domain near its C-terminus, which was proposed to be a putative RING finger domain with E3 ligase activity (Moriwaki et al., 2001; Saifi et al., 2005). However, unlike the typical RING finger domain, which usually consists of either six cysteines and two histidines (C3H2C3) or seven cysteines and one histidine (C3HC4), the C-rich domain of SIMPLE possesses ten cysteines with no histidine (Fig. 2A,B). Eight of the ten cysteine residues in the SIMPLE C-rich domain align well with those of the zinc finger domains that have no E3 ligase activity (Fig. 2A). Moreover, our sequence analyses using multiple prediction programs (Hirokawa et al., 1998; Tusnady and Simon, 2001; Rost et al., 2004) revealed a potential transmembrane domain (TMD) embedded within the SIMPLE C-rich domain (Fig. 2B), which distinguishes it from known RING finger, FYVE and other zinc finger domains. Together, these results argue against the possibility of the SIMPLE C-rich domain being a RING finger domain with E3 ligase activity.
Fig. 2.
SIMPLE is a transmembrane protein that requires its cysteine-rich (C-rich) domain for membrane association. (A) Sequence alignment of the SIMPLE C-rich domain with known RING finger domains and other C-rich domains. Residues implicated in zinc binding are highlighted in red, and two unaligned SIMPLE cysteine residues are colored in blue. (B) SIMPLE is predicted to be a single-spanning integral membrane protein. Cysteine residues are highlighted in red and residues mutated in CMT1C are highlighted in blue. Hydropathy plot (TMpred) indicates the presence of transmembrane domain (TMD) within the SIMPLE C-rich domain. (C) Post-nuclear supernatant (T) from HeLa cells was separated into cytosol (C) and membrane (M) fractions. Aliquots representing an equal percentage of each fraction were subjected to immunoblot analysis with antibodies against SIMPLE, STAM1 and LAMP2. (D,E) Membrane fractions from HeLa cells (D) or SH-SY5Y cells (E) were subjected to extraction by 1.5 M NaCl, 0.1 M Na2CO3 (pH 11.5), 4 M urea or 1% Triton X-100 (TX-100) and were then separated into supernatant (S) and pellet (P) fractions. Aliquots representing an equal percentage of each fraction were analyzed by immunoblotting with antibodies against SIMPLE, LAMP2 (D), SNAP-25 (E) and β-actin. (F) The membrane association of SIMPLE is palmitoylation independent. Post-nuclear supernatants of SH-SY5Y cells treated with 100 μM 2-bromohexadeconic acid (2-BA) or the vehicle control (CTRL) were separated into cytosol (C) and membrane (M) fractions. Aliquots representing an equal percentage of each fraction were subjected to immunoblot analysis with antibodies against SIMPLE, SNAP-25 and LAMP2. (G) Domain structures of SIMPLE and its deletion mutants (left). Post-nuclear supernatants (T) of HeLa cells expressing GFP-tagged SIMPLE, SIMPLEΔP, SIMPLEΔC or GFP were separated into cytosol (C) and membrane (M) fractions (right). Aliquots representing an equal percentage of each fraction were analyzed by immunoblotting with antibodies against GFP, SIMPLE and DJ-1.
To determine whether endogenous SIMPLE is associated with the membrane, post-nuclear supernatant of HeLa cells was separated into cytosol and membrane fractions and subjected to immunoblot analysis. SIMPLE was found exclusively in the membrane fraction, as confirmed by co-fractionation with the integral membrane protein marker LAMP2, but not with the cytosolic protein STAM1 (Fig. 2C). By contrast, the palmitoylated protein SNAP-25 was partitioned into both cytosol and membrane fractions (Fig. 2F), consistent with previous reports (Vogel and Roche, 1999; Li et al., 2001).
To investigate the nature of SIMPLE association with the membrane, the membrane fraction was extracted with 1.5 M NaCl, 0.1 M Na2CO3 (pH 11.5), 4 M urea, and 1% Triton X-100 (TX-100). We found that SIMPLE was resistant to extraction by high salt, high pH and urea, but was readily extracted when membranes were solubilized by the non-ionic detergent TX-100 (Fig. 2D). The SIMPLE extraction profile is similar to that of integral membrane proteins such as LAMP2 (Fig. 2D), suggesting that SIMPLE is not peripherally associated with the membrane. We also compared the extraction profile of SIMPLE with that of lipid-anchored proteins such as SNAP-25 (Fig. 2E) and the glycophosphatidylinositol (GPI)-anchored protein Thy-1 (supplementary material Fig. S2A). We found that, similarly to SIMPLE, SNAP-25 and Thy-1 were resistant to extraction by high salt, high pH and urea, as reported previously (Lu et al., 1994; Seaton et al., 2000; Li et al., 2001). However, unlike SIMPLE, Thy-1 and a pool of SNAP-25 were resistant to extraction by TX-100 (Fig. 2E; supplementary material Fig. S2A) because of their localization to lipid rafts (Turner and Shotton, 1989; Chamberlain et al., 2001).
Sequence analyses revealed that SIMPLE does not contain the consensus sequence for N-myristoylation (MGxxxS/T), prenylation (CAAx) or GPI-anchor attachment (Eisenhaber et al., 1999; Sorek et al., 2009). Consistent with these predictions, we found that addition of a Myc tag to the N-terminus of SIMPLE, a manipulation which is known to disrupt the N-myristoylation of proteins (Santonico et al., 2010), had no effect on the membrane association of SIMPLE (Fig. 4A), arguing against the involvement of N-myristoylation in anchoring SIMPLE to the membrane. Similarly, addition of a Myc tag to the C-terminus of SIMPLE, a manipulation that interferes with prenylation of proteins (Yamashita et al., 2009), had no effect on the membrane association of SIMPLE (supplementary material Fig. S2B), thus excluding the involvement of prenylation in the membrane attachment of SIMPLE. The detection of the C-terminal Myc-tagged SIMPLE in the membrane fraction by anti-Myc antibody (supplementary material Fig. S2B) indicated that SIMPLE did not undergo C-terminal cleavage associated with GPI-anchor attachment (White et al., 2000), thereby also arguing against the involvement of GPI-anchor attachment in the membrane association of SIMPLE. Finally, although sequence analysis identified four cysteine residues of SIMPLE (residues 95, 96, 131, and 132) as potential palmitoylated sites, analysis with the palmitoylation inhibitor 2-bromohexadeconic acid (2-BA) revealed that, unlike palmitoylated protein SNAP-25 which translocated to the cytosol after 2-BA treatment, SIMPLE remained exclusively in the membrane fraction (Fig. 2F), indicating that SIMPLE associates with the membrane by a palmitoylation-independent mechanism. Together, these results suggest that SIMPLE is an integral membrane protein rather than a lipid-anchored protein.
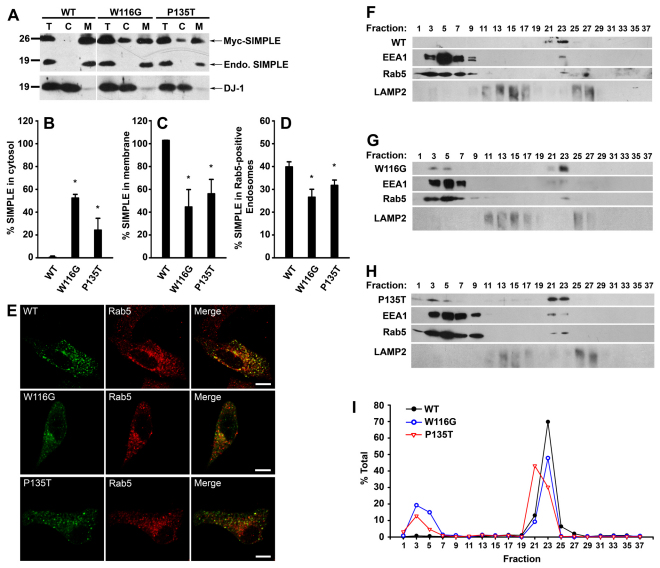
Fig. 4.
CMT1C-associated SIMPLE mutants are mislocalized from the early endosomal membrane to the cytosol. (A) Post-nuclear supernatants (T) from HeLa cells expressing Myc-tagged WT, W116G or P135T SIMPLE proteins were separated into cytosol (C) and membrane (M) fractions. Aliquots representing an equal percentage of each fraction were analyzed by immunoblotting with anti-DJ-1 antibody and anti-SIMPLE antibody, which recognized both Myc-tagged and endogenous (endo.) SIMPLE proteins. (B,C) The percentages of SIMPLE WT or mutant proteins in the cytosol (B) and membrane (C) fraction relative to the total amount in the corresponding post-nuclear supernatant (T) were quantified and shown as mean ± s.e.m. from three independent experiments. *P<0.05 compared with SIMPLE WT. (D) The percentages of SIMPLE WT or mutant proteins localized to Rab5-positive early endosomes in transfected HeLa cells were quantified as described in the Materials and Methods. Data represent mean ± s.e.m. from three independent experiments. *P<0.05 compared with SIMPLE WT. (E) Representative images from D. HeLa cells expressing Myc-tagged SIMPLE WT or mutant proteins were immunostained with antibodies against the Myc tag (green) and the early endosome marker Rab5 (red). Colocalization is indicated by the yellow color in the merged panels. Scale bars: 10 μm. (F–H) Post-nuclear supernatants from stably transfected HEK293 cells expressing Myc-tagged WT (F), W116G (G) and P135T (H) SIMPLE were fractionated on a 10–30% linear Optiprep gradient into 38 fractions, with fraction 1 corresponding to the top of the gradient. Equal volumes of each fraction were subjected to SDS-PAGE followed by immunoblot analysis. The lower band in the EEA1 immunoblots represents a EEA1 protein degradation product. (I) The level of SIMPLE WT or mutant proteins in each fraction was quantified and is shown as a percentage of the total level of the protein. Data are representative of at least three independent experiments.
Because sequence analyses predicted a TMD within the C-rich domain of SIMPLE (Fig. 2B), we performed deletion analysis to determine whether this C-rich domain is required for membrane association of SIMPLE. We generated two SIMPLE deletion mutants: SIMPLEΔP, which lacks the proline-rich region, and SIMPLEΔC, which lacks the C-rich domain (Fig. 2G). Membrane fractionation analysis revealed that, similarly to endogenous SIMPLE, GFP-tagged full-length SIMPLE and SIMPLEΔP mutant were present exclusively in the membrane fraction (Fig 2G). The membrane association of SIMPLE was disrupted by deletion of the C-rich domain, because GFP-tagged SIMPLEΔC mutant was found exclusively in the cytosol fraction similarly to GFP and the cytosolic protein DJ-1 (Fig. 2G). These results indicate that the C-rich domain of SIMPLE contains a membrane-anchoring region. Sequence analysis revealed that SIMPLE does not contain an ER-targeting signal sequence (Bendtsen et al., 2004). Moreover, the predicted TMD is localized near the C-terminus of SIMPLE (Fig. 2B). These characteristics suggest that SIMPLE is likely to undergo post-translational insertion as a C-tail-anchored membrane protein (Borgese et al., 2007; Borgese and Fasana, 2011).
Endogenous SIMPLE is localized to early endosome but not late endosome and lysosome
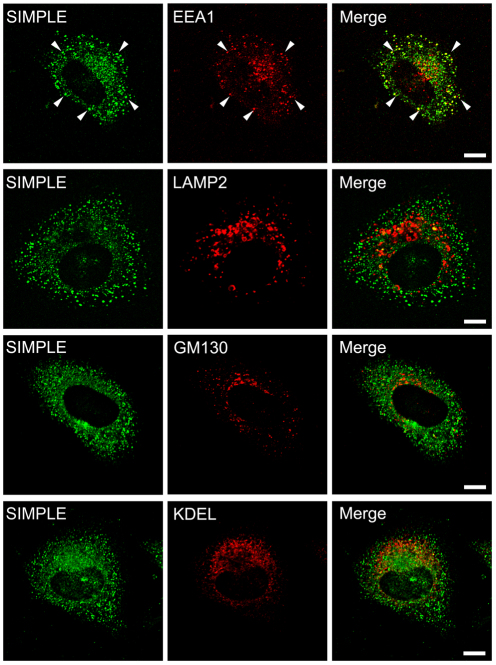
The subcellular localization of endogenous SIMPLE is poorly characterized. Although a previous study reported the presence of SIMPLE in the late endosome and lysosome, the SIMPLE immunostaining at the late endosome and lysosome may be non-specific because it used an anti-SIMPLE antibody that also recognized a non-specific band at the size of ~100 kDa on the immunoblot (Moriwaki et al., 2001). Furthermore, this previous study only examined the colocalization of SIMPLE with the late endosome and lysosome markers, but not colocalization with markers for other organelles. To clarify the subcellular localization of endogenous SIMPLE, we took advantage of our highly specific anti-SIMPLE antibody and performed double-labeling immunofluorescence confocal microscopic analysis to compare the intracellular distribution of endogenous SIMPLE with various organelle markers in HeLa cells. We found that endogenous SIMPLE exhibited substantial colocalization with the early endosome antigen 1 (EEA1) (Fig. 3) and Rab5 (supplementary material Fig. S3A), both of which are widely used markers for the early endosome (Li et al., 2002; Kirk et al., 2006). By contrast, there was very little colocalization of endogenous SIMPLE with the late endosome and lysosome marker LAMP2, the endoplasmic reticulum (ER) marker KDEL, or the Golgi marker GM130 (Fig. 3). The early endosomal localization of SIMPLE was further confirmed by the observation (supplementary material Fig. S3A) that a large population of SIMPLE was accumulated at the enlarged early endosomes induced by Rab5 Q79L, a constitutively active mutant of Rab5 (Ceresa et al., 2001). Together, these results indicate that SIMPLE is predominantly localized to early endosomes, but not the late endosome, lysosome, ER or Golgi.
Fig. 3.
Endogenous SIMPLE is localized to the early endosome but not to other organelles. HeLa cells were double-immunostained with antibodies against endogenous SIMPLE (green) and EEA1, LAMP2, GM130 or KDEL (red). Colocalization is indicated by the yellow color in the merge panel, and examples are indicated by arrowheads. Scale bars: 10 μm.
CMT1C-associated mutations cause mislocalization of SIMPLE from the early endosomal membrane to the cytosol
Our findings that all eight CMT1C-associated mutations identified so far are clustered within or around the TMD of SIMPLE (Fig. 2B) prompted us to investigate the effects of CMT1C-associated mutations on the membrane association and subcellular localization of SIMPLE. We focused on two representative CMT1C-associated SIMPLE mutations: W116G, which locates in the middle of a cluster of five mutations (A111G, G112S, T115N, W116G, and L122V) flanking the N-terminus of the TMD, and P135T, which is one of the two mutations (P135S and P135T) at the C-terminus of the TMD (Fig. 2B). Subcellular fractionation analysis revealed that, unlike endogenous SIMPLE and Myc-tagged SIMPLE WT, which were exclusively present in the membrane fraction, substantial amounts of SIMPLE W116G and P135T mutants were found in the cytosol fraction (Fig. 4A,B). Accordingly, the amounts of SIMPLE W116G and P135T mutants in the membrane fraction were significantly less than that of SIMPLE WT (Fig. 4A,C). We found that the membrane-associated SIMPLE W116G and P135T mutants, but not SIMPLE WT, were partially extracted by 0.1 M Na2CO3 (supplementary material Fig. S4A) and 4 M urea (data not shown), indicating that a subpopulation of SIMPLE W116G and P135T mutant proteins are peripherally associated with the membrane. Together, these data indicate that CMT1C-associated mutations cause a partial dislocation of SIMPLE from the membrane to the cytosol.
A potential mechanism for dislocating SIMPLE W116G and P135T mutant proteins from the membrane to the cytosol is via the AAA-ATPase-p97/VCP-dependent dislocation to the cytosol for ER-associated degradation (ERAD) (DeLaBarre et al., 2006). To address this possibility, we used p97/VCP H317A, a dominant-negative mutant that strongly inhibits p97/VCP-dependent dislocation and degradation of ERAD substrates (DeLaBarre et al., 2006). We found that, although co-expression of p97/VCP H317A increased the steady-state level of the ERAD substrate NHK, it did not affect the steady-state levels of SIMPLE W116G and P135T mutant proteins (supplementary material Fig. S4B,C). These results argue against the possibility that SIMPLE mutant proteins are ERAD substrates and suggest that p97/VCP-dependent dislocation is not involved in the detachment of SIMPLE mutant proteins from the membrane.
Immunofluorescence confocal microscopic analysis revealed that SIMPLE W116G and P135T mutants showed significant reductions in localization to Rab5-positive early endosomes compared with that of SIMPLE WT (Fig. 4D,E), consistent with the subcellular fractionation results, which showed less SIMPLE W116G and P135T mutants than SIMPLE WT in the membrane fraction (Fig. 4A,C). We also performed immunostaining experiments to address the possibility that SIMPLE mutants may be mislocalized to other membrane compartments. The results showed that, unlike other mutant membrane proteins such as CFTR (cystic fibrosis transmembrane conductance regulator) ΔF508 mutant (Lukacs et al., 1994), SIMPLE W116G and P135T mutant proteins were not retained at the ER (supplementary material Fig. S4D), nor were they mislocalized to LAMP2-positive late endosome and lysosome (supplementary material Fig. S4E).
We then performed density gradient fractionation experiments to further assess the effects of CMT1C-associated mutations on the subcellular localization of SIMPLE (Fig. 4F–I). As reported previously (Chin et al., 2001; Li et al., 2002), peripherally associated endosomal membrane proteins Rab5 and EEA1 fractionated into a membrane-bound pool (fractions 21–23) and a soluble pool (fractions 1–9) on the Optiprep gradient (Fig. 4F–H). We found that SIMPLE WT co-fractionated exclusively with the early endosomal membrane-associated Rab5 and EEA1 (fractions 21–23) (Fig. 4F,I). By contrast, SIMPLE W116G and P135T mutant proteins co-fractionated with both the early endosomal membrane-associated pool (fractions 21–23) and soluble pool (fractions 1–9) (Fig. 4G–I). Our analysis revealed no co-fractionation of SIMPLE WT and mutant proteins with LAMP2-positive membranous compartments (Fig. 4F–H). These results are consistent with the subcellular fractionation (Fig. 4A–C) and immunocytochemistry data (Fig. 4D,E; supplementary material Fig. S4E) and provide additional support for the CMT1C mutation-induced mislocalization of SIMPLE from the early endosome membrane to the cytosol.
CMT1C-associated mutations cause SIMPLE protein to be unstable
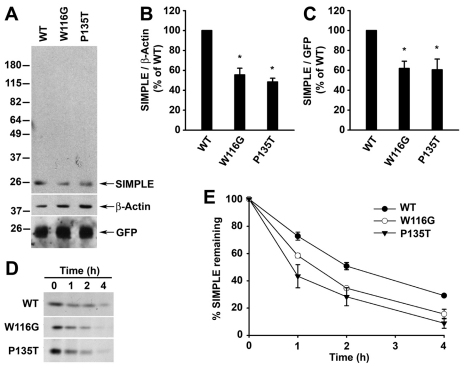
To determine whether CMT1C-associated mutations affect SIMPLE protein stability, we first assessed the steady-state levels of Myc-tagged SIMPLE WT and mutant proteins in HeLa cells. We found that the steady-state levels of SIMPLE W116G and P135T mutants were significantly lower compared with that of SIMPLE WT (Fig. 5A,B), suggesting that the SIMPLE mutant proteins were less stable than the WT protein. By using the cotransfected GFP as a reporter for transfection efficiency, we confirmed that the reductions in the steady-state protein levels of SIMPLE mutant proteins were not caused by the variability in the transfection efficiency (Fig. 5A,C). Moreover, our data indicated that the observed differences in the SIMPLE WT and mutant protein levels (Fig. 5A–C) were not due to the inability of the anti-SIMPLE antibody to detect aggregated forms of SIMPLE proteins, as this antibody was able to recognize cross-linked, oligomeric and aggregated species of SIMPLE proteins (data not shown).
Fig. 5.
CMT1C-associated mutations reduce the stability of SIMPLE protein in cells. (A) HeLa cells co-transfected with pEGFP vector and an equal amount of cDNAs encoding Myc-tagged WT, W116G, or P135T were analyzed by immunoblotting with antibodies against the Myc tag, β-actin and GFP. (B,C) Quantification of protein levels from A. After normalization to βactin (B) or GFP (C), SIMPLE W116G and P135T protein levels relative to that of SIMPLE WT are shown as mean ± s.e.m. from three independent experiments. *P<0.05 compared with SIMPLE WT. (D) The degradation of Myc-tagged SIMPLE WT, W116G or P135T proteins expressed in HEK293 cells were analyzed by [35S]Met/Cys pulse-chase assays. 35S-labeled SIMPLE WT or mutant proteins were immunoprecipitated from lysates with anti-Myc antibodies and detected by autoradiography. (E) The protein levels of WT and mutant SIMPLE from D were quantified and plotted relative to the corresponding protein levels at 0 hours. Data represent mean ± s.e.m. from three independent experiments.
Next, we performed pulse-chase experiments to examine the turnover of SIMPLE WT and mutant proteins in cells. The results revealed that SIMPLE W116G and P135T mutants were degraded significantly more than that of SIMPLE WT at 1, 2, and 4 hours (Fig. 5D,E). We found the half-life of SIMPLE WT was ~2 hours, whereas the half-life was reduced to ~1.3 hours for the SIMPLE W116G mutant and ~0.9 hours for the SIMPLE P135T mutant (Fig. 5E). Together, these results indicate that CMT1C-associated mutations destabilize SIMPLE protein in cells.
CMT1C-associated SIMPLE mutant proteins are prone to aggregation
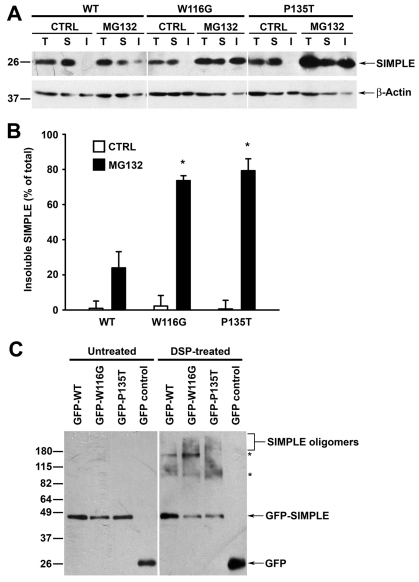
Our finding of CMT1C mutation-induced mislocalization of SIMPLE protein from the membrane to the cytosol (Fig. 4) raised the possibility that detachment of SIMPLE mutants from the membrane might liberate hydrophobic sequences and thus promote SIMPLE protein misfolding and aggregation. To examine this possibility, we performed detergent-insolubility assays (Johnston et al., 2002; Ross and Poirier, 2005) to assess the effects of CMT1C mutations on SIMPLE protein aggregation. The results showed that, under the normal cell culture conditions, SIMPLE WT and mutant proteins were predominantly found in the detergent-soluble fraction (Fig. 6A). However, upon proteasome inhibition, the amount of SIMPLE W116G and P135T mutants in the detergent-insoluble fraction was significantly greater than that of SIMPLE WT (Fig. 6A,B), indicating that CMT1C-associated mutations promote the accumulation of SIMPLE in detergent-insoluble aggregates.
Fig. 6.
CMT1C-associated mutations promote aggregation of SIMPLE. (A) Stably transfected HEK293 cells expressing Myc-tagged WT, W116G or P135T SIMPLE were treated for 24 hours with either 2 μM MG132 or vehicle control (CTRL). Cell lysates (T) were separated into TX-100-soluble (S) and -insoluble (I) fractions. Aliquots representing an equal percentage of each fraction were analyzed by immunoblotting with antibodies against SIMPLE and β-actin. (B) The relative level of WT or mutant SIMPLE proteins in the insoluble fraction was quantified and expressed as a percentage of the total SIMPLE protein level in the corresponding cell lysate. Data represent mean ± s.e.m. from three independent experiments. *P<0.05 compared with SIMPLE WT under the same treatment. (C) HEK293 cells expressing GFP or GFP-tagged WT or mutant SIMPLE were treated for 2 hours with 2 μM dithiobis(succinimidylpropionate) (DSP) or vehicle control (CTRL) and analyzed by immunoblotting with an anti-GFP antibody. Asterisk indicates cross-linked protein complexes formed between SIMPLE and its interacting proteins.
Misfolded proteins can form detergent-insoluble large aggregates or detergent-soluble small aggregates composed of misfolded protein oligomers (Ross and Poirier, 2005; Olzmann et al., 2008; Kubota, 2009). The apparent lack of detectable amounts of detergent-insoluble, SIMPLE mutant protein aggregates under normal conditions (Fig. 6A) prompted us to investigate whether SIMPLE mutant proteins aggregate into detergent-soluble, oligomeric species. To capture SIMPLE oligomeric species, we performed chemical cross-linking experiments with the cross-linker dithiobis(succinimidylpropionate) (DSP). We found that treatment of cells expressing SIMPLE WT with DSP resulted in the appearance of two SIMPLE-immunoreactive bands at ~95 kDa and ~160 kDa, both of which might represent the cross-linked protein complexes formed between SIMPLE and its yet-to-be-identified interacting partners (Fig. 6C). The levels of these two SIMPLE protein complexes were differentially affected by the W116G and P135T mutations (Fig. 6C), suggesting that CMT1C-associated mutations alter SIMPLE protein–protein interactions. In addition, cross-linking analysis revealed that both the W116G and P135T mutations caused a shift from the SIMPLE monomer to the higher molecular weight SIMPLE-immunoreactive protein smears, which probably represent mutant SIMPLE oligomeric species (Fig. 6C). Together, these results indicate that CMT1C-associated SIMPLE mutant proteins are more prone to aggregation compared with the SIMPLE WT protein.
CMT1C-associated mutations promote the formation of SIMPLE-positive aggresomes
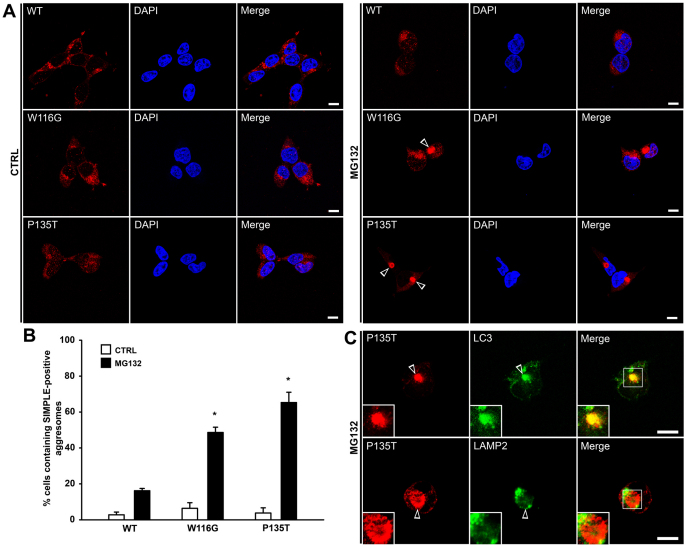
Next, we performed immunofluorescence confocal microscopic analysis to examine the nature of SIMPLE mutant protein aggregation in cells. We found that, under normal cell culture conditions, SIMPLE W116G and P135T mutant proteins did not form microscopically visible aggregates in cells (Fig. 7A,B), which is consistent with the detergent-insolubility data (Fig. 6A,B). Although the SIMPLE mutant oligomers formed under normal conditions (Fig. 6C) were microscopically undetectable, treatment of HEK293 cells (Fig. 7A,B) and primary Schwann cells (supplementary material Fig. S5D) with proteasome inhibitor MG132 caused SIMPLE W116G and P135T mutant proteins to accumulate in microscopically visible, perinuclear inclusion bodies that spatially and morphologically resemble aggresomes (Kopito, 2000; Olzmann et al., 2007; Olzmann et al., 2008). Further analyses confirmed that the mutant SIMPLE-positive inclusion bodies are bona fide aggresomes, because they were enriched with ubiquitin and Hsp70 and encaged by vimentin (supplementary material Fig. S5A,B) and their formation was blocked by the microtubuledepolymerizing drug nocodazole (supplementary material Fig. S6A,B). Quantitative analysis revealed that the percentage of cells containing SIMPLE W116G or SIMPLE P135T mutant-positive aggresomes upon proteasome inhibition was significantly greater than that of SIMPLE WT (Fig. 7B), indicating that CMT1C-associated mutations promote the formation of SIMPLE-positive aggresomes.
Fig. 7.
CMT1C-associated SIMPLE mutants are accumulated in aggresomes. (A) Stably transfected HEK293 cells expressing Myc-tagged WT, W116G or P135T SIMPLE were treated for 24 hours with 2 μM MG132 or vehicle control (CTRL) and then immunostained with anti-Myc antibody (red). Nuclei were visualized by DAPI stain. Aggresomes are indicated by open arrowheads. Scale bars: 10 μm. (B) The percentage of cells containing SIMPLE-positive aggresomes was quantified and shown as mean ± s.e.m. from three independent experiments. *P<0.05 compared with SIMPLE WT under the same treatment. (C) HEK293 cells expressing Myc-tagged P135T SIMPLE were treated with 2 μM MG132 for 24 hours and then double-immunostained with antibodies against the Myc tag (red) and LC3 or LAMP2 (green). Aggresomes are indicated by open arrowheads. Scale bars: 10 μm.
Increasing evidence suggests that aggresome formation serves as a mechanism for concentrating misfolded and aggregated proteins for subsequent clearance by autophagy (Fortun et al., 2003; Taylor et al., 2003; Chin et al., 2010). Autophagy is a bulk degradation process that involves the formation of a double-membrane structure called an autophagosome to engulf its cytoplasmic substrates, and the subsequent fusion of the autophagosome with the lysosome for the degradation of the substrates by lysosomal hydrolases (Mehrpour et al., 2010). Our immunofluorescence confocal microscopy analysis revealed that mutant SIMPLE-positive aggresomes were tightly encircled by the autophagosome marker LC3 (Fig. 7C, supplementary material Fig. S5C). Moreover, mutant SIMPLE-positive aggresomes were often found to be surrounded by the late endosome and lysosome marker LAMP2 (Fig. 7C, supplementary material Fig. S5C). These results suggest that SIMPLE mutant-containing aggresomes are substrates of autophagy.
CMT1C-associated SIMPLE mutant proteins are degraded by both the proteasome and autophagy pathways
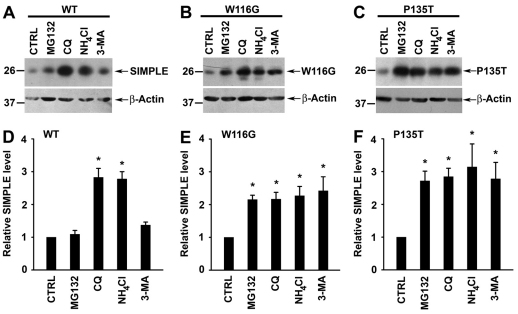
To determine the degradation pathway responsible for the clearance of WT and mutant SIMPLE proteins, we assessed the effects of proteasome, lysosome and autophagy inhibition on the steady-state levels of Myc-tagged WT and mutant SIMPLE proteins in stably transfected HEK293 cells. We found that SIMPLE WT protein level was significantly increased by treatment with the lysosome inhibitors chloroquine (CQ) and ammonium chloride (NH4Cl), but it was not affected by treatment with proteasome inhibitor MG132, autophagy inhibitor 3-methyladenine (3-MA) (Fig. 8A,D) or autophagy activator rapamycin (supplementary material Fig. S7A). In addition, immunostaining analysis showed that, although endogenous SIMPLE could not be detected in the LAMP2-positive late endosome and lysosome under normal conditions, a significant amount of SIMPLE protein was found in the late endosome and lysosome upon lysosome inhibition, indicating that SIMPLE is transiently present in the lysosome before its degradation (supplementary material Fig. S3B). Together, these results suggest that SIMPLE WT protein, similarly to many integral membrane proteins, mainly relies on the endosome-tolysosome sorting pathway for its degradation (Babst, 2004; Luzio et al., 2009). By contrast, we found that the protein levels of SIMPLE W116G and P135T mutants were significantly increased upon proteasome inhibition by MG132, autophagy inhibition by 3-MA or lysosome inhibition by CQ or NH4Cl (Fig. 8B,C,E,F). Conversely, SIMPLE mutant protein levels were significantly reduced by treatment with the autophagy activator rapamycin (supplementary material Fig. S7B,C). These data support the involvement of both the proteasome and autophagy pathways in selective clearance of CMT1C-associated SIMPLE mutant proteins.
Fig. 8.
Clearance of CMT1C-associated mutant SIMPLE proteins by both the proteasome and autophagy pathways. (A–C) Steady-state protein levels of WT (A), W116G (B), or P135T (C) SIMPLE in HEK293 cells treated with 20 μM MG132, 100 μM chloroquine (CQ), 50 μM NH4Cl, 10 mM 3-methyladenine (3-MA) or vehicle (CTRL) were analyzed by immunoblotting with antibodies against the Myc tag and β-actin. (D–F) The relative level of WT (D), W116G (E), or P135T (F) SIMPLE protein in the proteolysis inhibitor-treated cell lysates was normalized to the β-actin level and expressed relative to the normalized SIMPLE protein level in the corresponding vehicle-treated control lysate. Data represent mean ± s.e.m. from at least three independent experiments. *P<0.05 compared with vehicle-treated control.
Discussion
The identification of mutations in SIMPLE as the cause of demyelinating CMT1C underscores the importance of understanding the cellular function of this protein and the pathogenic mechanisms of its mutations. To elucidate the role of SIMPLE in normal physiology and in CMT pathogenesis, we generated a highly specific anti-SIMPLE antibody and characterized the expression and subcellular localization of endogenous SIMPLE protein by western blot analysis and immunocytochemistry. Our results showed that SIMPLE protein is highly expressed in peripheral nerves and Schwann cells, suggesting that SIMPLE participates in Schwann cell function and/or peripheral nerve function. Our finding that SIMPLE protein levels are considerably lower in the brain and muscle compared with peripheral nerves helps to explain why the primary defect caused by SIMPLE mutations is peripheral neuropathy rather than central nervous system dysfunction or muscular atrophy.
Our immunolocalization analyses with the highly specific anti-SIMPLE antibody revealed that, under normal physiological conditions, endogenous SIMPLE is predominantly localized to the early endosome but not the late endosome, lysosome, ER or Golgi. This result differs from the reported presence of SIMPLE at the late endosome and lysosome in a previous study which used an anti-SIMPLE antibody that also recognized a non-specific protein at ~100 kDa (Moriwaki et al., 2001). Our data clearly indicated that SIMPLE is not a resident protein of the lysosome and late endosome (Moriwaki et al., 2001). Instead, SIMPLE is a substrate protein for degradation by the lysosome, as we can only detect the presence of SIMPLE in the lysosome and late endosome upon lysosome inhibition. Our finding that SIMPLE is an early endosomal membrane protein is consistent with the reported interaction of SIMPLE with the endosomal sorting complex required for transport (ESCRT) subunit TSG101 (Shirk et al., 2005) and suggests that SIMPLE regulates protein sorting at the early endosome.
Our results strongly argue against the hypothesis that the SIMPLE C-rich domain is a RING finger domain with E3 ligase activity (Moriwaki et al., 2001; Saifi et al., 2005). We identified a TMD embedded within the SIMPLE C-rich domain and showed that SIMPLE is a membrane protein that requires the C-rich domain for its membrane association. Our results indicate that SIMPLE is an integral membrane protein rather than a lipid-anchored protein. The lack of an ER-targeting signal sequence and the location of the TMD close to the C-terminus of SIMPLE suggest that SIMPLE is likely to undergo post-translational insertion as a C-tail-anchored membrane protein (Borgese et al., 2007; Borgese and Fasana, 2011).
Our study is the first to examine the pathogenic effects of CMT1C-associated SIMPLE mutations. We found that CMT1C-associated SIMPLE W116G and P135T mutations cause dislocation of SIMPLE protein from the membrane to the cytosol. Interestingly, only a fraction of SIMPLE W116G and P135T mutant proteins is mislocalized to the cytosol, and a substantial amount of the mutant proteins remains attached to the membrane. Although our extraction analysis revealed that a subpopulation of the membrane-associated SIMPLE mutant proteins are peripherally attached to the membrane rather than being inserted as integral membrane proteins, the peripheral membrane association could not fully account for the observed partial dislocation of SIMPLE mutant proteins from the membrane to the cytosol. Moreover, we found that, unlike other mutant membrane proteins such as CFTR ΔF508 mutant (Lukacs et al., 1994), SIMPLE W116G and P135T mutant proteins were not retained at the ER or subjected to p97/VCP-dependent dislocation to the cytosol and ERAD. These results are consistent with the prediction that SIMPLE is a post-translationally inserted, C-tail-anchored membrane protein rather than a co-translationally inserted transmembrane protein (Borgese et al., 2007; Borgese and Fasana, 2011).
Although the molecular mechanisms underlying the post-translational insertion of C-tail-anchored membrane proteins remain poorly understood, it is clear that the C-terminal TMD constitutes the only membrane-targeting sequence (Borgese et al., 2007; Borgese and Fasana, 2011). Our findings that SIMPLE W116G and P135T mutant proteins are partially dissociated from the membrane is consistent with previous studies showing that mutations of Trp or Pro residue at the periphery of TMDs could cause a partial rather than a complete defect in membrane insertion (Hessa et al., 2005b; Hessa et al., 2005a). Our results, together with the fact that all CMT1C-associated mutations identified so far are clustered within or around the TMD of SIMPLE, suggest that impairment in the membrane insertion of SIMPLE protein could be a fundamental pathogenic event in CMT1C peripheral neuropathy.
CMT1C mutation-induced dislocation of SIMPLE protein from the membrane to the cytosol could cause peripheral neuropathy by at least two different mechanisms that are not mutually exclusive. First, mutation-induced mislocalization away from the early endosomal membrane would remove SIMPLE protein from its site of action, thereby contributing to CMT pathogenesis by a loss-of-function mechanism (Kim et al., 2002; Lupo et al., 2009; Roberts et al., 2010). Although the cellular function of SIMPLE is unknown, it has been proposed that SIMPLE might participate in protein sorting at the early endosome (Shirk et al., 2005). Loss of endosomal function of SIMPLE triggered by CMT1C mutations might cause deregulated trafficking of myelin proteins and/or other membrane proteins, resulting in Schwann cell dysfunction and demyelination. Second, mutation-induced detachment of SIMPLE protein from the membrane would expose its hydrophobic sequences and promote SIMPLE protein misfolding and aggregation, thereby contributing to CMT pathogenesis through a toxic gain-of-function mechanism. Protein misfolding and aggregation caused by genetic mutations or other factors has been shown to underlie the pathogenesis of many neurological diseases, including Parkinson disease and Alzheimer disease (Selkoe, 2004). Our findings that CMT1C mutations cause SIMPLE protein misfolding and aggregation, together with previous reports linking misfolded peripheral myelin protein 22 (PMP22) and myelin protein zero (MPZ) to other subtypes of demyelinating CMT (Shames et al., 2003; Myers et al., 2008; Mandich et al., 2009), suggest protein misfolding as a common cause of demyelinating peripheral neuropathies.
Understanding how cells handle and dispose of CMT-linked misfolded proteins are important because it might provide insights into strategies for combating the disease. Our study showed that CMT1C-associated SIMPLE mutant proteins, but not SIMPLE WT protein, are unstable and degraded by the proteasome. These results suggest that targeting the proteasome pathway might have therapeutic value in treating demyelinating CMT. However, the proteasome is capable of eliminating only the soluble, monomeric form of misfolded proteins, but is ineffective in degrading oligomeric and aggregated forms of misfolded proteins (Pickart and Cohen, 2004; Olzmann et al., 2008). Moreover, the proteasome function can even be directly inhibited by misfolded protein oligomers and aggregates (Kubota, 2009). Thus, targeting the proteasome alone might not be sufficient for halting CMT pathogenesis. Recently, impairment in proteasome function was found in a neuropathic mouse model expressing a CMT1A-linked PMP22 mutant protein (Fortun et al., 2005). In addition, the proteasome inhibitor bortezomib prescribed as a chemotherapeutic agent has been shown to cause dysfunction of the myelin sheath (Filosto et al., 2007) and peripheral neuropathy in human patients (Hamilton et al., 2005; Filosto et al., 2007). Together, these findings support a link between proteasome dysfunction and the pathogenesis of CMT demyelinating neuropathy.
The aggresome–autophagy pathway has emerged as a key cellular defense system against toxic build-up of misfolded proteins, particularly under the conditions of proteasome impairment (Fortun et al., 2003; Olzmann et al., 2007; Olzmann et al., 2008; Janen et al., 2010). Our finding of the selective targeting of misfolded SIMPLE mutant proteins to aggresomes upon proteasome inhibition supports the possibility that aggresome formation is a cytoprotective response serving to sequester potentially toxic misfolded proteins (Kopito, 2000; Olzmann et al., 2008). Consistent with recent reports suggesting the aggresomes as a staging area for the disposal of misfolded proteins by autophagy (Taylor et al., 2003; Olzmann et al., 2007), we found that SIMPLE mutant-containing aggresomes stained with the autophagosome marker LC3 and were tightly encircled by lysosomes. Moreover, the steady-state levels of SIMPLE mutants were significantly reduced by the autophagy activator rapamycin and were increased by the autophagy inhibitor 3-MA, or lysosome inhibitors CQ or NH4Cl. Together, our data support the involvement of the aggresome–autophagy pathway in the degradation of SIMPLE mutant proteins. Interestingly, aggresomes were also found in Schwann cells expressing misfolded PMP22 mutant proteins (Ryan et al., 2002; Fortun et al., 2006) and pharmacological activation of autophagy by rapamycin was shown to improve myelination in PMP22 mutant mice (Rangaraju et al., 2010). Thus, augmentation of the aggresome–autophagy pathway might be a viable therapeutic strategy for treating a number of demyelinating neuropathies.
Materials and Methods
Plasmids and antibodies
Conventional molecular biological techniques were used to generate expression vectors encoding N-terminal GST-, Myc- or GFP-tagged human SIMPLE WT (GenBank accession number NM_004862), W116G and P135T mutants, and SIMPLEΔP (residues 96–161) and SIMPLEΔC (residues 1–95) deletion mutants. The SIMPLE-targeting shRNA construct (NM_004862.1-397s1c1, Sigma) and the non-targeting shRNA control construct (SHC001, Sigma) were obtained commercially. A rabbit polyclonal anti-SIMPLE antibody was generated against a synthetic peptide corresponding to amino acid residues 50–64 of SIMPLE and affinity-purified as described previously (Chin et al., 2000). Other antibodies used in this study include anti-β-actin (C4, Sigma), anti-S-100 (S2644, Sigma), anti-DJ-1 (P7F), anti-LAMP2 (Iowa Developmental Studies Hybridoma Bank), anti-SNAP-25 (SMI81, Sternberger Monoclonals), anti-Thy-1 (AbD Serotec), anti-GFP (B2, Santa Cruz), anti-EEA1 (BD Transduction), anti-KDEL (Stressgen), anti-GM130 (BD Biosciences), anti-Rab5 (Sigma), anti-Myc (9E10), anti-Hsc70/Hsp70 (Stressgen), anti-ubiquitin (FL76, Santa Cruz), anti-vimentin (Sigma) and anti-LC3 (Sigma). All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories.
Cell culture and transfection
Primary Schwann cells were isolated from 3-month-old mice and cultured using established protocols (Pannunzio et al., 2005; Haastert et al., 2007). Primary cortical neuronal cultures were prepared from postnatal 0.5 day (P0.5) mice as described previously (Chen et al., 2010). All transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Stably transfected HEK293 cells were selected using 1000 μg/ml G418 (Sigma) as described (Hubbers et al., 2007).
GST-tagged protein purification
Expression of GST-tagged SIMPLE and GST proteins were induced in ArcticExpress competent cells (Agilent Technologies) and purified as described previously (Li et al., 2001).
Immunofluorescence confocal microscopy and quantification of endosomal localization
Cells were fixed in 4% paraformaldehyde and processed for immunofluorescence confocal microscopy as described previously (Olzmann et al., 2007). Endosomal localization of WT or mutant SIMPLE was quantified as the percentage of SIMPLE immunostaining that was colocalized with Rab5 immunostaining. Colocalization of SIMPLE and Rab5 was quantified as described (Giles et al., 2008; Webber et al., 2008) on unprocessed images of cells double-labeled for Myc-tagged WT or mutant SIMPLE and Rab5. Single cells were selected by manually tracing the cell outlines. The background was subtracted and the percentage of WT or mutant SIMPLE pixels overlapping with the Rab5 pixels was determined as described previously (Giles et al., 2008; Webber et al., 2008). For each experiment, 30–40 cells per group were randomly selected for analysis, and experiments were repeated at least three times.
Subcellular fractionation and membrane association analysis
Subcellular fractionations of HeLa, HEK293, or SH-SY5Y cells into membrane and cytosol fractions were performed as described (Giles et al., 2009). For membrane association analysis, membrane fractions were subjected to extraction with 1% Triton X-100 (TX-100), 4 M Urea, 1.5 M NaCl or 0.1 M Na2CO3 (pH 11.5) for 1 hour. Extracts were then subjected to centrifugation at 100,000 g to isolate the soluble and insoluble fractions as described previously (Li et al., 2001). For the inhibition of palmitoylation, SH-SY5Y cells were incubated for 24 hours at 37°C with of 2-bromohexadeconic acid (100 μM, Sigma) or vehicle (0.1% DMSO) as described (Chaudhury et al., 2009). The level of WT or mutant SIMPLE in each fraction relative to the total level in the post-nuclear supernatant was quantified by measuring the intensity of the SIMPLE band from the immunoblot image using the Scion image software as described previously (Giles et al., 2008). For density gradient fractionation analysis, post-nuclear supernatants were placed on a 10–30% linear Optiprep (Sigma) gradient and centrifuged for 20 hours at 125,000 g in a SW41 rotor (Beckman Coulter) as described previously (Chin et al., 2001; Li et al., 2002). Following centrifugation, the gradient was harvested into 250 μl fractions using an Auto Densi-Flow gradient harvester (Labconco).
[35S]Methionine pulse-chase analysis
Pulse-chase experiments were performed in stably transfected HEK293 cells expressing Myc-tagged WT or mutant SIMPLE as described previously (Olzmann et al., 2004; Giles et al., 2008). The protein levels of WT or mutant SIMPLE were quantified by measuring the intensity of the SIMPLE band from the image of the autoradiography films using the Scion image software.
Chemical crosslinking analysis
HEK293 cells were incubated for 2 hours on ice with the cross-linker DSP (2 mM, Pierce) or with vehicle (1× PBS) followed by 20 mM Tris-HCl to quench the cross-linking reaction as described previously (Cline and Mori, 2001). Cells were then lysed with 1% SDS, and an equal amount of total proteins from each lysate were subjected to immunoblot analyses.
Detergent insolubility assays
Stably transfected HEK293 cells were lysed and centrifuged at 100,000 g for 30 minutes to separate the TX-100-soluble and -insoluble fractions as described previously (Olzmann et al., 2007). Aliquots representing an equal percentage of each fraction were subjected to immunoblot analyses.
Analysis of aggresome formation
Stably transfected HEK293 cells expressing Myc-tagged WT or mutant SIMPLE were incubated in the presence and absence of 2 μM MG132 for 24 hours and processed for immunofluorescence confocal microscopic analysis of aggresome formation as described (Olzmann et al., 2007). An aggresome was defined as a single, perinuclear inclusion containing Myc-tagged WT or mutant SIMPLE proteins. For each experiment, 40–80 cells per group were randomly selected and scored for the presence of an aggresome in a blinded manner.
Treatment of cells with proteasome, lysosome and autophagy inhibitors and activators
Stably transfected HEK293 cells expressing WT or mutant SIMPLE were incubated for 24 hours at 37°C with protease inhibitor MG132 (20 μM, Sigma), lysosome inhibitor NH4Cl (50 mM, Sigma), lysosome inhibitor chloroquine (100 μM, Sigma), autophagy inhibitor 3-MA (10 mM, Sigma), autophagy activator rapamycin (100 nM, Sigma) or vehicle (0.1% DMSO) as described previously (Giles et al., 2008). Cells were then lysed with 1% SDS, and an equal amount of total proteins from each lysate were subjected to immunoblot analyses. The protein levels of WT or mutant SIMPLE were quantified as described (Giles et al., 2008) and then normalized to the corresponding β-Actin levels.
Statistical analysis
All experiments were repeated at least three times. Data were subjected to statistical analyses by ANOVA and appropriate post-hoc tests using SigmaPlot software (Systat Software). A P-value of less than 0.05 was considered statistically significant.
Acknowledgements
We thank Ron Kopito (Stanford University, Palo Alto, CA) for providing the NHK, p97/VCP WT and mutant constructs.
Footnotes
Funding
This work was supported by grants from National Institutes of Health [NS063501 (S.M.L.), NS050650, AG034126 (L.S.C.), ES015813, GM082828 (L.L.)]. The confocal imaging analysis was performed in Emory Neuroscience Core Facility supported in part by National Institutes of Health (NS055077). Deposited in PubMed for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.087114/-/DC1
References
- Babst M. (2004). GGAing ubiquitin to the endosome. Nat. Cell Biol. 6, 175-177 [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783-795 [DOI] [PubMed] [Google Scholar]
- Bennett C. L., Shirk A. J., Huynh H. M., Street V. A., Nelis E., Van Maldergem L., De Jonghe P., Jordanova A., Guergueltcheva V., Tournev I., et al. (2004). SIMPLE mutation in demyelinating neuropathy and distribution in sciatic nerve. Ann. Neurol. 55, 713-720 [DOI] [PubMed] [Google Scholar]
- Bolcato-Bellemin A. L., Mattei M. G., Fenton M., Amar S. (2004). Molecular cloning and characterization of mouse LITAF cDNA: role in the regulation of tumor necrosis factor-alpha (TNF-alpha) gene expression. J. Endotoxin Res. 10, 15-23 [DOI] [PubMed] [Google Scholar]
- Borgese N., Fasana E. (2011). Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta. 1808, 937-946 [DOI] [PubMed] [Google Scholar]
- Borgese N., Brambillasca S., Colombo S. (2007). How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368-375 [DOI] [PubMed] [Google Scholar]
- Ceresa B. P., Lotscher M., Schmid S. L. (2001). Receptor and membrane recycling can occur with unaltered efficiency despite dramatic Rab5(q79l)-induced changes in endosome geometry. J. Biol. Chem. 276, 9649-9654 [DOI] [PubMed] [Google Scholar]
- Chamberlain L. H., Burgoyne R. D., Gould G. W. (2001). SNARE proteins are highly enriched in lipid rafts in PC12 cells: implications for the spatial control of exocytosis. Proc. Natl. Acad. Sci. USA 98, 5619-5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A., He X. D., Goyal R. K. (2009). Role of PSD95 in membrane association and catalytic activity of nNOSalpha in nitrergic varicosities in mice gut. Am. J. Physiol. Gastrointest. Liver Physiol. 297, G806-G813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Li L., Chin L. S. (2010). Parkinson disease protein DJ-1 converts from a zymogen to a protease by carboxyl-terminal cleavage. Hum. Mol. Genet 19, 2395-2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L. S., Nugent R. D., Raynor M. C., Vavalle J. P., Li L. (2000). SNIP, a novel SNAP-25-interacting protein implicated in regulated exocytosis. J. Biol. Chem. 275, 1191-1200 [DOI] [PubMed] [Google Scholar]
- Chin L. S., Raynor M. C., Wei X., Chen H. Q., Li L. (2001). Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J. Biol. Chem. 276, 7069-7078 [DOI] [PubMed] [Google Scholar]
- Chin L. S., Olzmann J. A., Li L. (2010). Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem. Soc. Trans. 38, 144-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Mori H. (2001). Thylakoid DeltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154, 719-729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B., Christianson J. C., Kopito R. R., Brunger A. T. (2006). Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell 22, 451-462 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Bork P., Eisenhaber F. (1999). Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 292, 741-758 [DOI] [PubMed] [Google Scholar]
- Filosto M., Rossi G., Pelizzari A. M., Buzio S., Tentorio M., Broglio L., Mancuso M., Rinaldi M., Scarpelli M., Padovani A. (2007). A high-dose bortezomib neuropathy with sensory ataxia and myelin involvement. J. Neurol. Sci. 263, 40-43 [DOI] [PubMed] [Google Scholar]
- Fortun J., Dunn W. A., Jr, Joy S., Li J., Notterpek L. (2003). Emerging role for autophagy in the removal of aggresomes in Schwann cells. J. Neurosci. 23, 10672-10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortun J., Li J., Go J., Fenstermaker A., Fletcher B. S., Notterpek L. (2005). Impaired proteasome activity and accumulation of ubiquitinated substrates in a hereditary neuropathy model. J. Neurochem. 92, 1531-1541 [DOI] [PubMed] [Google Scholar]
- Fortun J., Go J. C., Li J., Amici S. A., Dunn W. A., Jr, Notterpek L. (2006). Alterations in degradative pathways and protein aggregation in a neuropathy model based on PMP22 overexpression. Neurobiol. Dis. 22, 153-164 [DOI] [PubMed] [Google Scholar]
- Gerding W. M., Koetting J., Epplen J. T., Neusch C. (2009). Hereditary motor and sensory neuropathy caused by a novel mutation in LITAF. Neuromuscul. Disord. 19, 701-703 [DOI] [PubMed] [Google Scholar]
- Giles L. M., Chen J., Li L., Chin L. S. (2008). Dystonia-associated mutations cause premature degradation of torsinA protein and cell-type-specific mislocalization to the nuclear envelope. Hum. Mol. Genet. 17, 2712-2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles L. M., Li L., Chin L. S. (2009). Printor, a novel torsinA-interacting protein implicated in dystonia pathogenesis. J. Biol. Chem. 284, 21765-21775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haastert K., Mauritz C., Chaturvedi S., Grothe C. (2007). Human and rat adult Schwann cell cultures: fast and efficient enrichment and highly effective non-viral transfection protocol. Nat. Protoc. 2, 99-104 [DOI] [PubMed] [Google Scholar]
- Hamilton A. L., Eder J. P., Pavlick A. C., Clark J. W., Liebes L., Garcia-Carbonero R., Chachoua A., Ryan D. P., Soma V., Farrell K., et al. (2005). Proteasome inhibition with bortezomib (PS-341): a phase I study with pharmacodynamic end points using a day 1 and day 4 schedule in a 14-day cycle. J. Clin. Oncol. 23, 6107-6116 [DOI] [PubMed] [Google Scholar]
- Hessa T., White S. H., von Heijne G. (2005a). Membrane insertion of a potassium-channel voltage sensor. Science 307, 1427 [DOI] [PubMed] [Google Scholar]
- Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. (2005b). Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377-381 [DOI] [PubMed] [Google Scholar]
- Hirokawa T., Boon-Chieng S., Mitaku S. (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378-379 [DOI] [PubMed] [Google Scholar]
- Hubbers C. U., Clemen C. S., Kesper K., Boddrich A., Hofmann A., Kamarainen O., Tolksdorf K., Stumpf M., Reichelt J., Roth U., et al. (2007). Pathological consequences of VCP mutations on human striated muscle. Brain 130, 381-393 [DOI] [PubMed] [Google Scholar]
- Janen S. B., Chaachouay H., Richter-Landsberg C. (2010). Autophagy is activated by proteasomal inhibition and involved in aggresome clearance in cultured astrocytes. Glia 58, 1766-1774 [DOI] [PubMed] [Google Scholar]
- Johnston J. A., Illing M. E., Kopito R. R. (2002). Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil. Cytoskeleton. 53, 26-38 [DOI] [PubMed] [Google Scholar]
- Kim B. E., Smith K., Meagher C. K., Petris M. J. (2002). A conditional mutation affecting localization of the Menkes disease copper ATPase. Suppression by copper supplementation. J. Biol. Chem. 277, 44079-44084 [DOI] [PubMed] [Google Scholar]
- Kirk E., Chin L. S., Li L. (2006). GRIF1 binds Hrs and is a new regulator of endosomal trafficking. J. Cell Sci. 119, 4689-4701 [DOI] [PubMed] [Google Scholar]
- Kopito R. R. (2000). Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524-530 [DOI] [PubMed] [Google Scholar]
- Kubota H. (2009). Quality control against misfolded proteins in the cytosol: a network for cell survival. J. Biochem. 146, 609-616 [DOI] [PubMed] [Google Scholar]
- Latour P., Gonnaud P. M., Ollagnon E., Chan V., Perelman S., Stojkovic T., Stoll C., Vial C., Ziegler F., Vandenberghe A., et al. (2006). SIMPLE mutation analysis in dominant demyelinating Charcot-Marie-Tooth disease: three novel mutations. J. Peripher. Nerv. Syst. 11, 148-155 [DOI] [PubMed] [Google Scholar]
- Li Y., Chin L. S., Weigel C., Li L. (2001). Spring, a novel RING finger protein that regulates synaptic vesicle exocytosis. J. Biol. Chem. 276, 40824-40833 [DOI] [PubMed] [Google Scholar]
- Li Y., Chin L. S., Levey A. I., Li L. (2002). Huntingtin-associated protein 1 interacts with hepatocyte growth factor-regulated tyrosine kinase substrate and functions in endosomal trafficking. J. Biol. Chem. 277, 28212-28221 [DOI] [PubMed] [Google Scholar]
- Lu C. F., Kurjan J., Lipke P. N. (1994). A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol. Cell Biol. 14, 4825-4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs G. L., Mohamed A., Kartner N., Chang X. B., Riordan J. R., Grinstein S. (1994). Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 13, 6076-6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo V., Galindo M. I., Martinez-Rubio D., Sevilla T., Vilchez J. J., Palau F., Espinos C. (2009). Missense mutations in the SH3TC2 protein causing Charcot-Marie-Tooth disease type 4C affect its localization in the plasma membrane and endocytic pathway. Hum. Mol. Genet 18, 4603-4614 [DOI] [PubMed] [Google Scholar]
- Luzio J. P., Parkinson M. D., Gray S. R., Bright N. A. (2009). The delivery of endocytosed cargo to lysosomes. Biochem. Soc. Trans. 37, 1019-1021 [DOI] [PubMed] [Google Scholar]
- Mandich P., Fossa P., Capponi S., Geroldi A., Acquaviva M., Gulli R., Ciotti P., Manganelli F., Grandis M., Bellone E. (2009). Clinical features and molecular modelling of novel MPZ mutations in demyelinating and axonal neuropathies. Eur. J. Hum. Genet. 17, 1129-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrpour M., Esclatine A., Beau I., Codogno P. (2010). Overview of macroautophagy regulation in mammalian cells. Cell Res. 20, 748-762 [DOI] [PubMed] [Google Scholar]
- Mestre-Escorihuela C., Rubio-Moscardo F., Richter J. A., Siebert R., Climent J., Fresquet V., Beltran E., Agirre X., Marugan I., Marin M., et al. (2007). Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood 109, 271-280 [DOI] [PubMed] [Google Scholar]
- Moriwaki Y., Begum N. A., Kobayashi M., Matsumoto M., Toyoshima K., Seya T. (2001). Mycobacterium bovis Bacillus Calmette-Guerin and its cell wall complex induce a novel lysosomal membrane protein, SIMPLE, that bridges the missing link between lipopolysaccharide and p53-inducible gene, LITAF(PIG7), and estrogen-inducible gene, EET-1. J. Biol. Chem. 276, 23065-23076 [DOI] [PubMed] [Google Scholar]
- Myers J. K., Mobley C. K., Sanders C. R. (2008). The peripheral neuropathy-linked Trembler and Trembler-J mutant forms of peripheral myelin protein 22 are folding-destabilized. Biochemistry 47, 10620-10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naef R., Suter U. (1999). Impaired intracellular trafficking is a common disease mechanism of PMP22 point mutations in peripheral neuropathies. Neurobiol. Dis. 6, 1-14 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Yoshikawa H., Fujimura H., Sakoda S., Yanagihara T. (1996). Accumulation of peripheral myelin protein 22 in onion bulbs and Schwann cells of biopsied nerves from patients with Charcot-Marie-Tooth disease type 1A. Acta. Neuropathol. 92, 454-460 [DOI] [PubMed] [Google Scholar]
- Notterpek L., Ryan M. C., Tobler A. R., Shooter E. M. (1999). PMP22 accumulation in aggresomes: implications for CMT1A pathology. Neurobiol. Dis. 6, 450-460 [DOI] [PubMed] [Google Scholar]
- Olzmann J. A., Brown K., Wilkinson K. D., Rees H. D., Huai Q., Ke H., Levey A. I., Li L., Chin L. S. (2004). Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 279, 8506-8515 [DOI] [PubMed] [Google Scholar]
- Olzmann J. A., Li L., Chudaev M. V., Chen J., Perez F. A., Palmiter R. D., Chin L. S. (2007). Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178, 1025-1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J. A., Li L., Chin L. S. (2008). Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr. Med. Chem. 15, 47-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzio M. E., Jou I. M., Long A., Wind T. C., Beck G., Balian G. (2005). A new method of selecting Schwann cells from adult mouse sciatic nerve. J. Neurosci. Methods 149, 74-81 [DOI] [PubMed] [Google Scholar]
- Patzko A., Shy M. E. (2011). Update on Charcot-Marie-Tooth disease. Curr. Neurol. Neurosci. Rep. 11, 78-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Cohen R. E. (2004). Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177-187 [DOI] [PubMed] [Google Scholar]
- Rangaraju S., Verrier J. D., Madorsky I., Nicks J., Dunn W. A., Jr, Notterpek L. (2010). Rapamycin activates autophagy and improves myelination in explant cultures from neuropathic mice. J. Neurosci. 30, 11388-11397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. C., Peden A. A., Buss F., Bright N. A., Latouche M., Reilly M. M., Kendrick-Jones J., Luzio J. P. (2010). Mistargeting of SH3TC2 away from the recycling endosome causes Charcot-Marie-Tooth disease type 4C. Hum. Mol. Genet. 19, 1009-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. A., Poirier M. A. (2005). Opinion: What is the role of protein aggregation in neurodegeneration? Nat. Rev. Mol. Cell Biol. 6, 891-898 [DOI] [PubMed] [Google Scholar]
- Rost B., Yachdav G., Liu J. (2004). The PredictProtein server. Nucleic Acids Res. 32, W321-W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M. C., Shooter E. M., Notterpek L. (2002). Aggresome formation in neuropathy models based on peripheral myelin protein 22 mutations. Neurobiol. Dis. 10, 109-118 [DOI] [PubMed] [Google Scholar]
- Saifi G. M., Szigeti K., Wiszniewski W., Shy M. E., Krajewski K., Hausmanowa-Petrusewicz I., Kochanski A., Reeser S., Mancias P., Butler I., et al. (2005). SIMPLE mutations in Charcot-Marie-Tooth disease and the potential role of its protein product in protein degradation. Hum. Mutat. 25, 372-383 [DOI] [PubMed] [Google Scholar]
- Santonico E., Belleudi F., Panni S., Torrisi M. R., Cesareni G., Castagnoli L. (2010). Multiple modification and protein interaction signals drive the Ring finger protein 11 (RNF11) E3 ligase to the endosomal compartment. Oncogene 29, 5604-5618 [DOI] [PubMed] [Google Scholar]
- Scarpini E., Meola G., Baron P., Beretta S., Velicogna M., Scarlato G. (1986). S-100 protein and laminin: immunocytochemical markers for human Schwann cells in vitro. Exp. Neurol. 93, 77-83 [DOI] [PubMed] [Google Scholar]
- Seaton G. J., Hall L., Jones R. (2000). Rat sperm 2B1 glycoprotein (PH20) contains a C-terminal sequence motif for attachment of a glycosyl phosphatidylinositol anchor. Effects of endoproteolytic cleavage on hyaluronidase activity. Biol. Reprod. 62, 1667-1676 [DOI] [PubMed] [Google Scholar]
- Selkoe D. J. (2004). Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat. Cell Biol. 6, 1054-1061 [DOI] [PubMed] [Google Scholar]
- Shames I., Fraser A., Colby J., Orfali W., Snipes G. J. (2003). Phenotypic differences between peripheral myelin protein-22 (PMP22) and myelin protein zero (P0) mutations associated with Charcot-Marie-Tooth-related diseases. J. Neuropathol. Exp. Neurol. 62, 751-764 [DOI] [PubMed] [Google Scholar]
- Shirk A. J., Anderson S. K., Hashemi S. H., Chance P. F., Bennett C. L. (2005). SIMPLE interacts with NEDD4 and TSG101: evidence for a role in lysosomal sorting and implications for Charcot-Marie-Tooth disease. J. Neurosci. Res. 82, 43-50 [DOI] [PubMed] [Google Scholar]
- Sorek N., Bloch D., Yalovsky S. (2009). Protein lipid modifications in signaling and subcellular targeting. Curr. Opin. Plant Biol. 12, 714-720 [DOI] [PubMed] [Google Scholar]
- Street V. A., Bennett C. L., Goldy J. D., Shirk A. J., Kleopa K. A., Tempel B. L., Lipe H. P., Scherer S. S., Bird T. D., Chance P. F. (2003). Mutation of a putative protein degradation gene LITAF/SIMPLE in Charcot-Marie-Tooth disease 1C. Neurology 60, 22-26 [DOI] [PubMed] [Google Scholar]
- Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K. H. (2003). Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 12, 749-757 [DOI] [PubMed] [Google Scholar]
- Tobler A. R., Notterpek L., Naef R., Taylor V., Suter U., Shooter E. M. (1999). Transport of Trembler-J mutant peripheral myelin protein 22 is blocked in the intermediate compartment and affects the transport of the wild-type protein by direct interaction. J. Neurosci. 19, 2027-2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C. E., Shotton D. M. (1989). Effects of capping on the non-ionic detergent solubility of rat thymocyte glycoproteins. Eur. J. Cell Biol. 50, 324-332 [PubMed] [Google Scholar]
- Tusnady G. E., Simon I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849-850 [DOI] [PubMed] [Google Scholar]
- Vogel K., Roche P. A. (1999). SNAP-23 and SNAP-25 are palmitoylated in vivo. Biochem. Biophys. Res. Commun. 258, 407-410 [DOI] [PubMed] [Google Scholar]
- Wang D., Liu J., Tang K., Xu Z., Xiong X., Rao Q., Wang M., Wang J. (2009). Expression of pig7 gene in acute leukemia and its potential to modulate the chemosensitivity of leukemic cells. Leuk. Res. 33, 28-38 [DOI] [PubMed] [Google Scholar]
- Webber E., Li L., Chin L. S. (2008). Hypertonia-associated protein Trak1 is a novel regulator of endosome-to-lysosome trafficking. J. Mol. Biol. 382, 638-651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White I. J., Souabni A., Hooper N. M. (2000). Comparison of the glycosylphosphatidylinositol cleavage/attachment site between mammalian cells and parasitic protozoa. J. Cell Sci. 113, 721-727 [DOI] [PubMed] [Google Scholar]
- Yamashita A., Tanaka K., Kamata R., Kumazawa T., Suzuki N., Koga H., Waku K., Sugiura T. (2009). Subcellular localization and lysophospholipase/transacylation activities of human group IVC phospholipase A2 (cPLA2gamma). Biochim. Biophys. Acta. 1791, 1011-1022 [DOI] [PubMed] [Google Scholar]