Abstract
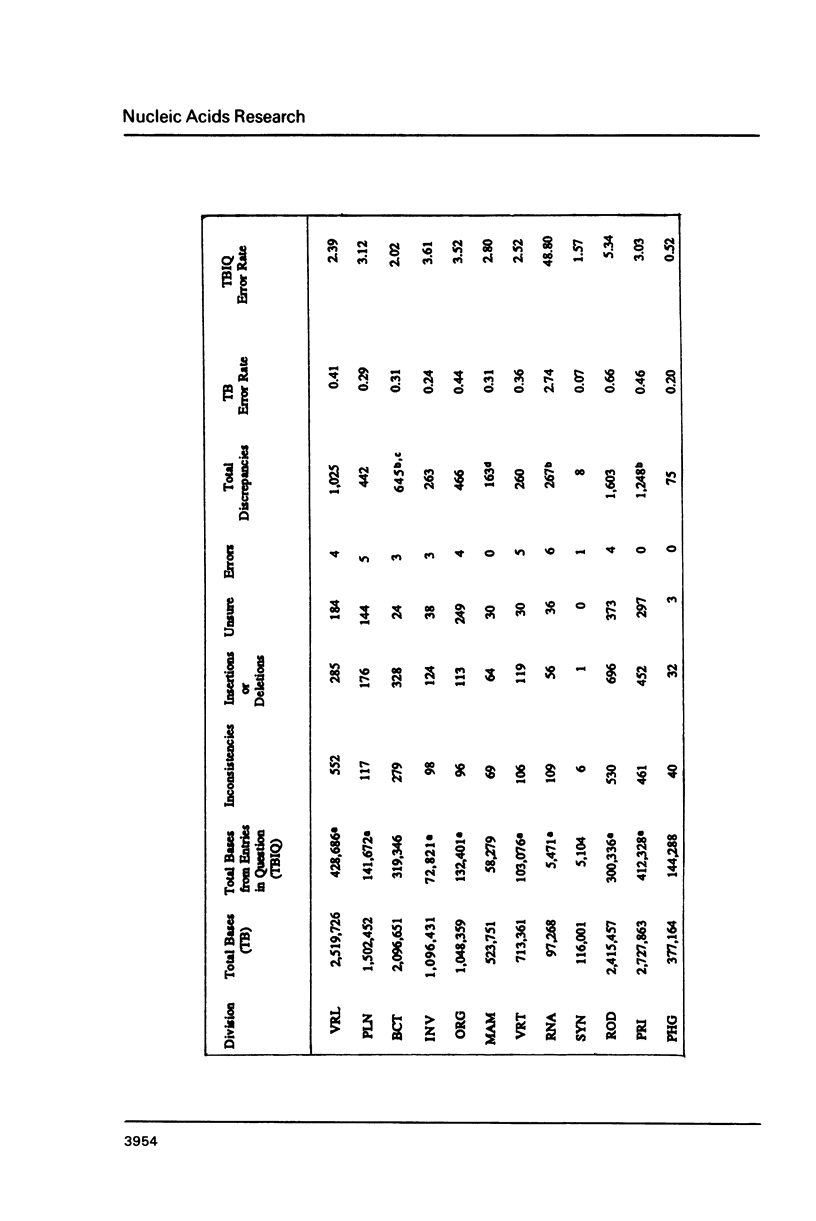
The accuracy of nucleic acid sequence data interpretation was determined by assessing and quantifying the discrepancies reported in the GenBank database. This permitted the calculation of an Error Rate (ER) for nucleic acid sequence determination. If one assumes that most entries (TB, Total Bases) were independently verified or those without reported discrepancies were correct, the ER is 0.368 errors per 1000 bases. However, if one assumes that only those sequences with reported discrepancies (TBIQ, Total Bases from entries In Question) were verified and are thus correct, the ER is 2.887 errors per 1000 bases. This establishes the first set of limit boundaries of the ER for sequence interpretation and sequence errors within the GenBank database and provides the foundation for future assessments and the monitoring of sequence data accumulation. In addition, the ER measure provides a basis to evaluate the efficiency and merit of present and future automated nucleic acid sequencing technologies which will have a direct impact upon the final outcome of the "Human Genome Initiative".
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron G. N. The EMBL data library. Nucleic Acids Res. 1988 Mar 11;16(5):1865–1867. doi: 10.1093/nar/16.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J. K., Green D. K., Southern E. M. Automatic reading of DNA sequencing gel autoradiographs using a large format digital scanner. Nucleic Acids Res. 1986 Jan 10;14(1):417–424. doi: 10.1093/nar/14.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan T. P., Krawetz S. A. Computer video acquisition and analysis system for biological data. Comput Appl Biosci. 1988 Mar;4(1):203–210. doi: 10.1093/bioinformatics/4.1.203. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Loeb L. A. Fidelity of mammalian DNA polymerases. Science. 1981 Aug 14;213(4509):765–767. doi: 10.1126/science.6454965. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall K. R., Kunkel T. A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988 Aug 9;27(16):6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]


