Abstract
The Major Histocompatibility Complex (MHC) plays an important role in the human immune system. The MHC is involved in the antigen presentation system assisting T cells to identify foreign or pathogenic proteins. However, an MHC molecule binding a self-peptide may incorrectly trigger an immune response and cause an autoimmune disease, such as multiple sclerosis. Understanding the molecular mechanism of this process will greatly assist in determining the aetiology of various diseases and in the design of effective drugs. In the present study, we have used the Fresno semi-empirical scoring function and modify the approach to the prediction of peptide-MHC binding by using open-source and public domain software. We apply the method to HLA class II alleles DR15, DR1, and DR4, and the HLA class I allele HLA A2. Our analysis shows that using a large set of binding data and multiple crystal structures improves the predictive capability of the method. The performance of the method is also shown to be correlated to the structural similarity of the crystal structures used. We have exposed some of the obstacles faced by structure-based prediction methods and proposed possible solutions to those obstacles. It is envisaged that these obstacles need to be addressed before the performance of structure-based methods can be on par with the sequence-based methods.
Introduction
Multiple sclerosis (MS) is a neurological disease characterised by inflammation and demyelination in the central nervous system. MS is regarded as an autoimmune disease by many researchers [1]–[5], however, the pathogenesis of the disease is not well understood. Genetic linkage analyses of MS patients have identified the DRB1*1501 and DQB1*0602 alleles of the Major Histocompatibility Complex (MHC) molecule as definite genetic risk factors [2], [5]. This has been confirmed in more recent genome wide association studies [6]. The MHC molecule is involved in the antigen presentation system and assists the T cells to identify pathogenic proteins. While the overall antigen presentation mechanism is reasonably well understood, the specificity and sensitivity of peptide binding to MHC molecules, and the binding of T-cells to the resultant complex, required to elicit an immune response, is not well defined. Deeper knowledge of the peptide binding process may help to isolate the cause of the disease and detect peptides with therapeutic potential.
Currently, there are three schools of MHC-peptide binding prediction methods based on the information and approach used in the prediction: sequence-motif (PSSM-) based, artificial intelligence- (AI-) based, and structure-based. The first two schools examine the patterns exhibited by the sequences of binding peptides, whereas structure-based methods study the relationship between the binding affinities and the structures of MHC-peptide complexes.
Early work on peptides that bind to MHC molecules observed patterns in the peptide sequences. Systemic analyses of the effects of amino acids on the peptide binding affinities provide the basis for position-specific scoring matrices to predict binding affinity [7]–[10]. More recently, many studies introduced artificial intelligence algorithms in the attempt to understand the subtle underlying patterns [11]–[14]. Due to the type of input, PSSM- and AI-based methods are sometimes generalised as sequence-based prediction methods [14].
In addition to sequence information, structure-based methods also incorporate additional structural information from experimental crystal structures of MHC-peptide complexes [15]–[21]. Usually the atomic coordinates of the MHC molecule are extracted from an experimental crystal structure as the frame template, and the atomic coordinates of the peptide from the same structure are used as the template for fitting new peptides. Once a structure fitted with a new peptide is constructed, the structure may be subjected to energy minimisation. Using the new structure, the distance between two atoms and the physiochemical properties of the atoms are used to determine if the interaction is beneficial or not to the binding.
Much effort has been put into developing sequence-based methods, which have shown considerable performance [8], [11], [14], [22]. On the other hand, the availability of experimentally determined structures allows structure-based methods to study the precise relationship between the structure and peptide binding specificity. The inclusion of structural information may reveal properties affecting the binding not obvious on the sequence level. Furthermore, the recent increase in the number of experimentally determined structures for MHC-peptide complexes is expected to provide further data to improve the performance of structure-based methods. A more detailed and comprehensive review of computational methods for predicting peptide binding to the MHC, particularly structure-based methods, has been written by Liao and Arthur [23].
Despite considerable research into the development of computational techniques for determining peptide binding to the MHC and successful predictions for some alleles, the performance of various binding prediction algorithms for MHC class II alleles, including DRB1*1501, is still relatively poor. Previously, Rognan et al. [24] had some success in predicting the binding affinity of peptides for the HLA A*0201 allele using a structure-based method. In the present study, we adopt the Fresno semi-empirical scoring function developed by Rognan et al. to study peptide binding to MHC class I and II alleles in order to improve the computational prediction of peptide binding to DRB1*1501.
Results
Validation of the prediction method
In this study, we adapted the semi-empirical method for predicting peptide binding affinity for MHC class I molecules originally proposed by Rognan et al. [24]. The public domain software packages MolProbity [25] and SCWRL 4 [26] were used instead of SYBYL BIOPOLYMER to add hydrogen atoms to the crystal structures and predict peptide side chain atomic positions. The modelling algorithm was implemented in PERL and R was used to perform the partial-least-square regression analysis with leave-one-out cross-validation.
The open source adaptation of the protocol was tested using the original five HLA-A0201 (A2) structures (the Madden structures) used by Rognan et al. Table 1 compares the experimental free energy of binding with the theoretical values of Rognan et al. and our analysis. In each case, our prediction more accurately estimates the experimental free energy of binding. The cross-validation correlation score, q2, was excellent at 0.971 and the standard error of prediction, Spress, was appropriately low at 0.727. In comparison, Rognan et al. achieved a q2 value of 0.895 and a Spress value of 3.448. Thus, we established that our approach, using open source equivalents and our own PERL implementation of the Fresno scoring function, performs better than the original implementation.
Table 1. Comparison of the free energies for five HLA-A*0201 structures.
| Peptide | PDB ID | ΔGbind, kJ/mol | ||
| Experimentala | Rognanb | Predictedc | ||
| TLTSCNTSV | 1HHG | −37.32 | −36.85 (−0.47) | −37.19 (−0.13) |
| FLPSDFFPSV | 1HHH | −48.45 | −48.56 (+0.11) | −48.41 (−0.04) |
| GILGFVFTL | 1HHI | −46.94 | −47.03 (+0.09) | −47.01 (+0.07) |
| ILKEPVHGV | 1HHJ | −37.60 | −38.96 (+1.36) | −37.74 (+0.14) |
| LLFGYPVYV | 1HHK | −45.48 | −45.57 (−0.09) | −45.43 (−0.05) |
Experimental values from the original publications.
Predictions made by Rognan et al. in the original Fresno implementation; the deviations from the experimental values are included in parentheses.
Our predictions; the deviations from the experimental values are included in parentheses.
Validation of our open source adaption of the method is crucial to ensure the integrity of our PERL implementation of the technique and the alternate use of open source applications. By repeating the analysis of Rognan et al., we were able to show that our open source adaptation of the method reproduces the results of the original analysis, thus validating our adaptation. In fact, our approach generates slightly more accurate predictions than the original method.
Prediction of peptide binding to HLA-DR15
Having validated the prediction method, we applied the procedure to the prediction of the free energy of peptide binding in HLA-DRB1*1501 (DR15). The HLA-DR15 allele of the MHC is a major genetic risk factor for MS. Our aim here was to use the method developed and validated above to predict peptide binding in this allele as a step to understanding the role this allele plays in the pathogenesis of MS.
There are only two experimentally determined structures for HLA-DR15: 1YMM and 1BX2. 1YMM was chosen as a reference structure as it was the most recently published crystal structure. The AntiJen database contains 188 entries of peptides with peptide binding data for HLA-DR15. Of these, only twenty peptides were fourteen amino acids in length as required to match the length of the peptide in the 1YMM reference structure. These peptides are shown in Table 2.
Table 2. All twenty 14-mer peptides with experimental binding data in regard to HLA-DR15 extracted from AntiJen.
| Peptide | IC50 (nmol) | Temp (°C) |
| ADTISSYFVGKMYF [40] | 160 | 37 |
| DENPVVHFFKNIVT [41] | 4.6 | 37 |
| DTISSYFVGKMYFN [41] | 780 | 37 |
| ENPVVHFFKNIVTA [41] | 12 | 37 |
| FNLIDTKCYKLEHP [41] | 35000 | 37 |
| GKMYFNLIDTKCYK [41] | 33000 | 37 |
| HFFKNIVTPRTPPY [41] | 405 | 37 |
| ISSYFVGKMYFNLI [41] | 1600 | 37 |
| KMYFNLIDTKCYKL [41] | 68000 | 37 |
| KNSADTISSYFVGK [41] | 210 | 37 |
| MYFNLIDTKCYKLE [41] | 6500 | 37 |
| NLIDTKCYKLEHPV [41] | 40000 | 37 |
| NPVVHFFKNIVTPR [41] | 6.8 | 37 |
| NSADTISSYFVGKM [41] | 330 | 37 |
| SADTISSYFVGKMY [41] | 230 | 37 |
| SSYFVGKMYFNLID [41] | 1600 | 37 |
| SYFVGKMYFNLIDT [41] | 400 | 37 |
| TISSYFVGKMYFNL [41] | 190 | 37 |
| YFNLIDTKCYKLEH [41] | 15000 | 37 |
| YFVGKMYFNLIDTK [41] | 33000 | 37 |
Each peptide was modelled in the binding groove of the MHC molecule and the resulting structure used to determine the terms of Fresno scoring function (equation 2). The resulting equations for all twenty peptides were then subjected to the statistical analysis to determine the regression coefficients. These regression coefficients are then used to predict the theoretical binding free energy for each peptide for comparison with the experimental data. After the cross-validation analysis, the q2 value for the analysis was 0.243 and Spress was 6.429 confirming the prediction method was unable to accurately reproduce binding free energies for peptides in HLA-DR15.
To confirm this result was not due to an anomaly with the 1YMM structure, we also repeated the analysis with the 1BX2 structure. Similar results were obtained (data not shown).
Thus, the success of the scoring function in reproducing, and slightly improving, the results of Rognan et al. with the class I A*0201 allele, was not seen when working with the class II DRB1*1501 allele. This prompted us to a detailed examination of the Rognan et al. scoring function and its applications to assess the efficacy of the method in different circumstances.
Effect of data quantity on prediction accuracy
One possible explanation for the failure to adequately predict binding free energies in HLA-DR15 compared to the success in predictions with HLA-A2 may relate to class II MHC molecules requiring a larger set of binding data to better predict peptide binding. However, as noted above, only twenty peptides of appropriate size are contained in the AntiJen database for HLA-DR2.
In order to test this hypothesis, we considered HLA-DRB1*0101 (DR1) and HLA-DRB1*0401 (DR4): the two most studied class II alleles. Multiple PDB entries can be found for both HLA-DR1 and HLA-DR4 alleles. The most recently published structures with the best resolution were used as reference structures (1FTY and 1J8H). Both these alleles have more peptides with experimental binding data in the AntiJen database than HLA-DR15 with 74 peptides from 11 studies meeting the selection criteria for HLA-DR1 and 58 usable peptides from the same study for HLA-DR4.
The calculated q2 and Spress values for HLA-DR1 were 0.275 and 7.795 respectively. The calculated q2 and Spress values for HLA-DR4 were 0.283 and 6.390. Thus, using larger peptide binding data reference sets results in a modest improvement in both the cross-validation correlation score and the standard error of prediction to DR-15. However, the former remains low, and the latter high, indicating that the predictive capacity of the method remains poor. This suggests that while the quantity of peptide binding data does have an impact on the predictive ability of the scoring function, it is not the primary factor.
Effect of MHC class on prediction accuracy
Another possible factor affecting the prediction may be the class of MHC molecule used as the reference structure. The original method of Rognan et al. was developed and tested on MHC class I, and allele A*0201 in particular. It is possible that the more open topology of the MHC class II structure means the approach is not suitable, at least in its current form, for class II molecules. To explore this possibility, we attempted to duplicate our experiments above, but with class I molecules, and the A*0201 allele in particular.
As a reference structure, we chose 2GTW for the HLA-A2 allele [27]. This structure is not one of the five Madden structures, has a high resolution, and was published recently. A list of 174 peptides from 22 studies was extracted from the AntiJen database. Thus, our selection replicates the selection we made previously for a class II allele.
The calculated q2 and Spress values using the structure 2GTW were 0.01974 and 6.037. Thus, even using a class I structure, with a large set of peptide binding data, the technique does not achieve good predictive capability. To confirm this, we repeated the experiment using one of the five Madden structures as a reference. Since the peptide in the structure 1HHH is longer (decamer) than the other structures (nonamers), the 1HHH data was incorporated in two ways. The peptide of 1HHH was either truncated at the N-terminal or the C-terminal of the peptide in order to fit into the other structures with nonamers, or the peptide was excluded from the analysis completely. The MHC structure of 1HHH structure was not used at all, since peptides from the other structures will not fit. The procedure was repeated for each of the four structures (1HHG, 1HHI, 1HHJ, and 1HHK).
When the peptide from 1HHH was not used (i.e. only four peptides were used as input data), 1HHG, 1HHI, and 1HHJ returned low q2 values suggesting no predictive capability for the technique. The q2 and Spress values for 1HHK, however, were significantly better at 0.7897 and 1.75, although still not nearly as good as the values seen in the validation study. When the peptide from 1HHH was used, none of the reference structures was able to return a good result.
The favourable result for 1HHK presented a possible reason for the performance of validation study. 1HHK was therefore used as the reference structure in a further analysis under the same conditions used for 2GTW. However, this analysis gave q2 and Spress values of 0.002 and 6.083.
Predictive capability is dependent on quantity of structural data
The previous experiments consistently showed poor predictive capability for the approach, despite the remarkable success of the approach in the validation study. A final point of difference between the experiments is that the validation study uses five reference structures i.e., in calculating the terms and thence the regression coefficients, the atomic distances used are those of the peptide in its native crystal structure. In contrast, the other studies use peptides modeled in a single reference crystal structure.
Since calculation of the free energy of binding is based on the reference structure, if the predicted structure is different from how the peptide binds the MHC molecule natively, it may damage the predictive performance of the method. Thus, using a large set of reference structures simultaneously may provide more structural information and thus lead to better predictions.
To test the hypothesis, we searched PDB for HLA-A2 structures with one of the 174 peptides previously collected from AntiJen database, and found 17 structures, including 1HHJ (Table 3). Fourteen of them share one of three common peptides (ILKEPVHGV, NLVPMVATV, and SLLMWITQC) with other structures. Thus, we used various combinations of 6 structures, consisting of the 3 unique structures and a combination of three structures chosen from the 14 structures sharing the three common peptides, such that only one structure with each peptide was used.
Table 3. List of PDB entries and corresponding peptide binding data.
| Peptide | Temperature (°C) | IC50 (nmol) | PDB |
| AAGIGILTV [42] | 4 | 0.00008 | 2GUO |
| FLWGPRALV [42] | 4 | 0.0000021 | 1QEW |
| ILKEPVHGV [43] | 4 | 0.000008 | 1AKJ |
| ILKEPVHGV | 4 | 0.000008 | 1HHJ |
| ILKEPVHGV | 4 | 0.000008 | 1P7Q |
| ILKEPVHGV | 4 | 0.000008 | 2X4U |
| IMDQVPFSV [44] | 26 | 0.00000654 | 1TVH |
| NLVPMVATV [45] | 4 | 0.0000125 | 2X4R |
| NLVPMVATV | 4 | 0.0000125 | 3GSN |
| NLVPMVATV | 4 | 0.0000125 | 3GSO |
| SLLMWITQC [46] | 37 | 0.00002107 | 1S9W |
| SLLMWITQC | 37 | 0.00002107 | 2BNR |
| SLLMWITQC | 37 | 0.00002107 | 2F53 |
| SLLMWITQC | 37 | 0.00002107 | 2F54 |
| SLLMWITQC | 37 | 0.00002107 | 2P5E |
| SLLMWITQC | 37 | 0.00002107 | 2P5W |
| SLLMWITQC | 37 | 0.00002107 | 2PYE |
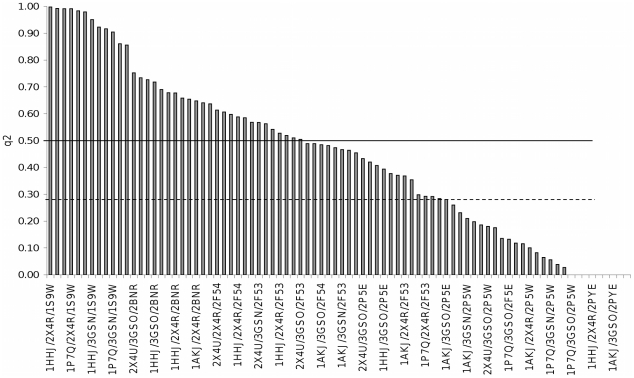
The q2 and Spress values varied between 0.998 to complete randomness. However, most combinations (57 combinations) showed improvement over the best of the previous analyses using a single reference structure (q2 value of 0.283) and nearly half of the combinations (37 combinations) achieved a q2 value greater than 0.5 (Fig. 1). This supports our hypothesis that using multiple reference structures will boost the prediction performance.
Figure 1. Spread of q2 values for different combinations of reference structures.
37 out of 84 combinations of reference structures (44%) achieved a q2 value greater than 0.5 and 57 (68%) achieved a q2 value greater than 0.283. which was the best predictive performance for analyses using only one reference structure.
Yet, the effect is not definitive. While most sets of reference structures generate better results than a single reference structure, the predictive capability still varies depending on the reference set chosen, with many reference sets still showing less than adequate predictive capability, despite improvements over single reference structure methods.
To examine the potential impact of different structural characteristics on the predictive performance, we explored the correlation between q2 and Spress values and various characteristics of the structures (Table 4). The first of these was the average root mean square deviation (RMSD) of the reference structures. The RMSD was calculated for all the combinations used in the analysis using the atoms from the MHC molecule alone, the peptide alone, and the whole structure. The RMSD scores were calculated for all pairs of structures in the set of reference structures and the results averaged to give a mean RMSD score for the set The RMSD scores were compared to the corresponding q2 and Spress values using Spearman's rank correlation. Secondly, the q2 and Spress values were also compared to the average resolution of the structures using Spearman's rank correlation. A correlation coefficient of 1 (or −1) indicates perfect correlation in the same (or opposite) direction. A value of 0 indicates no correlation.
Table 4. Comparison between q2 and Spress to the RMSD score and the resolution of structures.
| q2 | Spress | |
| RMSD | −0.607 | 0.604 |
| RMSDMHC | −0.579 | 0.577 |
| RMSDpeptide | 0.076 | −0.080 |
| Average resolution | −0.103 | 0.105 |
Three RMSD scores were calculated based on the use of the structures. RMSDMHC is the RMSD for the structure of the MHC molecule alone, RMSDpeptide is the RMSD for the structure of peptide alone, and the RMSD for the whole structure.
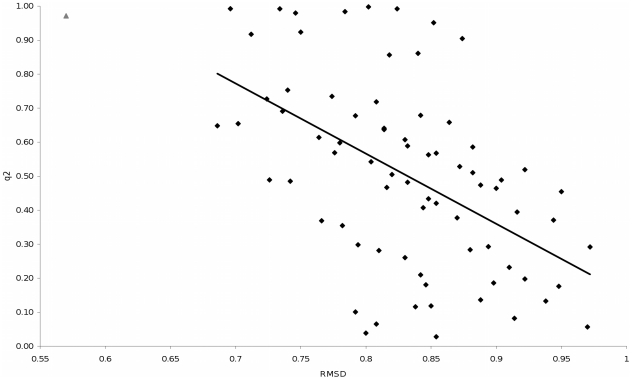
The Spearman's coefficient between the q2 values and the RMSD scores shows an intermediate correlation between average RMSD score and q2 value with a small average RMSD between the structures giving rise to a high q2 value, and thus better predictive performance for the approach (Fig. 2). This is also the case for the five Madden structures used in the original Fresno study. The average RMSD score of the five Madden structures was 0.57, which is better than all of the combinations used in the analysis, giving rise to the high q2 value and predictive performance in the original study. The correlation between the q2 values and the RMSDMHC scores suggests that the correlation is primarily attributed to the structure of the MHC molecule.
Figure 2. The comparison of q2 values and RMSD scores shows a general negative correlation.
The point for the Madden structures is the grey triangle located towards the top left of the figure.
On the other hand, little correlation was seen between the average resolution of the structures and the q2 values. This suggests that depth of resolution of the reference structures is not critical to the predictive performance of the method.
Discussion
In this series of experiments, we have shown that our implementation of the Fresno scoring function, using open source/free software, reproduces the results of Rognan et al. and, in fact, performs slightly better than the original implementation. However, when the number of reference structures used is reduced to one, the performance of the scoring function is greatly diminished, even if a large set of peptide binding data is used. This indicates that either MHC molecules assume quite different positions whilst binding to different peptides or that the theoretical approach used to predict peptide binding is quite sensitive to small changes in MHC structure. If a MHC molecule binds to the peptides more or less in the same way, the differences between structures should be minimal, and the scoring function should still be able to predict the binding affinity albeit with a less satisfying performance. On the other hand, if the MHC molecule assume different positions when binding to different peptides, multiple structures will be required to effectively sample all possible confirmations used as a basis for the semi-empirical model. Our experiments demonstrate this to be the case. When only one of the five structures used in the original Fresno study was used to analyse the binding affinities of all the peptides, only one structure could be used to achieve a good performance. Nonetheless, this performance was still worse than using all five reference structures. We also showed that even when one of the best structures for HLA-A2 is used as the reference structure, the prediction performance was still less than ideal, but when more reference structures were employed the q2 value can reach over 0.9. It is therefore important to consider various binding confirmations when constructing a free energy scoring function.
The best solution is to determine the structure for all binding peptides used in both establishing the regression coefficients for the scoring function as well as those whose binding free energy is to be predicted. However, this need for structural information for each peptide being considered makes it effectively impossible to use the method in large scale computational studies, such as an exhaustive scan of all possible peptides to predict potential epitopes for the MHC molecule.
Two further approaches offer a potential solution to this problem. The first is to obtain a large set of structures and use the structure with the most similar peptide for the peptides that do not have an experimentally determined structure. The other approach is to derive a “consensus structure” by averaging all the available structures. A consensus structure may sacrifice accuracy for some peptides but will hopefully be able to fit most peptides within a tolerable error level. Due to the nature of these approaches, the first may provide higher accuracy, however, the second approach should be easier to implement.
Another obstacle for structure-based methods is the reduced set of binding data. While sequence-based methods can simply categorise peptides into binders or non-binders based on the IC50 values, structure-based methods often rely on precise input, which excludes implicit values, such as strong, intermediate, and weak binding. Moreover, there is discrepancy in the binding data for many peptides due to various experimental settings. Any slight change in the input can produce a different result, and a large inconsistency in the input can render the result useless. However, discrepancies may be introduced in two areas: the detection method and the choice of competing peptide in the competitive assay. There are two detection methods based on the labelling tag, either fluorescence or radioactive isotopes, used to label the target peptide. While the two methods share the same principle, the readings can vary greatly and a difference is observed between two studies using different labelling method. In addition to the detection methods, the choice of competing peptide is also an important factor in determining the IC50 value. When two competing assays are performed using the same detection method and same experimental conditions but different competing peptides, the relative binding affinity of the two competing peptides will affect the resulting binding affinity of the target peptide. If the first competing peptide is a better binder than the second competing peptide, there will be a difference in the resulting IC50 values. This may be the reason why two studies may arrive at different IC50 values even though all the other experimental conditions appear to be the same.
It is possible to include the implicit values if the scoring function is classification-based, where input is classified into weak, intermediate, or strong binders. Although this will inevitably reduce the information used to deduce the scoring function and reduce the accuracy of the scoring function, using a classification-based approach will allow more input data. This may compensate for the loss of specific binding information. Unfortunately, it is impossible to resolve the discrepancy introduced by using different competing peptides; prior knowledge will be required to be able to choose one IC50 value over another.
In conclusion, the present study implemented the Fresno scoring function using open source and free software. We have also looked at some of the obstacles faced by researchers in the attempt to develop free energy scoring functions. Currently, sequence-based methods exploring binding motif or utilising artificial intelligence are leading the race to accurately predict peptide binding affinity. However, sequence-based methods do not face the same obstacles as structure-based methods, as they do not utilise structural information and tend to be classification based. While structure-based methods are not so far behind, it is foreseeable that these obstacles need to be addressed before the performance of structure-based methods can be on par with the sequence-based methods.
Materials and Methods
Preparation of MHC structures
A list of experimentally determined structures of the MHC-peptide complex for alleles HLA-A*0201, HLA-DRB1*0101, HLA-DRB1*0401, and HLA-DRB1*1501 (Table 5) were collected from the Protein Data Bank [28]. For analyses where only one structure was used the most recent structure with best resolution was used. Structures, referred to as the Madden structures hereafter, used by Rognan et al. in their study (1HHG, 1HHH, 1HHI, 1HHJ, 1HHK) were also obtained from the PDB [29].
Table 5. Experimental crystal structures used in the present study.
| Allele | PDB ID |
| HLA-A*0201 (Madden structures)* | 1HHG, 1HHH, 1HHI, 1HHJ,1HHK [29] |
| HLA-A*0201 | 1AKJ, 1B0R, 1OGA, 1P7Q, 1QEW, 1S9W, 1TVH, 2BNR, 2BNQ, 2F53, 2F54, 2GTW, 2GT9, 2GUO, 2P5E, 2P5W, 2PYE, 2X4U, 2X4R, 3GSN, 3GSO [27], [47]–[57] |
| HLA-DRB1*0101 | 1FYT [58] |
| HLA-DRB1*0401 | 1J8H [59] |
| HLA-DRB1*1501 | 1YMM, 1BX2 [60]–[61] |
The Madden structures were the five structures used in the original Fresno study.
Each crystal structure gives the positional information of the atoms of the MHC molecule and a peptide of particular sequence bound to the MHC molecule. In order to study the binding affinity of other peptides, the structure of a new peptide, bound to the same MHC molecule, is determined from the existing structure by using the same positions for the backbone atoms and rebuilding the side chains in the context of the MHC molecule. In the present study, the side chain rebuilding was performed using SCWRL 4 [26]. SCWRL 4 preserves the positions of the backbone atoms for the new peptide. It then attempts to predict the positions of the side-chain atoms for the new peptide while considering steric effects of the surrounding framework: in this case, the MHC molecule. Once a structure with the new peptide was constructed, hydrogen atoms were added using MolProbity 3.14 [30].
Preparation of peptide binding data
When the concentration of the binding peptide is sufficiently low, the dissociation constant can be represented by the inhibitory concentration (IC50): the concentration of inhibitor required to halve the level of binding of the substrate to the enzyme in a competitive assay. The free energy of binding can be calculated from the experimental temperature in Kelvin (T), the IC50 value, and the gas constant (R) according to equation 1.
| (1) |
A list of peptides with known binding affinity was extracted from the AntiJen database for each allele [31]–[32]. The AntiJen database contains experimental binding data for peptides known to bind to MHC molecules. Only peptides with the same length as the peptide in the reference crystal structure were used; typically, these were nine amino acids long. Inconsistencies or implicit values in the data set, such as multiple IC50 values for individual peptides due to different experimental settings, were resolved by manual reference to the original citations. If there is inexplicable discrepancy, the peptides in question were excluded from the analysis. The experimental data for the five structures used in Rognan et al. were taken from their original publication [33].
Calculation of the Scoring Function Terms
The Fresno free energy scoring function was previously described by Rognan et al. [24]. Briefly, there are five terms used by the Fresno scoring function (equation 2). Each term attempts to model the contribution to the binding energy made by a different atomic interaction.
| (2) |
The first three terms describe the energies associated with hydrogen bonds (HB), the interactions between lipophilic atoms in the MHC molecule and the peptide (LIPO), and the unfavourable interactions between polar and lipophilic atoms (BP). The rotational term (ROT) estimates the loss of energy due to the freezing of the rotatable bonds of the peptide upon binding. Lastly, the desolvation term (DESOLV) considers the energies required to solvate the MHC molecule, the peptide, and the MHC-peptide complex. The equations and related details for calculating each term are given in Rognan et al [24] and Eldrige et al. [34].
Calculation of the Regression Coefficients
The HB, LIPO, ROT, and BP terms were calculated using an adaptation of the Fresno scoring function developed in PERL. If the reference PDB file contained a bound T-cell receptor, this part of the file was removed prior to the analysis. The DESOLV term for all peptides was estimated using the DelPhi program [35]–[36]. The parameters were similar to those used by Rognan et al. The only difference being the atomic radii and the charges. Atomic radii and charges used in this study were taken from PARSE [37].
The values of all terms and the experimental free energy are used in a partial least square analysis using R and the pls package [38]–[39]. Regression coefficients were derived for each term and optimised for each dataset. The theoretical free energy was predicted using these regression coefficients. The cross-validation correlation score (q2) and standard error of prediction (Spress) were calculated from leave-one-out cross-validation using the built-in functions (R2 and RMSEP) from the pls package. q2 estimates the accuracy of the model and Spress estimates the error rate of the prediction. Thus a good prediction model should have q2 close to 1 and a low Spress value.
Correlation Studies
The difference between protein structures was calculated in terms of root mean square deviation (RMSD) scores. Structures were superimposed and the RMSD values were calculated using the Discovery Studio Visualizer 3.0 by Accelrys. Correlation between the q2 values and the RMSD scores were analysed using Spearman's rank correlation in R. Correlation between the q2 values and the average resolution of structures, obtained from the PDB structure files, was calculated similarly.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: WWPL receives an Australian Postgraduate Award from the Australian Government, Department of Innovation, Industry, Science, and Research (http://www.innovation.gov.au/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hafler DA, Slavik JM, Anderson DE, O'Connor KC, Jager PD, et al. Multiple sclerosis. Immunological Reviews. 2005;204:208–231. doi: 10.1111/j.0105-2896.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- 2.Westall FC. Molecular mimicry revisited: gut bacteria and multiple sclerosis. Journal of Clinical Microbiology. 2006;44:2099–2104. doi: 10.1128/JCM.02532-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serafini B, Rosicarelli B, Franciotta D, Magliozzi R, Reynolds R, et al. Dysregulated Epstein-Barr virus infection in the multiple sclerosis brain. Journal of Experimental Medicine. 2007;204:2899–2912. doi: 10.1084/jem.20071030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang HLE, Jacobsen H, Ikemizu S, Andersson C, Harlos K, et al. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nature Immunology. 2002;3:940–943. doi: 10.1038/ni835. [DOI] [PubMed] [Google Scholar]
- 5.Levin MC, Lee SM, Kalume F, Morcos Y, Dohan FCJ, et al. Autoimmunity due to molecular mimicry as a cause of neurological disease. Nature Medicine. 2002;8:509–513. doi: 10.1038/nm0502-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, et al. Risk alleles for multiple sclerosis identified by a genomewide study. The New England Journal Of Medicine. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, et al. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Research. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordner AJ, Mittelmann HD. Prediction of the binding affinities of peptides to class II MHC using a regularized thermodynamic model. BMC Bioinformatics. 2010;11:41. doi: 10.1186/1471-2105-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P, Sidney J, Kim Y, Sette A, Lund O, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lata S, Bhasin M, Raghava GPS. Application of machine learning techniques in predicting MHC binders. Methods in Molecular Biology. 2007;409:201–215. doi: 10.1007/978-1-60327-118-9_14. [DOI] [PubMed] [Google Scholar]
- 13.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Research. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundegaard C, Hoof I, Lund O, Nielsen M. State of the art and challenges in sequence based T-cell epitope prediction. Immunome Research. 2010;6(Suppl 2):S3. doi: 10.1186/1745-7580-6-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattotuwagama CK, Doytchinova IA, Flower DR. Toward the prediction of class I and II mouse major histocompatibility complex-peptide-binding affinity: in silico bioinformatic step-by-step guide using quantitative structure-activity relationships. Methods in Molecular Biology. 2007;409:227–245. doi: 10.1007/978-1-60327-118-9_16. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Wu S, Chen Z, Ye N, Yang S, et al. Structural parameterization and functional prediction of antigenic polypeptome sequences with biological activity through quantitative sequence-activity models (QSAM) by molecular electronegativity edge-distance vector (VMED). Science in China Series C: Life Sciences. 2007;50:706–716. doi: 10.1007/s11427-007-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimitrov I, Garnev P, Flower DR, Doytchinova I. Peptide binding to the HLA-DRB1 supertype: a proteochemometrics analysis. European Journal of Medicinal Chemistry. 2010;45:236–243. doi: 10.1016/j.ejmech.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 18.Kumar N, Mohanty D. MODPROPEP: A program for knowledge-based modeling of protein–peptide complexes. Nucleic Acids Research. 2007;35:W549–W555. doi: 10.1093/nar/gkm266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiewe AJ, Haworth IS. Structure-based prediction of MHC-peptide association: algorithm comparison and application to cancer vaccine design. Journal of Molecular Graphics and Modelling. 2007;26:667–675. doi: 10.1016/j.jmgm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Aldulaijan S, Platts JA. Theoretical prediction of a peptide binding to major histocompatibility complex II. Journal of Molecular Graphics and Modelling. 2010;29:240–245. doi: 10.1016/j.jmgm.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Bordner AJ. Towards universal structure-based prediction of class II MHC epitopes for diverse allotypes. PLoS ONE. 2010;5:e14383. doi: 10.1371/journal.pone.0014383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Science. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao WW, Arthur JW. Predicting peptide binding to Major Histocompatibility Complex molecules. Autoimmunity Reviews. 2011 doi: 10.1016/j.autrev.2011.02.003. in publication. [DOI] [PubMed] [Google Scholar]
- 24.Rognan D, Lauemoller SL, Holm A, Buus S, Tschinke V. Predicting binding affinities of protein ligands from three-dimensional models: application to peptide binding to class I major histocompatibility proteins. Journal of Medicinal Chemistry. 1999;42:4650–4658. doi: 10.1021/jm9910775. [DOI] [PubMed] [Google Scholar]
- 25.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Research. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krivov GG, Shapovalov MV., Jr. RLD Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borbulevych OY, Insaidoo FK, Baxter TK, Powell DJ, Johnson LA, et al. Structures of MART-1(26/27–35) peptide/HLA-A2 complexes reveal a remarkable disconnect between antigen structural homology and T cell recognition. Journal of Molecular Biology. 2007;372:1123–1136. doi: 10.1016/j.jmb.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. The Protein Data Bank Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madden DR, Garboczi DN, Wiley DC. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell. 1993;75:693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- 30.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain. Journal of Molecular Biology. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 31.Blythe MJ, Doytchinova IA, Flower DR. JenPep: A database of quantitative functional peptide data for immunology. Bioinformatics. 2002;18:434–439. doi: 10.1093/bioinformatics/18.3.434. [DOI] [PubMed] [Google Scholar]
- 32.McSparron H, Blythe MJ, Zygouri C, Doytchinova IA, Flower DR. JenPep: a novel computational information resource for immunobiology and vaccinology. Journal of Chemical Information and Computer Science. 2003;43:1276–1287. doi: 10.1021/ci030461e. [DOI] [PubMed] [Google Scholar]
- 33.Altuvia Y, Schueler O, Margalit H. Ranking potential binding peptides to MHC molecules by a computational threading approach. Journal of Molecular Biology. 1995;249:244–250. doi: 10.1006/jmbi.1995.0293. [DOI] [PubMed] [Google Scholar]
- 34.Eldridge M, Murray CW, Auton TA, Paolini GV, Lee RP. Empirical scoring functions: I. the development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. Journal of Computer-Aided Molecular Design. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 35.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 36.Rocchia W, Alexov E, Honig B. Extending the applicability of the nonlinear Poisson-Boltzmann equation: Multiple dielectric constants and multivalent Ions. Journal of Physical Chemistry B. 2001;105:6507–6514. [Google Scholar]
- 37.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. Journal of Physical Chemistry. 1994;98:1978–1988. [Google Scholar]
- 38.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 39.Mevik B-H, Wehrens R. The pls package: Principal component and partial least squares regression in R. Journal of Statistical Software. 2007;18:1–24. [Google Scholar]
- 40.Texier C, Pouvelle S, Busson M, Herve M, Charron D, et al. HLA-DR restricted peptide candidates for bee venom immunotherapy. Journal of Immunology. 2000;164:3177–3184. doi: 10.4049/jimmunol.164.6.3177. [DOI] [PubMed] [Google Scholar]
- 41.Wucherpfennig KW, Sette A, Southwood S, Oseroff C, Matsui M, et al. Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. Journal of Experimental Medicine. 1994;179:279–290. doi: 10.1084/jem.179.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Elsas A, van der Burg SH, van der Minne CE, Borghi M, Mourer JS, et al. Peptide-pulsed dendritic cells induce tumoricidal cytotoxic T lymphocytes from healthy donors against stably HLA-A*0201-binding peptides from the Melan-A/MART-1 self antigen. European Journal of Immunology. 1996;26:1683–1689. doi: 10.1002/eji.1830260803. [DOI] [PubMed] [Google Scholar]
- 43.Wilson CC, McKinney D, Anders M, MaWhinney S, Forster J, et al. Development of a DNA vaccine designed to induce cytotoxic T lymphocyte responses to multiple conserved epitopes in HIV-1. Journal of Immunology. 2003;171:5611–5623. doi: 10.4049/jimmunol.171.10.5611. [DOI] [PubMed] [Google Scholar]
- 44.Dionne SO, Smith MH, Marincola FM, Lake DF. Functional characterization of CTL against gp100 altered peptide ligands. Cancer Immunology and Immunotherapy. 2003;52:199–206. doi: 10.1007/s00262-002-0358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solache A, Morgan CL, Dodi AI, Morte C, Scott I, et al. Identification of three HLA-A*0201-restricted cytotoxic T cell epitopes in the cytomegalovirus protein pp65 that are conserved between eight strains of the virus. Journal of Immunology. 1999;163:5512–5518. [PubMed] [Google Scholar]
- 46.Zeng G, Li Y, El-Gamil M, Sidney J, Sette A, et al. Generation of NY-ESO-1-specific CD4+ and CD8+ T cells by a single peptide with dual MHC class I and class II specificities: a new strategy for vaccine design. Cancer Research. 2002;62:3630–3635. [PMC free article] [PubMed] [Google Scholar]
- 47.Gras S, Saulquin X, Reiser JB, Debeaupuis E, Echasserieau K, et al. Structural bases for the affinity-driven selection of a public TCR against a dominant human Cytomegalovirus epitope. Journal of Immunology. 2009;183:430–437. doi: 10.4049/jimmunol.0900556. [DOI] [PubMed] [Google Scholar]
- 48.Celie PHN, Toebes M, Rodenko B, Ovaa H, Perrakis A, et al. UV-Induced ligand exchange in MHC class I protein crystals. Journal of the American Chemical Society. 2009;131:12298–12304. doi: 10.1021/ja9037559. [DOI] [PubMed] [Google Scholar]
- 49.Borbulevych OY, Baxter TK, Yu ZY, Restifo NP, Baker BM. Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: Implications for vaccine design. Journal of Immunology. 2005;174:4812–4820. doi: 10.4049/jimmunol.174.8.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb AI, Dunstone MA, Chen WS, Aguilar MI, Chen QY, et al. Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. Journal of Biological Chemistry. 2004;279:23438–23446. doi: 10.1074/jbc.M314066200. [DOI] [PubMed] [Google Scholar]
- 51.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nature Immunology. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 52.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nature Immunology. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 53.Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, et al. Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2. Nature. 1997;387:630–634. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 54.Bouvier M, Guo HC, Smith KJ, Wiley DC. Crystal structures of HLA-A*0201 complexed with antigenic peptides with either the amino- or carboxyl-terminal group substituted by a methyl group. Proteins. 1998;33:97–106. doi: 10.1002/(sici)1097-0134(19981001)33:1<97::aid-prot9>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 55.Chen JL, Stewart-Jones G, Bossi G, Lissin NM, Wooldridge L, et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. Journal of Experimental Medicine. 2005;201:1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sami M, Rizkallah PJ, Dunn S, Molloy P, Moysey R, et al. Crystal structures of high affinity human T-cell receptors bound to peptide major histocompatibility complex reveal native diagonal binding geometry. Protein Engineering, Design & Selection. 2007;20:397–403. doi: 10.1093/protein/gzm033. [DOI] [PubMed] [Google Scholar]
- 57.Dunn SM, Rizkallah PJ, Baston E, Mahon T, Cameron B, et al. Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity. Protein Science. 2006;15:710–721. doi: 10.1110/ps.051936406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hennecke J, Carfi A, Wiley DC. Structure of a covalently stabilized complex of a human alpha beta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO Journal. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hennecke J, Wiley DC. Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, Influenza Hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity. Journal of Experimental Medicine. 2002;195:571–581. doi: 10.1084/jem.20011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith KJ, Pyrdol J, Gauthier L, Wiley DC, Wucherpfennig KW. Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein. Journal of Experimental Medicine. 1998;188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nature Immunology. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]