Abstract
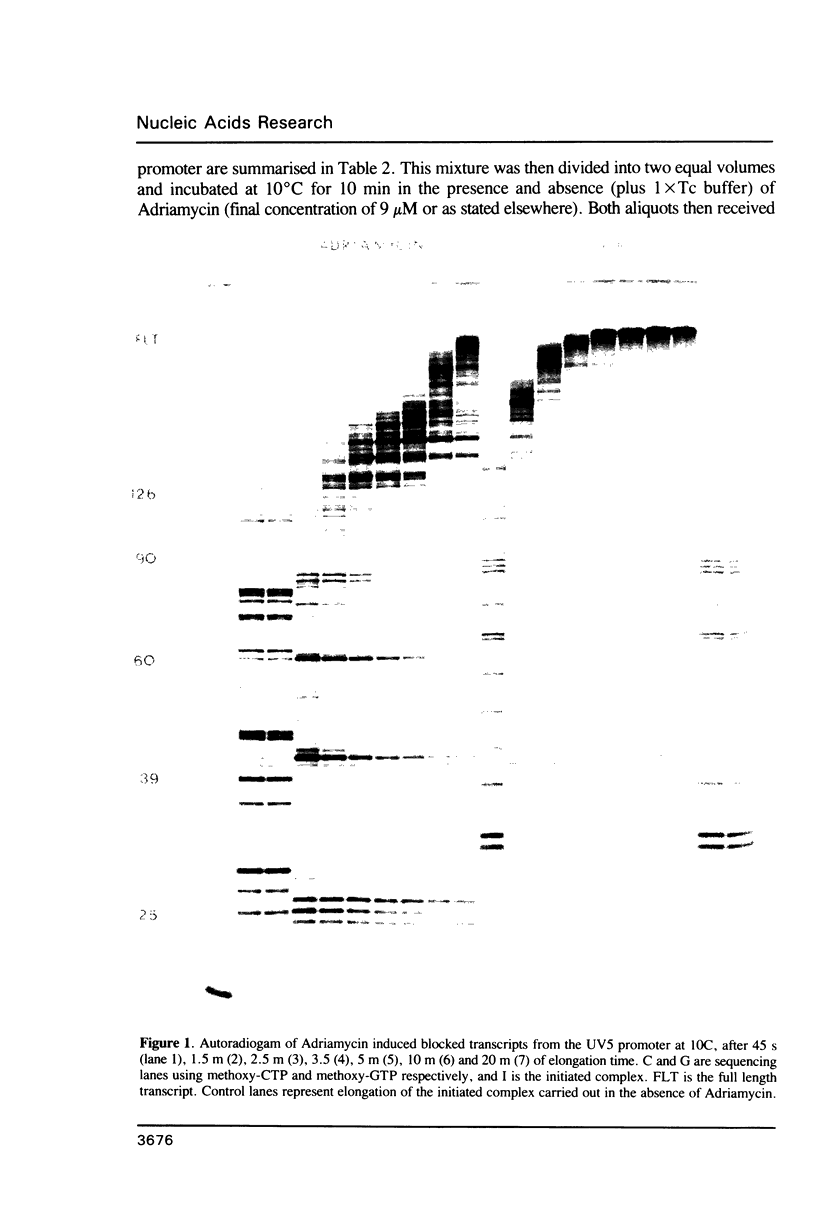
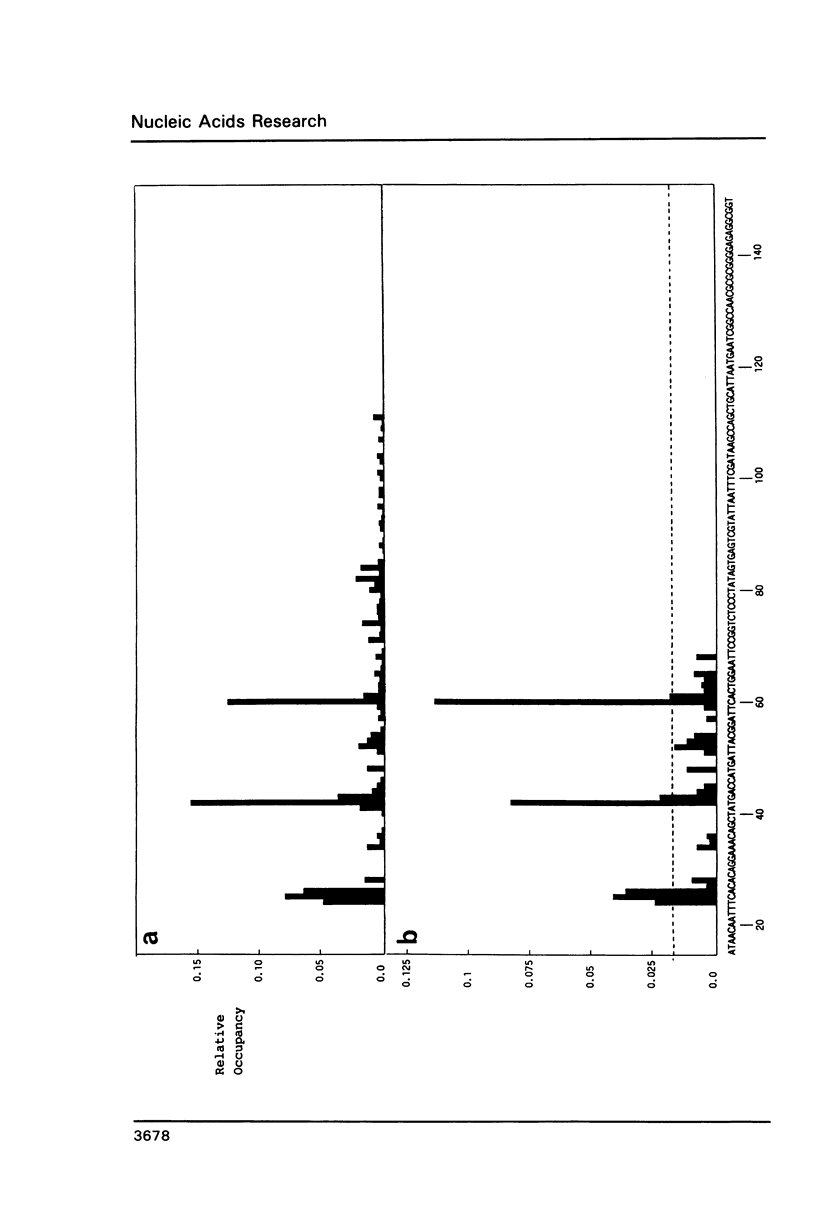
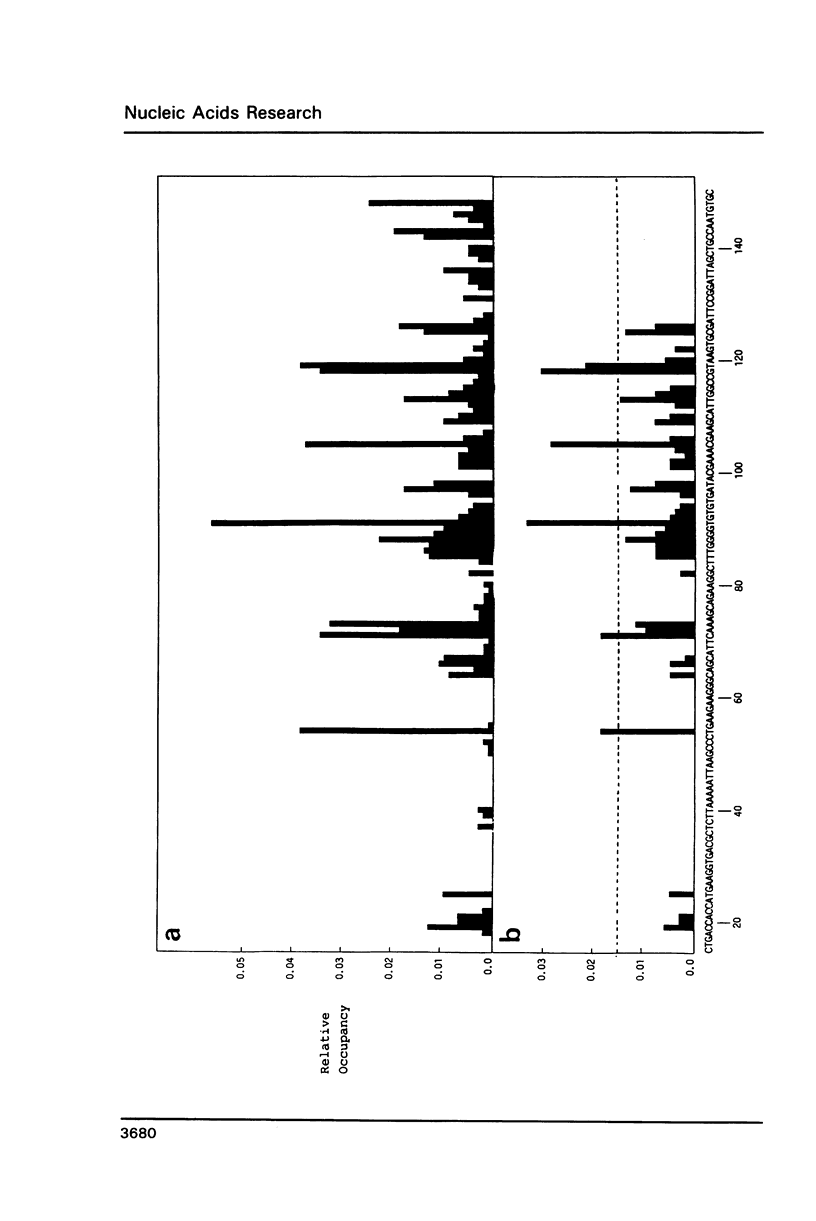
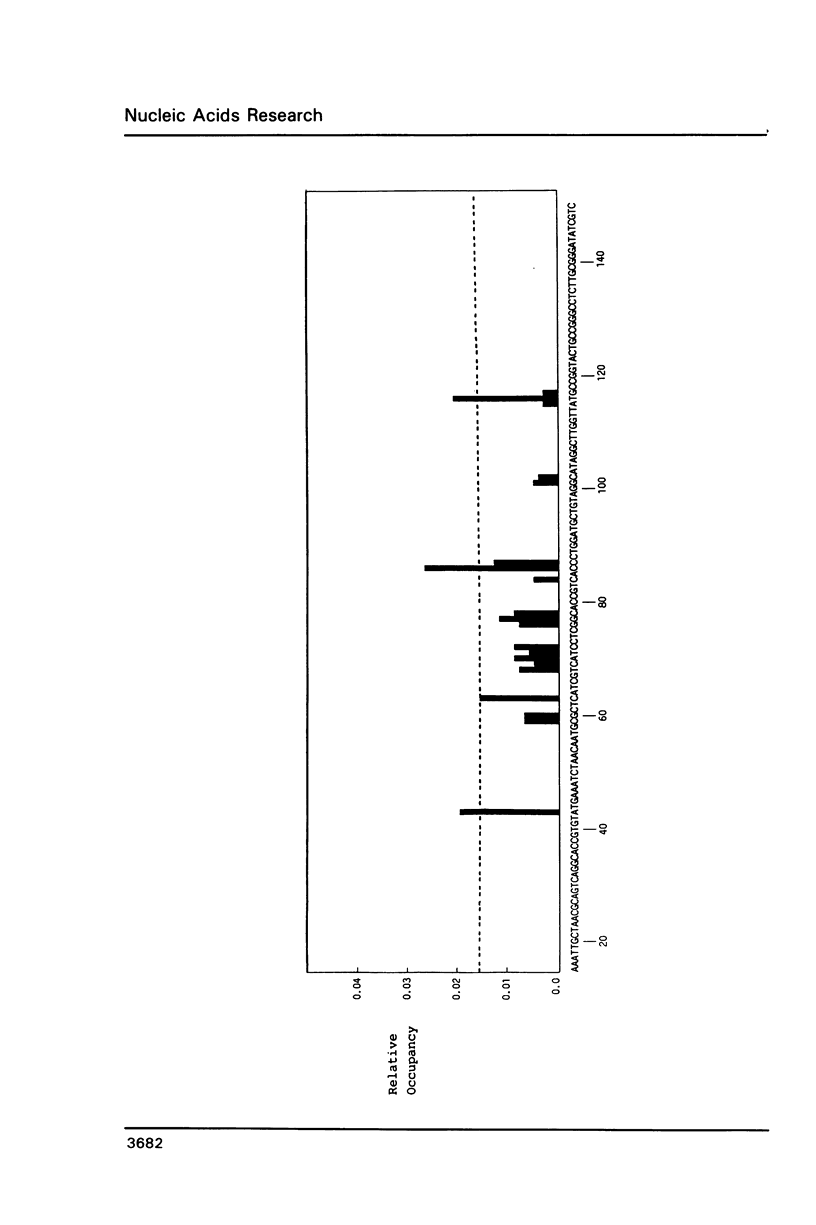
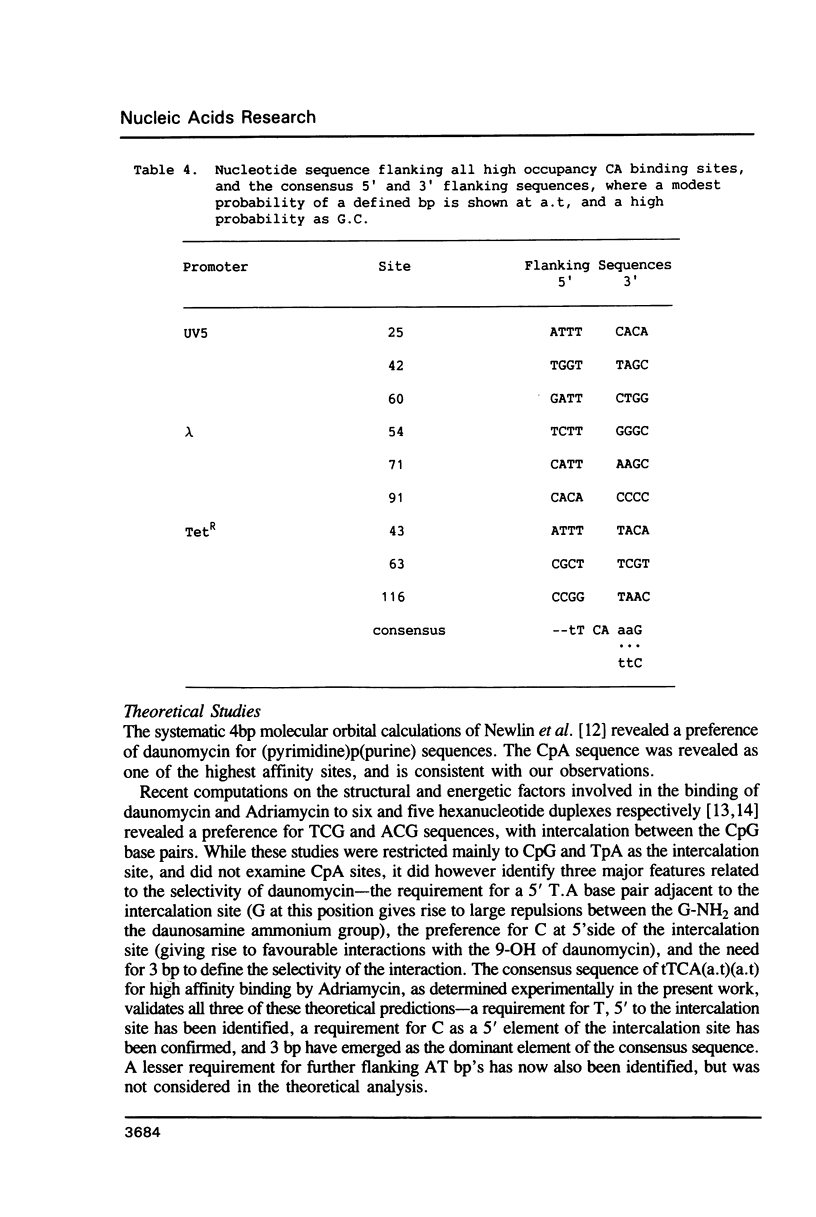
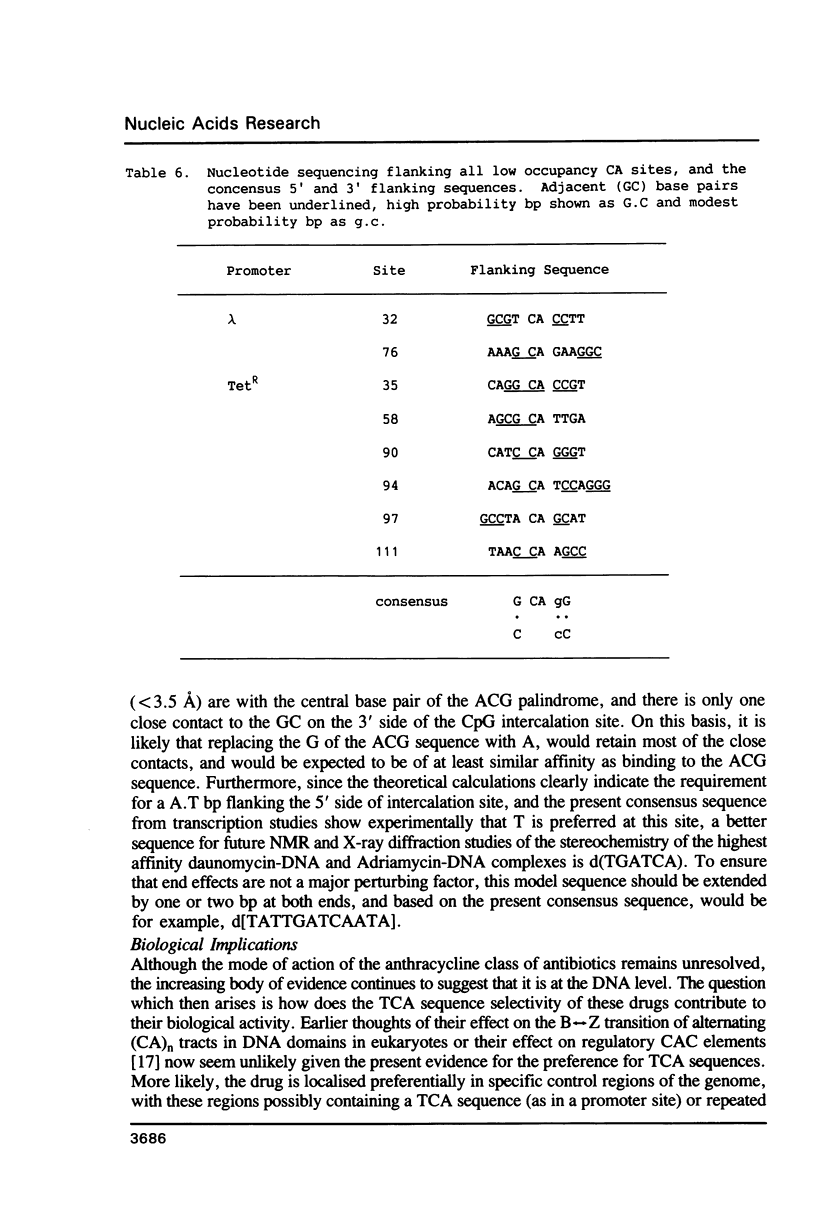
An in vitro transcription assay has been used to define twelve high occupancy transcriptional blockage sites in 260 bp of heterogenous DNA on three promoter-containing DNA fragments. Transcription proceeded to immediately upstream of CpA sequences in nine of these sites, and this defines the most preferred intercalation site as CpA. In almost all cases, this sequence was flanked by T on the 5' end. The consensus sequence for the highest affinity Adriamycin site is therefore 5'-TCA, with some evidence for preference of AT base pairs flanking both ends of this trinucleotide [i.e., (t)TCA(a.t)(a.t)]. In contrast, lower occupancy CpA sites were flanked on the 5' end by a GC base pair, with a preference for up to three GC base pairs on the 3' end. The low affinity consensus sequence is therefore (G.C)CA(g.c)(g.c)(g.c).
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chaires J. B., Dattagupta N., Crothers D. M. Kinetics of the daunomycin--DNA interaction. Biochemistry. 1985 Jan 15;24(2):260–267. doi: 10.1021/bi00323a004. [DOI] [PubMed] [Google Scholar]
- Chaires J. B., Fox K. R., Herrera J. E., Britt M., Waring M. J. Site and sequence specificity of the daunomycin-DNA interaction. Biochemistry. 1987 Dec 15;26(25):8227–8236. doi: 10.1021/bi00399a031. [DOI] [PubMed] [Google Scholar]
- Chen K. S., Gresh N., Pullman B. A theoretical investigation on the sequence selective binding of adriamycin to double-stranded polynucleotides. Nucleic Acids Res. 1986 Mar 11;14(5):2251–2267. doi: 10.1093/nar/14.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. X., Gresh N., Pullman B. A theoretical investigation on the sequence selective binding of daunomycin to double-stranded polynucleotides. J Biomol Struct Dyn. 1985 Dec;3(3):445–466. doi: 10.1080/07391102.1985.10508434. [DOI] [PubMed] [Google Scholar]
- DuVernay V. H., Jr, Pachter J. A., Crooke S. T. Deoxyribonucleic acid binding studies on several new anthracycline antitumor antibiotics. Sequence preference and structure--activity relationships of marcellomycin and its analogues as compared to adriamycin. Biochemistry. 1979 Sep 4;18(18):4024–4030. doi: 10.1021/bi00585a028. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Brassett C., Waring M. J. Kinetics of dissociation of nogalamycin from DNA: comparison with other anthracycline antibiotics. Biochim Biophys Acta. 1985 Jul 5;840(3):383–392. doi: 10.1016/0304-4165(85)90219-3. [DOI] [PubMed] [Google Scholar]
- Grant M., Phillips D. R. Dissociation of polydeoxynucleotide-daunomycin complexes. Mol Pharmacol. 1979 Jul;16(1):357–360. [PubMed] [Google Scholar]
- Graves D. E., Krugh T. R. Adriamycin and daunorubicin bind in a cooperative manner to deoxyribonucleic acid. Biochemistry. 1983 Aug 2;22(16):3941–3947. doi: 10.1021/bi00285a033. [DOI] [PubMed] [Google Scholar]
- Graves D. E., Krugh T. R. Adriamycin and daunorubicin bind in a cooperative manner to deoxyribonucleic acid. Biochemistry. 1983 Aug 2;22(16):3941–3947. doi: 10.1021/bi00285a033. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Austin R. H. Importance of DNA stiffness in protein-DNA binding specificity. Nature. 1987 Sep 17;329(6136):263–266. doi: 10.1038/329263a0. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Wang A. H., Rich A., Kim S. H. Local mobility of nucleic acids as determined from crystallographic data. III. A daunomycin-DNA complex. J Mol Biol. 1988 Jan 20;199(2):349–357. doi: 10.1016/0022-2836(88)90318-x. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy C. R., Yen S. F., Smith J. C., Lown J. W., Wilson W. D. Stopped-flow kinetic analysis of the interaction of anthraquinone anticancer drugs with calf thymus DNA, poly[d(G-C)].poly[d(G-C)], and poly[d(A-T)].poly[d(A-T)]. Biochemistry. 1986 Oct 7;25(20):5933–5940. doi: 10.1021/bi00368a015. [DOI] [PubMed] [Google Scholar]
- Lown J. W., Krowicki K., Bhat U. G., Skorobogaty A., Ward B., Dabrowiak J. C. Molecular recognition between oligopeptides and nucleic acids: novel imidazole-containing oligopeptides related to netropsin that exhibit altered DNA sequence specificity. Biochemistry. 1986 Nov 18;25(23):7408–7416. doi: 10.1021/bi00371a024. [DOI] [PubMed] [Google Scholar]
- Maizels N. M. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlin D. D., Miller K. J., Pilch D. F. Interactions of molecules with nucleic acids. VII. Intercalation and T.A specificity of daunomycin in DNA. Biopolymers. 1984 Jan;23(1):139–158. doi: 10.1002/bip.360230111. [DOI] [PubMed] [Google Scholar]
- Phillips D. R., Crothers D. M. Kinetics and sequence specificity of drug-DNA interactions: an in vitro transcription assay. Biochemistry. 1986 Nov 18;25(23):7355–7362. doi: 10.1021/bi00371a017. [DOI] [PubMed] [Google Scholar]
- Phillips D. R., DiMarco A., Zunino F. The interaction of daunomycin with polydeoxynucleotides. Eur J Biochem. 1978 Apr 17;85(2):487–492. doi: 10.1111/j.1432-1033.1978.tb12264.x. [DOI] [PubMed] [Google Scholar]
- Phillips D. R., Greif P. C., Boston R. C. Daunomycin-DNA dissociation kinetics. Mol Pharmacol. 1988 Feb;33(2):225–230. [PubMed] [Google Scholar]
- Quigley G. J., Wang A. H., Ughetto G., van der Marel G., van Boom J. H., Rich A. Molecular structure of an anticancer drug-DNA complex: daunomycin plus d(CpGpTpApCpG). Proc Natl Acad Sci U S A. 1980 Dec;77(12):7204–7208. doi: 10.1073/pnas.77.12.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorobogaty A., White R. J., Phillips D. R., Reiss J. A. Elucidation of the DNA sequence preferences of daunomycin. Drug Des Deliv. 1988 Jul;3(2):125–151. [PubMed] [Google Scholar]
- Skorobogaty A., White R. J., Phillips D. R., Reiss J. A. The 5'-CA DNA-sequence preference of daunomycin. FEBS Lett. 1988 Jan 25;227(2):103–106. doi: 10.1016/0014-5793(88)80877-9. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell. 1985 Dec;43(2 Pt 1):449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- Tsou K. C., Yip K. F. Effect of deoxyribonuclease on adriamycin-polynucleotide complexes. Cancer Res. 1976 Sep;36(9 PT1):3367–3373. [PubMed] [Google Scholar]
- Valentini L., Nicolella V., Vannini E., Menozzi M., Penco S., Arcamone F. Association of anthracycline derivatives with DNA: a fluorescence study. Farmaco Sci. 1985 Jun;40(6):377–390. [PubMed] [Google Scholar]
- Wang A. H., Ughetto G., Quigley G. J., Rich A. Interactions between an anthracycline antibiotic and DNA: molecular structure of daunomycin complexed to d(CpGpTpApCpG) at 1.2-A resolution. Biochemistry. 1987 Feb 24;26(4):1152–1163. doi: 10.1021/bi00378a025. [DOI] [PubMed] [Google Scholar]
- White R. J., Phillips D. R. Transcriptional analysis of multisite drug-DNA dissociation kinetics: delayed termination of transcription by actinomycin D. Biochemistry. 1988 Dec 27;27(26):9122–9132. doi: 10.1021/bi00426a009. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]



