Abstract
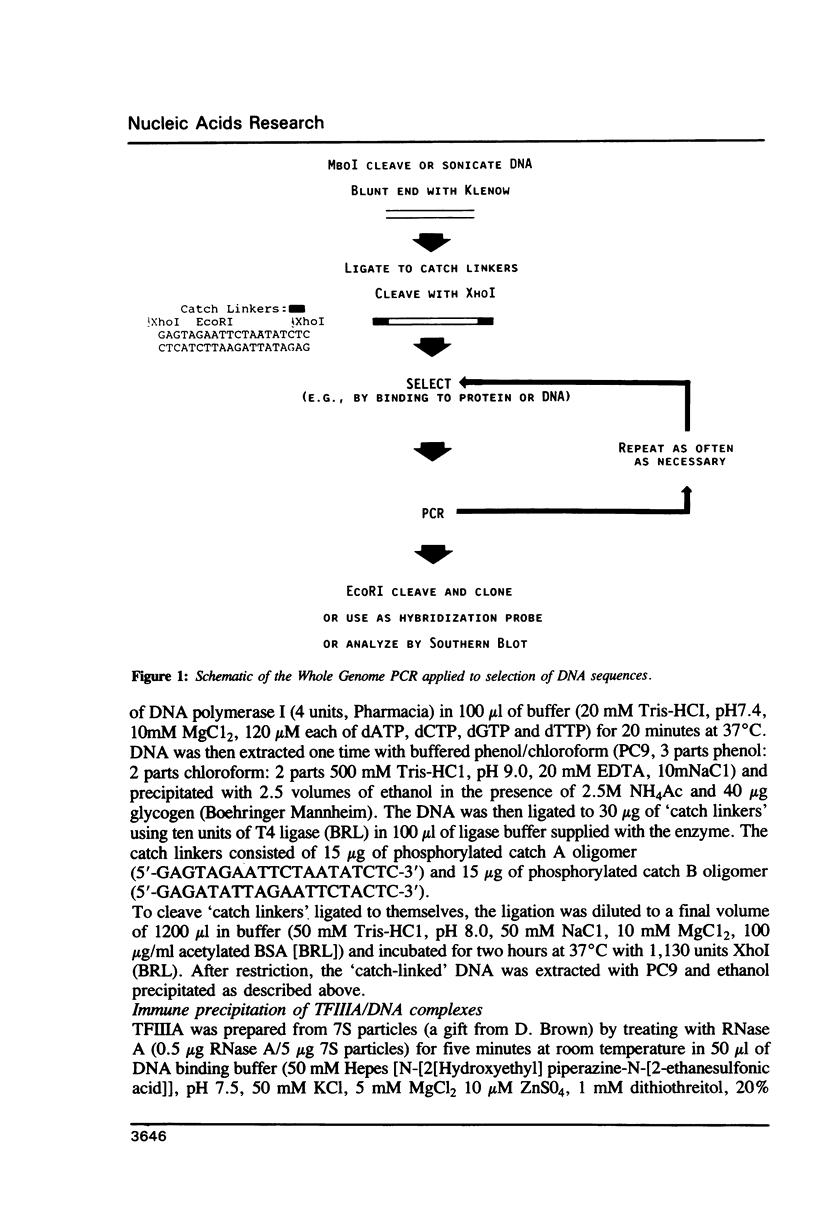
A strategy is described that allows the isolation of DNA sequences that can bind to gene regulatory proteins. Total genomic DNA is first converted to a form that is suitable for amplification by the polymerase chain reaction (Whole Genome PCR), and the DNA sequences of interest are selected by binding to the regulatory protein and immune precipitation. Because sequences recovered from the selection step can be amplified by PCR, the selection process can be designed for maximum enrichment with little concern about recovery. Furthermore, the selection process can be repeated as often as necessary. Sequences recovered after amplification can be cloned and/or used as hybridization probes. As a test of this strategy, we selected human sequences that bound to Xenopus transcription factor IIIA (TFIIIA). Seven clones were isolated that were on the average 94% identical to the previously described 61 bp binding site of TFIIIA. This strategy could be adapted to isolate sequences that can be selected by any physical or biological method.
Full text
PDF
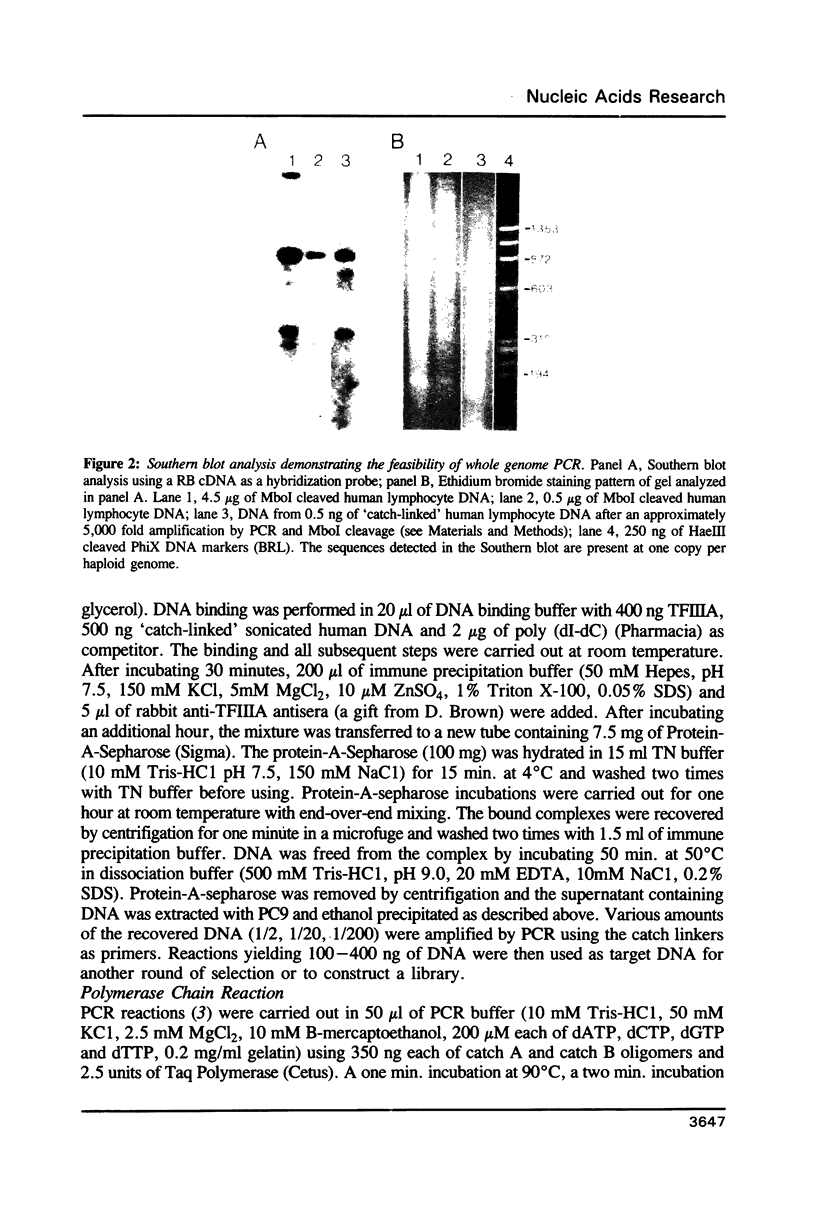






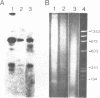
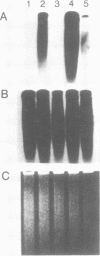
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biedenkapp H., Borgmeyer U., Sippel A. E., Klempnauer K. H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988 Oct 27;335(6193):835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Buluwela L., Forster A., Boehm T., Rabbitts T. H. A rapid procedure for colony screening using nylon filters. Nucleic Acids Res. 1989 Jan 11;17(1):452–452. doi: 10.1093/nar/17.1.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Bogenhagen D. F., Wu C. W. Cooperative model for the binding of Xenopus transcription factor A to the 5S RNA gene. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2142–2145. doi: 10.1073/pnas.80.8.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlen L., Attardi G. Proportion of HeLa cell genome complementary to transfer RNA and 5 s RNA. J Mol Biol. 1971 Mar 28;56(3):535–553. doi: 10.1016/0022-2836(71)90400-1. [DOI] [PubMed] [Google Scholar]
- Haymerle H., Herz J., Bressan G. M., Frank R., Stanley K. K. Efficient construction of cDNA libraries in plasmid expression vectors using an adaptor strategy. Nucleic Acids Res. 1986 Nov 11;14(21):8615–8624. doi: 10.1093/nar/14.21.8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Kunkel L. M., Monaco A. P., Middlesworth W., Ochs H. D., Latt S. A. Specific cloning of DNA fragments absent from the DNA of a male patient with an X chromosome deletion. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4778–4782. doi: 10.1073/pnas.82.14.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. H., Bookstein R., Hong F., Young L. J., Shew J. Y., Lee E. Y. Human retinoblastoma susceptibility gene: cloning, identification, and sequence. Science. 1987 Mar 13;235(4794):1394–1399. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- Pollwein P., Wagner S., Knippers R. Application of an immunoprecipitation procedure to the study of SV40 tumor antigen interaction with mouse genomic DNA sequences. Nucleic Acids Res. 1987 Dec 10;15(23):9741–9759. doi: 10.1093/nar/15.23.9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert J. M., Kinzler K. W., Wong A. J., Bigner S. H., Kao F. T., Law M. L., Seuanez H. N., O'Brien S. J., Vogelstein B. The GLI-Kruppel family of human genes. Mol Cell Biol. 1988 Aug;8(8):3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sealey P. G., Whittaker P. A., Southern E. M. Removal of repeated sequences from hybridisation probes. Nucleic Acids Res. 1985 Mar 25;13(6):1905–1922. doi: 10.1093/nar/13.6.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E. Compilation of transcription regulating proteins. Nucleic Acids Res. 1988 Mar 25;16(5):1879–1902. doi: 10.1093/nar/16.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. Developmental regulation of two 5S ribosomal RNA genes. Science. 1988 Sep 23;241(4873):1626–1632. doi: 10.1126/science.241.4873.1626. [DOI] [PubMed] [Google Scholar]