Abstract
Across the evolutionary spectrum, living organisms depend on high-fidelity DNA replication and recombination mechanisms to maintain genome stability and thus to avoid mutation and disease. The repair of severe lesions in the DNA such as double-strand breaks or stalled replication forks requires the coordinated activities of both the homologous recombination and DNA replication machineries. Growing evidence indicates that so-called “accessory proteins” in both systems are essential for the effective coupling of recombination to replication that is necessary to restore genome integrity following severe DNA damage. In this article we review the major processes of homology-directed DNA repair (HDR), including the double Holliday Junction (dHJ), synthesis-dependent strand annealing (SDSA), break-induced replication (BIR), and error-free lesion bypass pathways. Each of these pathways involves the coupling of a homologous recombination event to DNA synthesis. We highlight two major classes of accessory proteins in recombination and replication that facilitate HDR: Recombination mediator proteins exemplified by T4 UvsY, S. cerevisiae Rad52, and human BRCA2; and DNA helicases/translocases exemplified by T4 Gp41/Gp59, E. coli DnaB and PriA, and eukaryotic Mcm2-7, Rad54, and Mph1. We illustrate how these factors help to direct the flow of DNA and protein-DNA intermediates on the pathway from a double-strand break or stalled replication fork to a high-fidelity recombination-dependent replication apparatus that can accurately repair the damage.
Keywords: Recombination, Replication, DNA repair, Genome stability, Recombinase, Mediator, Helicase
Maintaining the integrity of the genome is critical to maintaining proper cell function and survival. The genome can be damaged from both exogenous and endogenous agents. Damage to DNA can interfere with DNA replication, which is required for cell division, growth and development of organisms. DNA damage can also affect gene expression and thus the physiology of non-replicating cells. Cells have evolved many strategies to avoid DNA damage and to repair damage when it occurs. A particularly deleterious form of DNA damage is the double-strand break (DSB). Double-strand breaks can occur as a result of ionizing radiation, chemical insults, or errors in DNA metabolism. DSBs if not repaired can hamper replication and transcription and lead either to gross mutations or to apoptosis.
Most cells have two primary pathways used to repair DSBs [Kass and Jasin, 2010]. The first is non-homologous end joining (NHEJ) which mainly functions to ligate two double-stranded ends of DNA together. Repair of DSBs by NHEJ allows replication to occur and avoids possible apoptosis as a result of accumulating stalled replication forks, however it may also lead to loss of critical genetic information and to translocations. Cells can also repair DSBs by homologous recombination (HR) (Figure 1). This pathway uses homologous sequences in intact DNA (potentially a sister chromatid or homologous chromosome) as a template to repair the damaged portion of the genome. This type of DSB repair is referred to as homology-directed repair or HDR. HDR is essentially error-free, therefore it can repair a DSB without loss of genetic information although it can result in gene conversion [Haber, 1999].
Figure 1.
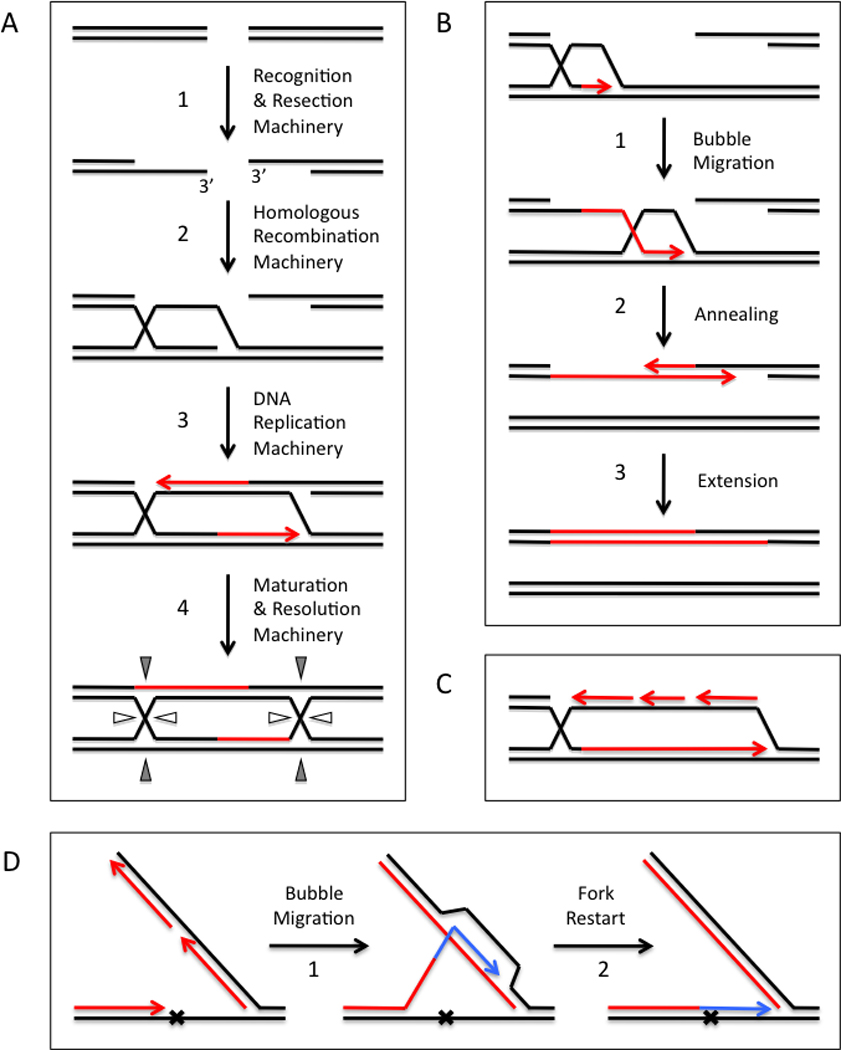
High-fidelity, homology-directed repair of DNA double-strand breaks and lesions requires the coupling of homologous recombination events to DNA synthesis. (A) A version of the double Holliday Junction (dHJ) model involves DNA synthesis that initiates from both ends of the DSB. Newly synthesized DNA is shown in red. Step 1- Double-strand breaks are nucleolytically resected to generate DNA ends with 3’ ssDNA tails. Recombination proteins assemble on the exposed ssDNA. Step 2 – A search for homology ensues in which the first end is paired with its homologous sequence in an intact dsDNA molecule. Strand invasion creates a D-loop intermediate. Step 3 - The 3’-OH of the invading end primes DNA synthesis which extends the D-loop in the 5’ → 3’ direction. This exposes ssDNA that is complementary to the second end, which is captured by annealing and extended by DNA synthesis. Step 4 – The events in Steps 1–3 generate a double Holliday Junction that can be resolved in different geometries to yield either crossover or non-crossover products. (B) Synthesis-dependent strand annealing (SDSA) also involves DNA synthesis that initiates from both ends of the DSB. Newly synthesized DNA is shown in red. Step 1 – Like dHJ, invasion of duplex by the first end initiates DNA synthesis. Step 2 – Unlike dHJ, the D-loop intermediate physically translocates along the dsDNA template, a process known as “bubble migration”. Bubble migration is driven by 5’ → 3’ DNA synthesis at the leading edge followed by branch migration at the trailing edge of the D-loop that displaces the extended first end from the template. Collapse of the migrating D-loop allows the first end to anneal to the second end. Step 3 – DNA synthesis extends both ends and fills in the gaps left over from the annealing reaction. Non-crossover products are generated in which both strands of the homologous dsDNA are conserved. (C) Break-induced replication (BIR) takes place when DNA synthesis initiates from only one end, as may occur when DSBs are generated in regions of incomplete homology. Newly synthesized DNA is shown in red. DNA synthesis is extensive and requires both leading and lagging strand replication machinery. In contrast, both dHJ and SDSA pathways require leading strand replication machinery but are independent of lagging strand machinery. (D) Error-free lesion bypass by a DNA replication fork involves intra-fork template switching by homologous recombination. Bypass of a lesion on the leading strand template is depicted. New DNA formed by semi-conservative and bubble migration synthesis reactions are shown in red and blue, respectively. When the replication fork stalls at a lesion, lagging strand synthesis is uncoupled from leading strand synthesis, allowing the synthesis of an Okazaki fragment that is homologous to the site of the lesion and its flanking sequences. Step 1 – Recombination promotes template switching from the damaged parental strand to the undamaged, homologous sequences present in the overlapping Okazaki fragment. Bubble migration synthesis extends the daughter strand beyond the site of damage. No mutation is introduced due to the templating mechanism of bubble migration synthesis. Step 2 – The extended daughter strand re-anneals to the damaged template. Extension of the 3’ end is no longer blocked by damage, which allows the replication fork to restart.
Homology-directed repair pathways
To initiate DSB repair by homologous recombination, DSBs are recognized and resected by nuclease complexes to generate DNA molecules with 3’ ssDNA tails (Figure 1A, Step 1). The single-stranded region of a resected DSB is recognized and bound by recombination proteins, leading to the assembly of a recombinase-ssDNA presynaptic filament (Figure 2). The filament catalyzes invasion of the ssDNA into the homologous dsDNA and displaces one of the strands, forming a D-loop structure in DNA containing the 3’ end of the invading ssDNA now annealed to its complement in the targeted dsDNA (Figure 1A, Step 2). The 3’ end is subsequently used as a primer by a DNA polymerase to synthesize DNA and allow for the repair to be completed. This coupling of a homologous recombination event to DNA synthesis is referred to as recombination-dependent replication (RDR), and it is a hallmark of all homology-directed repair of DNA double-strand breaks (Figure 1) [Haber, 1999].
Figure 2.
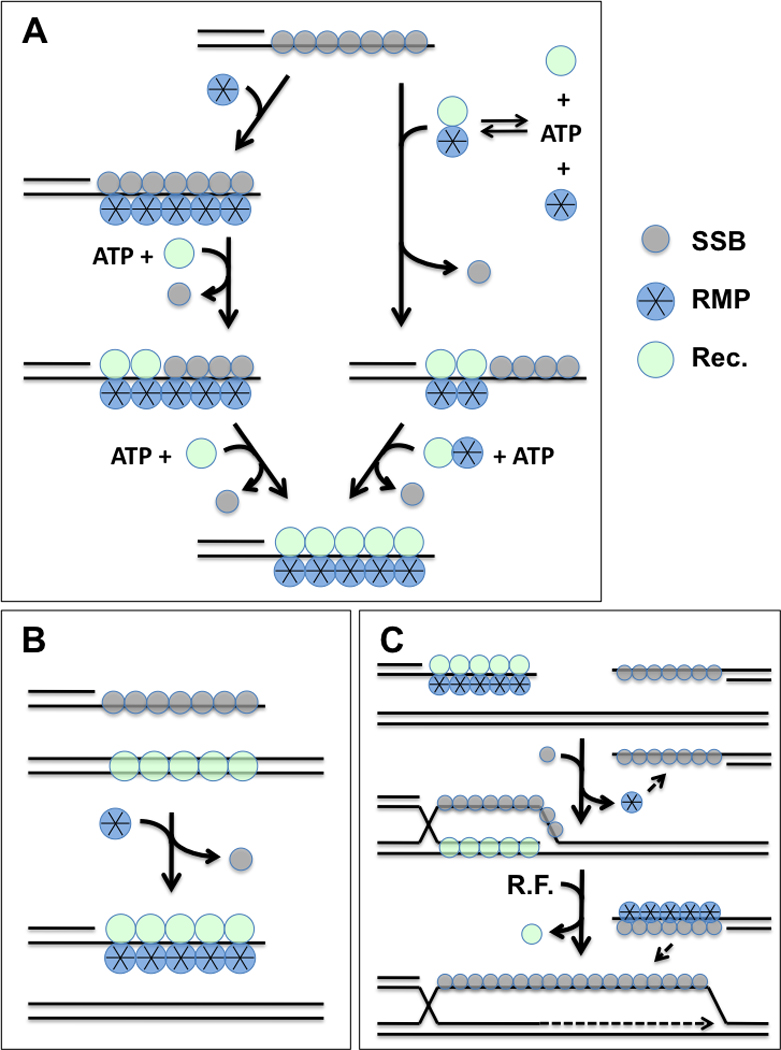
Role of recombination mediator proteins (RMP) in presynaptic filament assembly, stabilization, and second-end capture reactions during homologous recombination. “Rec.” denotes RecA/Rad51 family recombinase; SSB denotes ssDNA-binding protein; R.F. denotes replication factors. (A) Presynaptic filament assembly occurs on resected DSB ends covered with SSB and requires cognate RMP. Two possible mechanisms are depicted: Left, RMP binds to SSB-ssDNA and promotes SSB-recombinase exchange. Right, RMP binds to recombinase and the complex competes off SSB from ssDNA. Both mechanisms require ATP binding by recombinase. RMP stabilizes the resulting recombinase-ssDNA presynaptic filament. (B) RMP overcomes dsDNA inhibition of recombinase by selectively promoting filament assembly on ssDNA in the presence of excess heterologous dsDNA. (C) Following strand invasion by the presynaptic filament and D-loop extension by replication factors, RMP facilitates second end capture by promoting annealing between complementary ssDNA segments covered with SSB.
HDR events may occur by several different but related mechanisms depending on such factors as the degree of homology between the damaged and intact DNA molecules. Figure 1A shows the classic DSB repair model of Szostak et al. [Szostak et al., 1983], also known as the double Holliday Junction or dHJ model, in which both resected ends of the DSB are homologous to regions of the intact DNA molecule, and both 3’ ends are extended by semi-conservative DNA synthesis. Following strand invasion by the first ssDNA tail, DNA synthesis is primed from its 3’ end leading to replicative extension of the D-loop in the 5’ → 3’ direction (Figure 1A, Step 3). D-loop extension eventually exposes ssDNA on the displaced strand that is complementary to the second ssDNA tail, leading to second end capture by annealing. The annealed 3’ end primes DNA synthesis to fill in the gap. Ligation creates a double Holliday Junction (Figure 1A, Step 4) that can translocate by branch migration. Resolution by structure-specific endonucleases in different orientations leads to either crossover or non-crossover of flanking markers. The net result is that the structural integrity of the broken chromosome is restored by high-fidelity RDR events using the intact DNA molecule as template.
Synthesis-dependent strand annealing (SDSA) (Figure 1B) is closely related to the dHJ pathway but differs in some of the details of the replicative steps. Both mechanisms occur when the two ends of the resected DSB are homologous to the intact DNA molecule. The major difference is that in SDSA the extension of the first strand by RDR occurs within a translocating D-loop (Figure 1B, Step 1). As DNA synthesis extends the first strand at the leading edge of the D-loop, the first strand is simultaneously displaced at the trailing edge of the D-loop by branch migration (both in the 5’ → 3’ direction). This mode of recombination-dependent replication is referred to as “bubble migration” DNA synthesis because of the net movement of the D-loop “bubble” [Formosa and Alberts, 1986]. A unique feature of SDSA is that D-loop translocation causes replication to be conservative rather than semi-conservative, since the elongating first strand is displaced from its template strand by branch migration. Thus the intact duplex is conserved after transiently serving as template for the RDR process (Figure 1B, Step 1). Second end capture in SDSA occurs when D-loop translocation exposes ssDNA at the 3’ end of the extended first strand that is complementary to the second ssDNA tail. Following D-loop collapse, the two 3’ ends can anneal and prime DNA synthesis to fill the gaps (Figure 1B, Steps 2–3). Because of the conservative bubble migration process, newly synthesized DNA appears only in the repaired chromosome and there is no crossover of flanking markers (Figure 1B, Step 3). Alternatively, second end capture could occur by annealing to the migrating D-loop (not shown) which would lead to the formation of a double Holliday Junction and to crossover of flanking markers similar to dHJ.
A third type of HDR is break-induced replication (BIR), which occurs when only one end of the resected DSB is homologous to the intact dsDNA molecule (Figure 1C). In this pathway, strand invasion by the ssDNA tail initiates a semi-conservative DNA replication fork with both leading and lagging strand synthesis machineries [Lydeard et al., 2010]. This distinguishes BIR from dHJ and SDSA, because the latter two pathways use only the leading strand synthesis machinery to extend both the first and second ends [Wang et al., 2004]. BIR may continue for many tens of kilobases leading to extensive copying of sequences from the intact DNA molecule.
HDR also provides an error-free mechanism for lesion bypass by replication forks (Figure 1D). This mechanism is distinct from translesion DNA synthesis reactions carried out by error-prone DNA polymerases [Livneh et al., 2010]. When leading strand replication stalls at a lesion in the template, a mechanism exists to uncouple lagging from leading strand synthesis so that an Okazaki fragment can be synthesized with sequences homologous to the site of the lesion and its flanking regions. Recombination causes the 3’ end of the lesion-stalled daughter strand to switch templates by invading the homologous portion of the lagging strand DNA. This initiates a bubble-migration synthesis reaction using the Okazaki fragment sequences as template (Figure 1D, Step 1). The daughter strand is extended past the site of the lesion, but no mutation is introduced since the homologous template is undamaged. The daughter strand eventually re-anneals to the damaged template, but the lesion has been bypassed so leading strand synthesis can restart (Figure 1D, Step 2).
Key proteins required for HDR
The pivotal event in all HDR pathways is homologous pairing and DNA strand exchange, which is catalyzed by recombinase enzymes of the RecA/Rad51 family. All recombinases share the property of forming presynaptic filaments on single-stranded DNA in the presence of ATP, which is a prerequisite for homologous pairing and strand exchange. The assembly, stability, and turnover of presynaptic filaments is closely regulated to ensure that recombination does not occur at inappropriate times or places in the genome, and that when it does occur the recombination events can be correctly coordinated with DNA synthesis and repair.
Recombination mediator proteins (RMPs) are an important class of proteins that promote the recombination activity of RecA/Rad51 recombinases [Beernink and Morrical, 1999]. These proteins facilitate and regulate homologous recombination by regulating recombinase access to ssDNA. The DNA synthesis portion of HDR also requires several accessory factors to allow for DNA polymerase function. Most importantly, the recombination proteins still complexed with D-loop intermediates must undergo disassembly to allow for correct assembly of the DNA synthesis machinery [Li and Heyer, 2009]. DNA motors such as helicases and translocases are essential for this processing, which involves the removal of recombination proteins from the invading 3’ end, recruitment of a DNA polymerase holoenzyme to the resulting primed template, and unwinding of the duplex ahead of the polymerase to allow DNA synthesis to occur. Like presynaptic filament assembly, specific loading mechanisms likely exist that regulate the access of DNA synthesis machinery to recombination intermediates, to ensure that DNA synthesis does not occur at inappropriate times or places in the genome.
This article will focus on some of the proteins working at the DNA level to control both the strand invasion and DNA synthesis portions of homology-directed DNA repair. A repeating theme in HDR is the occurrence at each stage of ATPase proteins belonging to the AAA+ and RecA superfamilies, including recombinases, DNA helicases and translocases, and clamp-loader components of DNA polymerase holoenzyme among others. These enzymes generally use the ATP binding and hydrolysis cycle to control conformational changes in the protein in order to do work. In HDR, each ATPase typically works in tandem with its accessory protein(s) to assemble on an appropriate DNA structure, and then to modify that structure so that it may be recognized by the next ATPase in the pathway working in tandem with its accessory protein(s). Thus HDR proceeds as a series of DNA hand-off transactions involving recombinase, motor, and replicase components and coordinated by accessory proteins of each machine. We begin with a detailed examination of the role of the recombination mediator proteins.
Recombination Mediator Proteins
To repair a DSB using homologous recombination, the site must first be recognized and processed by an exonuclease to generate 3’ single-stranded DNA tails [Krogh and Symington, 2004]. The recombinase must then form a multimeric helical filament on the ssDNA. The filament then carries out a search for homologous regions of dsDNA in the sister chromosome. Several accessory proteins control access of the recombinase to the single-stranded DNA and regulate its function. Proteins that promote filament assembly fall into two general classes, single-stranded DNA binding proteins (SSBs) and recombination mediator proteins (RMPs) respectively. SSBs prevent the formation of secondary structure in ssDNA during various stages of DNA metabolism. Secondary structures in ssDNA are inhibitory to many DNA modifying enzymes including recombinases. In this way SSBs have a stimulatory effect on recombinases. However SSBs themselves also inhibit recombinase activity by blocking access to the ssDNA substrate. Recombination mediator proteins are a class of proteins that promote recombinase binding to SSB-covered ssDNA [Beernink and Morrical, 1999]. RMPs accomplish this by enhancing recombinase-ssDNA interactions while promoting SSB displacement (Figure 2A). Other presynaptic functions of RMPs include stabilization of presynaptic filaments, modulation of filament catalytic activities, and prevention of inappropriate filament formation on dsDNA (Figure 2B). Post-synaptically, certain RMPs appear to be important for the DNA annealing reactions involved in second end capture (Figure 2C). Examples of RMPs include the T4 UvsY protein, bacterial RecF, RecO, and RecR proteins, eukaryotic Rad52 and BRCA2 proteins, and eukaryotic Rad51 paralogs. Here we review the well-characterized UvsY and Rad52 proteins as well as summarize recent progress towards understanding the function of BRCA2.
T4 UvsY Protein
The homologous recombination system of Bacteriophage T4 is relatively simple but functionally conserved. The properties of this system have been recently reviewed [Liu and Morrical, 2010; Liu and Morrical, 2011]. The T4 core recombination machinery includes a recombinase, UvsX, a recombination mediator protein, UvsY, and a ssDNA-binding protein, Gp32. UvsX and UvsY are both required for recombination in vivo and under physiological salt concentrations in vitro. Phage lacking functional UvsY exhibit a small plaque phenotype due to their inability to initiate replication via homologous recombination.
UvsY is the prototypical RMP [Beernink and Morrical, 1999]. Its major biochemical function is to promote the formation of UvsX-ssDNA presynaptic filaments while displacing Gp32 from ssDNA (Figure 2A) [Liu and Morrical, 2010; Liu and Morrical, 2011]. The resulting filaments catalyze homologous pairing and strand invasion by the ssDNA substrate into the dsDNA substrate (Figure 2C). The recombination mediator activity of UvsY hinges on its ability to alter ssDNA structure. UvsY has been shown to destabilize Gp32-ssDNA interactions and to stabilize UvsX-ssDNA interactions. The ssDNA binding activity of UvsY is required for both activities. UvsY forms a hexamer in solution. Studies suggest that UvsY binds to ssDNA much like a spindle, wrapping the DNA around the surface of the hexamer. This induces conformational changes in the ssDNA which may be responsible for destabilizing the Gp32-ssDNA interactions and stabilizing UvsX-ssDNA interactions. ssDNA wrapping by UvsY is responsible for its large affinity preference for ssDNA over dsDNA, which helps to promote productive UvsX filament assembly on ssDNA and to avoid unproductive binding to dsDNA (Figure 2B) [Xu et al., 2010].
Yeast Rad52 Protein
Eukaryotes have several recombination mediator proteins that collectively perform the same role as UvsY in the cell. The Rad52 protein of the budding yeast Saccharomyces cerevisiae is the best-studied and is conserved in higher eukaryotes including humans. Recently proteins which have some sequence homology and exhibit some functional similarities to Rad52 have been identified in bacteriophage [Ploquin et al., 2008].
Yeast Rad52 possesses ssDNA and dsDNA binding activity as well as the ability to detect homology and anneal complementary ssDNA strands. The N-terminal portion of the proteins contains a self-association domain as well as a DNA binding domain. The N-terminal domain alone can form an undecameric ring [Kagawa et al., 2002]. The central portion of the protein contains the RPA interaction domain. The C-terminal portion of the protein contains the Rad51 binding domain as well as another DNA binding domain. Full length Rad52 forms a ring shaped heptamer and is thought to wrap ssDNA around the exterior of the ring [Shinohara et al., 1998; Ranatunga et al., 2001].
In vitro Rad52 acts as a classical recombination mediator by promoting Rad51 presynaptic filament assembly and RPA displacement [reviews in Beernink and Morrical, 1999; San Filippo et al., 2008; Liu et al., 2011]. The mediator activity of Rad52 requires physical interactions with Rad51 and RPA. Recombination defects in Rad52 mutants can be ameliorated by overexpression of Rad51 indicating that the main function of Rad52 in DNA recombination is to load and stabilize Rad51 filaments.
In addition to acting as a recombination mediator protein, Rad52 possesses single-strand annealing activity. ssDNA is wrapped around the ringed Rad52 multimer and facilitates annealing with other complimentary ssDNAs including those bound by RPA [Wu et al., 2006; Nimonkar et al., 2009]. This single-strand annealing capacity of Rad52 is thought to contribute to homology directed repair in two potential ways. The first is that Rad52 may be important for capturing and annealing the second DNA end to the displaced strand of the D-loop structure during dHJ (Figure 1A) (Figure 2C). Rad52 may also participate in synthesis dependent strand annealing in which the second end is captured by annealing either to the extended first strand (Figure 1B) or to the displaced strand of the translocating D-loop (not shown).
S. cerevisiae rad52 mutants exhibit severe recombination and DNA repair deficiencies, consistent with the dual roles of Rad52 protein in presynaptic filament assembly and in DNA annealing [Krogh and Symington, 2004]. Elimination of RAD52 function in the human recombination system seems to have less significant effects than its elimination from the yeast recombination system. This may be because RAD52 functions slightly differently in the human system, i.e. it may be specialized for second end capture [McIlwraith and West, 2008; Grimme et al., 2010]. In addition the human system apparently has several other recombination mediator proteins (e.g. BRCA2 and potentially RAD51 paralogs) that may replace or overlap with the RMP functions of RAD52 [Liu et al., 2011].
Human BRCA2 Protein
Breast cancer susceptibility protein BRCA2 was identified when certain mutations were correlated with familial breast cancer [Wooster et al., 1995]. BRCA2-like proteins are only found in some fungi, vertebrates and higher eukaryotes. BRCA2-like proteins have not been identified in yeast. Knockout of the BRCA2 gene in mice is embryonic lethal [Sharan et al., 1997]. The human protein is 3418 amino acids in length. Until recently, the large size of the protein has made recombinant expression and in vitro study difficult [however see Jensen et al. 2010; Liu et al., 2010; Thorslund et al., 2010]. Sub-domains of the protein have been analyzed in vitro to determine the functional significance of this protein [Yang et al., 2005; Shivji et al., 2006]. BRCA2 has direct contact with RAD51 through 2 separate domains. The first of these is the BRC repeat domain. The human protein has 8 BRC repeats 6 of which have been found to interact with RAD51. The structure of the RAD51-BRC4 repeat complex indicates that a BRC repeat interacts with a RAD51 monomer in the absence of ssDNA [Pellegrini et al., 2002]. The BRC repeats of BRCA2 mimic the RAD51 motif used for homo-polymerization. It has been hypothesized that this domain of BRCA2 might facilitate the nucleation of RAD51 presynaptic filaments [Yang et al., 2005; Shivji et al., 2006], or conversely that BRCA2 could facilitate RAD51 filament disassembly [Pellegrini et al., 2002].
The second domain of BRCA2 that interacts with RAD51 is near the C-terminus. This domain interacts with the multimeric form of RAD51 through the ATPase domains of adjacent subunits, and is postulated to protect RAD51 filaments against disassembly promoted by the BRC repeats [Davies and Pellegrini, 2007]. BRCA2 also has interactions with ssDNA through its DNA binding domain. This consists of three oligonucleotide/oligosaccharide (O/B)-fold like domains and two helix-turn-helix domains.
BRCA2 has been found to modulate RAD51 activity and facilitate stand exchange through direct interaction [Carreira and Kowalczykowski, 2009]. BRC4 of BRCA2 has been found to support presynaptic filament formation by facilitating RAD51 interactions with ssDNA, destabilizing nucleation on dsDNA and slowing ATP hydrolysis by RAD51 thereby maintaining the ATP bound form of the protein with the highest affinity for ssDNA. Recent studies of the full-length protein [Liu et al., 2010; Jensen et al., 2010; Thorslund et al., 2010] corroborate the idea that BRCA2 behaves like a classic recombination mediator protein by promoting RAD51 filament assembly onto RPA-covered ssDNA, stabilizing presynaptic filaments, and avoiding filament nucleation on dsDNA. BRCA2 effects are modulated by other recombination/repair proteins such as DSS1, which further enhances BRCA2-dependent assembly of RAD51 onto RPA-covered ssDNA [Liu et al., 2010]. BRCA2 also promotes the specific assembly of RAD51 filaments on ssDNA over dsDNA [Jensen et al., 2010; Thorslund et al., 2010]. Thus BRCA2 appears to stimulate DNA strand exchange by a mechanism similar to classic recombination mediators such as T4 UvsY and yeast Rad52. Although a fungal ortholog of BRCA2, the Brh2 protein of Ustilago maydis, promotes single-strand annealing and second end capture [Mazloum and Holloman, 2009], full-length human BRCA2 does not anneal ssDNA molecules complexed with RPA, suggesting that it does not play a role in recombination processes that involve ssDNA annealing [Jensen et al., 2010; Thorslund et al., 2010]. It seems likely that in vertebrates and higher eukaryotes BRCA2 has replaced RAD52 as the major mediator protein regulating presynaptic filament assembly, whereas RAD52 performs specialized functions such as second end capture.
Common Themes in Recombination Mediator Protein Activity
In all systems, RMPs promote the timely assembly of presynaptic filaments on recombinagenic ssDNA that exists only transiently in the cell. They do so by overcoming the inhibitory effects of SSBs and excess dsDNA, thereby enforcing the specific assembly of recombinase filaments on ssDNA. Inducing structural changes in ssDNA through binding to multiple subunits or domains appears to be a common theme in the mechanisms of RMPs such as UvsY, Rad52, and BRCA2. By promoting the saturation of ssDNA with recombinase and the displacement of the SSB component, RMPs appear to play a key role in directing where subsequent events such as helicase assembly can occur on recombination intermediates. This has important effects on the trafficking of DNA during DSB repair, as we will see in the next section.
DNA Motors - Helicases and Translocases
DNA translocases and helicases are motor proteins that couple the energy derived from nucleoside triphosphate hydrolysis to unidirectional translocation along DNA. In the case of DNA helicases, translocation is coupled to the unwinding of double-helical DNA structures. In the case of DNA translocases, translocation occurs without unwinding. Both types of motor proteins play diverse roles in DNA metabolism, including homologous recombination and homology-directed repair. DNA motors can promote recombination/repair by stimulating branch migration, by helping to convert recombination intermediates into replication forks, and by promoting the replicative extension of D-loops [Colavito et al., 2010; Liu and Morrical, 2010]. DNA motors also play negative regulatory roles in recombination/repair through anti-recombination, e.g. by disrupting presynaptic filaments prior to strand invasion, or by resolving D-loops before they can be extended and converted into replication forks [Liu et al., 2011]. Here we will focus on DNA motors that promote the coupling of recombination to replication for purposes of DNA repair. Examples include the Gp41/Gp59 helicase complex of bacteriophage T4, DnaB and PriA helicases of E. coli, the eukaryotic Mcm2-7/Cdc45/GINS helicase complex, and the eukaryotic Rad54 DNA translocase.
Gp41 Helicase and Gp59 Helicase Loader in T4 Recombination-Dependent Replication
Bacteriophage T4 is a laboratory for studying the coordination of recombination and replication since the phage relies on RDR for most of its genomic DNA replication. Biochemical studies established that D-loops generated by UvsX, UvsY, and Gp32 can be extended by DNA synthesis, and that this coupling of recombination to replication requires the activity of a DNA helicase (Figure 3) [reviewed in Bleuit et al. 2001; Liu and Morrical, 2010]. The main helicase affecting this pathway in vivo is Gp41, which is the major replicative helicase of T4 phage.
Figure 3.
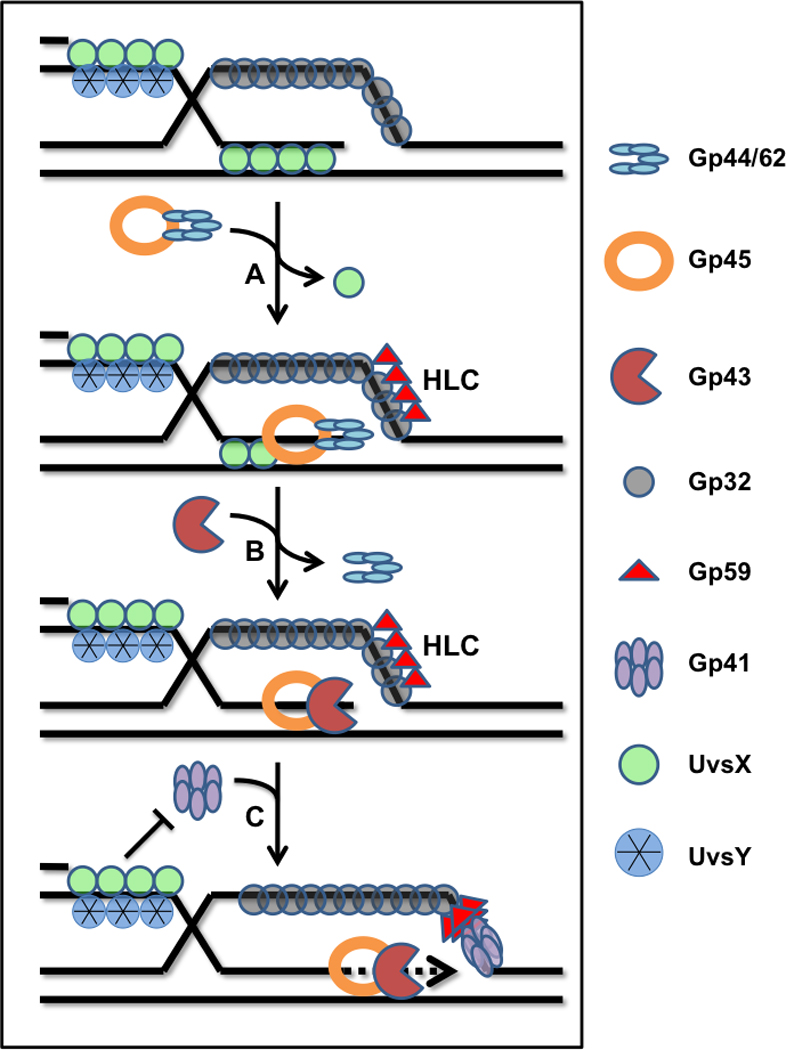
D-loop extension in the T4 recombination-dependent replication system is coupled to assembly of the replicative DNA helicase. The displaced strand of a D-loop formed by UvsX recombinase is coated with Gp32, the T4 SSB. (A) Helicase loading protein Gp59 is recruited to the displaced strand-Gp32 complex, where it forms a helicase loading complex (HLC). Spontaneous dissociation of UvsX from the heteroduplex allows clamp-loader (Gp44/62) to load a sliding clamp (Gp45) onto the 3’ end of the invading strand. Alternatively, UvsX may actively promote polymerase holoenzyme assembly. (B) Recruitment of Gp43 polymerase reconstitutes the leading strand DNA polymerase holoenzyme. However synthesis is blocked by the presence of HLC on the lagging strand ssDNA. (C) HLC recruits the replicative helicase, Gp41, which processively translocates 5’ → 3’ on the lagging strand ssDNA. Polymerase blockage is simultaneously relieved, allowing D-loop extension to occur. Subsequent recruitment of primase by Gp41 reconstitutes the full BIR mode of T4 recombination-dependent replication (not shown).
Gp41 is a member of the hexameric DNA helicase family. It translocates processively in a 5’ → 3’ direction on the lagging strand of replication forks. Gp41 activation requires initial binding to ssDNA, which induces hexamerization and significantly increases its rate of ATP hydrolysis. However, Gp41 has a low affinity for ssDNA in the presence of Gp32, the single stranded DNA binding protein [Morrical et al., 1994]. Consequently, Gp41 relies on Gp59, which functions as a helicase loader, to assist in its correct positioning at the replication fork. The loading of Gp41 by Gp59, which greatly enhances helicase unwinding activity, occurs on a variety of replication sites including origins, replication forks in regions distant from origins, and recombination intermediates [Jones et al., 2001]. In fact, Gp59 is absolutely required for D-loop extension in the presence of Gp41 helicase, recombination proteins, and T4 DNA polymerase holoenzyme (Figure 3).
The structural and biochemical properties of Gp59 have been reviewed [Jones et al., 2001]. Gp59 plays a critical role in the strand-specific loading of Gp41 onto D-loop intermediates generated by the actions of UvsX, UvsY, and Gp32. As shown in Figure 3, helicase loading potentially could occur at two different ssDNA sites within D-loops, resulting in two different outcomes. Gp41 loading on the displaced strand of the D-loop is compatible with recombination-dependent replication mechanisms in DSBR, SDSA, and BIR pathways since this strand becomes the lagging strand as strand-displacement synthesis extends the D-loop. Conversely, Gp41 loading on the invading strand would result in anti-recombination, since 5’→ 3’ translocation would resolve the D-loop and prevent its extension by RDR. Biochemical studies show that Gp59 in combination with UvsX, UvsY, and Gp32 enforces the strand-specific loading of Gp41 onto the displaced strand of the D-loop [Bleuit et al., 2001]. Gp59 binds tightly to branched DNA structures such as forks and D-loops, and also binds very tightly to Gp32. Gp59 targets Gp41 assembly onto clusters of Gp32 that are cooperatively bound to ssDNA. Gp32 clusters do not occur on the invading ssDNA, which is saturated by a stable filament of UvsX and UvsY. Therefore Gp41/Gp59 are excluded from this strand. Instead, Gp32 rapidly sequesters the displaced strand of the D-loop, where it forms a target for Gp59-dependent helicase assembly. Gp41 assembly on this strand leads to efficient reconstitution of the DNA synthesis apparatus that extends the D-loop (Figure 3). This in turn leads either to second end capture by a dHJ mechanism (Figure 1A), second end capture by a bubble migration or SDSA mechanism (Figure 1B), or to the reconstitution of the intact lagging strand synthesis apparatus for break-induced replication (Figure 1C) [Kreuzer, 2000].
An additional function of Gp59 in the BIR mode of recombination-dependent replication is the coordination of leading and lagging strand DNA synthesis [Dudas and Kreuzer, 2005; Xi et al. 2005]. Gp59 binding to fork or D-loop DNA blocks replication by the leading strand polymerase until Gp41 helicase is assembled on the lagging strand. This ensures that the initiation of leading strand synthesis does not occur until the lagging strand synthesis machinery is in place.
DnaB and PriA Helicases in E. coli Stable DNA Replication
Stable DNA Replication (SDR) is a recombination-dependent replication pathway in Escherichia coli [Masai et al., 1994]. SDR requires the activity of RecA recombinase but is independent of the replication initiator protein DnaA and of the chromosomal origin OriC. SDR is important for replication fork restart and for the extensive BIR that initiates from RecA-catalyzed strand invasion events (D-loop or R-loop formation) when origin-dependent replication is compromised. A key event in SDR is the loading of DnaB, the replicative 5’ → 3’ helicase, onto the displaced strand of the D- or R-loop to generate a replication fork. This loading requires elements of the primosome assembly apparatus including PriA, which is itself a 3’ → 5’ helicase. DnaB together with DnaG primase reconstitutes the E. coli primosome which supports extensive leading + lagging strand synthesis in a recombination-initiated mode of replication, similar to Gp41/Gp59-dependent BIR in the T4 system.
The role of PriA in DnaB helicase loading during E. coli SDR is intriguing. PriA contains both a helicase/motor domain with 3’ → 5’ translocation activity, and a domain that specifically recognizes D-loop and fork DNA structures [Liu and Marians, 1999]. Through its structure-specific DNA binding activity PriA initiates a series of recruitments leading to the assembly of DnaB on the displaced/lagging strand of the D-loop/replication fork. The helicase or ssDNA translocation activity of PriA also appears to be important for SDR [Tanaka et al., 2003]. Possible roles include: Melting of ssDNA secondary in the invading or displaced strand (to promote strand invasion or primosome assembly); Participation with DnaB in duplex unwinding at the fork, by simultaneously translocating on the opposite strand; or, translocation to facilitate loading of a second primosome necessary for bidirectional replication.
Mcm2-7 and Associates in Eukaryotic BIR
Break-induced replication in yeast (S. cerevisiae) requires both leading and lagging strand DNA synthesis machinery including the replicative DNA helicase (Mcm2-7, Cdc45, GINS) [Lydeard et al., 2010]. A reasonable model is that assembly of the helicase apparatus on the displaced strand of D-loops, which likely depends on Cdt1 and Cdc7 kinase, drives the extensive leading + lagging strand synthesis that is observed during BIR, reminiscent of the T4 model and the role of Gp41/Gp59. In contrast, dHJ and SDSA processes in yeast appear to require only the leading strand replication apparatus and are not dependent on Mcm2-7 [Wang et al., 2004]. This leaves open the question of what helicase, if any, drives D-loop extension in these pathways. One possibility is that no helicase is required for D-loop extension, based on the observation that DNA polymerase δ can carry out significant strand displacement synthesis in the absence of Mcm2-7, which might be sufficient to promote D-loop extension and second end capture. A second possibility is that a yet-to-be-identified 5’ → 3’ helicase activity substitutes for Mcm2-7 during D-loop extension. Additional possibilities are that a DNA translocase such as Rad54, or one of a suite of 3’ → 5’ helicase activities are required to promote D-loop extension (see below).
Multiple Functions of Eukaryotic Rad54 DNA Translocase
Rad54 has been called the “Swiss army knife of recombination” because of its multifaceted roles in promoting DNA strand exchange by Rad51 [Heyer et al., 2006]. A member of the Swi2/Snf2 family of chromatin remodeling factors, Rad54 also exhibits ATP-dependent DNA translocase activity in vitro. Yeast have two Rad54 like proteins, Rad54 and Rdh54. Mammals also have two, Rad54 and Rad54B. The yeast Rad54 crystal structure has been solved and contains many motifs typical of chromatin remodeling and helicase proteins [Thoma et al., 2005]. Rad54 exhibits typical recombination mediator activity in vitro much like Rad52 in that it stimulates Rad51 binding to ssDNA in the presence of RPA [Wolner et al., 2003]. Rad54 also stimulates joint molecule formation in vitro and requires specific contacts with Rad51 [Solinger et al., 2001]. After joint molecule formation, Rad54 increases the rate of branch migration in an ATP dependent fashion [Solinger and Heyer, 2001; Mazina and Mazin, 2008]. Rad54 also exhibits chromatin remodeling activity and is required for D-loop formation by Rad51 on nucleosomal substrates [Sinha and Peterson, 2008]. The human protein has been found to exhibit many of the properties found in the yeast protein. In addition, human Rad54 mutants with altered properties have been identified in cancer cells [Zhang et al., 2007].
The DNA translocation activity of Rad54 has been shown to disrupt Rad51 filaments bound to double-stranded DNA [Solinger et al., 2002]. Filament disruption requires specific Rad54-Rad51 protein-protein interactions, as well as ATP hydrolysis by both Rad54 and Rad51. Two physiological roles have been proposed for Rad54-catalyzed depolymerization of Rad51-dsDNA: First, this activity is important to reverse inappropriate Rad51-dsDNA interactions that inhibit DNA strand exchange. Rad51 must assemble into presynaptic filaments on ssDNA in order to carry out recombination. Paradoxically, Rad51 has substantial affinity for dsDNA, which leads to inhibition of DNA strand exchange by excess non-homologous dsDNA, as would normally occur in the cell. Rad54 functions in part to disrupt non-productive Rad51-dsDNA interactions, so that Rad51 subunits can be recruited into a productive presynaptic filament on dsDNA.
Second, Rad54 has been shown to displace Rad51 from the newly formed heteroduplex DNA in the D-loop following strand invasion, in a reaction that requires species-specific interactions between Rad54 and Rad51 as well as the ATPase activity of Rad54 [Li and Heyer, 2009]. This activity is thought to be important for exposing the 3’ end of the invading strand to allow for the assembly of the leading strand DNA polymerase (Figure 4). In fact, a recent study demonstrated that Rad54’s DNA translocase activity is necessary for the loading of DNA polymerase δ onto D-loops generated by Rad51 in the presence of RPA [Li et al., 2009]. This reaction also requires PCNA and RFC in addition to Rad51, RPA, Rad54, and Pol δ. Therefore Rad54 promotes the assembly of the intact, processive leading strand DNA polymerase at Rad51-generated D-loops, consistent with genetic models of dHJ, SDSA, and BIR in yeast [Wang et al., 2004; Lydeard et al., 2010].
Figure 4.
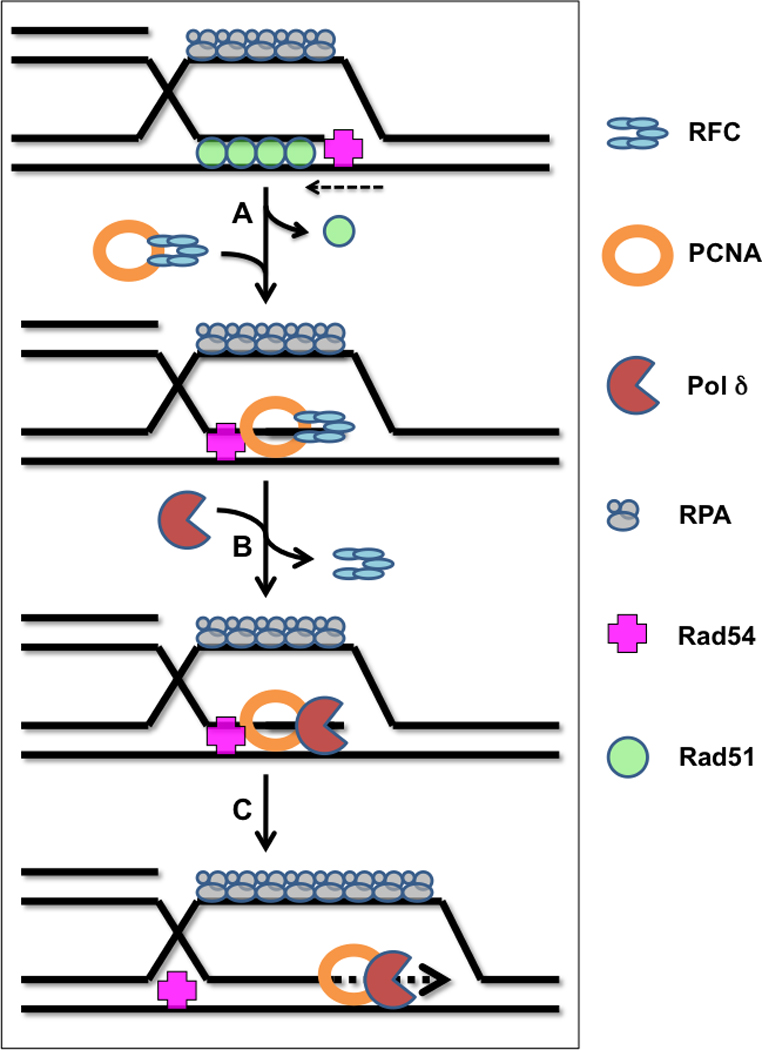
D-loop extension in eukaryotic systems requires Rad54 and the leading strand replication machinery. (A) Following D-loop formation, Rad51 recombinase remains in a stable post-synaptic filament on the heteroduplex DNA while RPA stabilizes the displaced strand. The DNA translocation activity of Rad54 disrupts the post-synaptic filament, allowing the clamp loader complex, RFC, to load the sliding clamp, PCNA, onto the primer-template formed by the invading strand. (B) Recruitment of Pol δ reconstitutes the leading strand DNA polymerase holoenzyme. (C) Strand displacement DNA synthesis occurs to extend the D-loop, possibly assisted by Rad54 or other motor proteins (dHJ and SDSA pathways). The lagging strand synthesis machinery is not required except in the special case of break-induced replication, BIR.
It is interesting to speculate that Rad54 could continue to influence the DNA synthesis component of HDR subsequent to the polymerase loading step. The translocation of Rad54 on DNA has been proposed to alter DNA topology ahead of and behind the moving motor protein [Petukhova et al., 1999]. Even though Rad54 is not formally a DNA helicase, the topology changes it induces could make it easier for Pol δ to carry out strand displacement synthesis, thus facilitating D-loop extension during dHJ or SDSA processes.
Role of Mph1 Helicase in Error-free Lesion Bypass
A number of eukaryotic 3’→ 5’ helicases have been identified that influence homologous recombination and HDR, including members of the RecQ and FANCM families, several of which are associated with human disease states (e.g. BLM, WRN, FANCM, and others) [Whitby, 2010; Bernstein et al., 2010]. These helicases play diverse roles in recombination metabolism, such as promoting DSB resection, stimulating strand invasion or branch migration reactions, or crossover control (anti-recombination via presynaptic filament dissolution, D-loop resolution, etc.). An interesting example is the yeast Mph1 helicase, an ortholog of FANCM. Mph1 is proposed to play an important role in error-free lesion bypass by replication forks, which involves transient template switching by an RDR mechanism [Panico et al., 2009; Ede et al., 2011] (Figure 1D). Mph1 is reported to be a 3’ → 5’ helicase and has been demonstrated to disrupt Rad51-generated D-loops, suggestive of an anti-recombination function. This cannot be the sole function of Mph1, however, since mph1 mutants have a mutator phenotype and reduced sister chromatid interactions [Ede et al., 2011]. Instead, Mph1 is proposed to stabilize and/or promote unwinding of the D-loop to allow Pol δ or η to extend the 3’ end of the invading strand. Stabilization of the D-loop may be sufficient since Pol δ and η are able to carry out some strand displacement synthesis without a helicase. Alternatively, interactions of Mph1 with RPA on the displaced strand may bring about an activity change that results in unwinding of the D-loop at its leading edge, which would promote DNA synthesis. It is intriguing to speculate that Mph1 or a related helicase could promote D-loop extension by a similar mechanism during dHJ or SDSA.
Discussion
The error-free repair of DNA double-strand breaks and stalled replication forks requires the coordinated activities of two machines of enormous complexity—the homologous recombination apparatus and the DNA replication apparatus. Homology-directed repair involves the generation of DNA structures such as single-stranded DNA and D-loops that if improperly handled would lead either to apoptosis or to mutation and possible tumorigenesis. Therefore it is logical to expect that ssDNA and D-loops would rarely if ever be allowed to exist as free structures in the cell. Indeed, evidence suggests that DNA structures arising at repair foci are carefully shepherded through the pre-synapsis, synapsis, post-synapsis, and synthesis stages of HDR, via sequential handoff transactions that efficiently pass each DNA intermediate to the next enzyme in the pathway while keeping toxic intermediates sequestered.
The concept of sequential DNA handoffs during the pre-synaptic phase of recombination is well-established. The formation of ssDNA through resection of DSBs may be coupled directly to presynaptic filament assembly as demonstrated elegantly for the E. coli RecBCD and RecA enzymes [Kowalczykowski, 2000]. Otherwise presynaptic filament assembly proceeds through sequential handoffs of ssDNA from ssDNA-binding protein to mediator to recombinase as shown by studies of the T4 recombination system [Liu and Morrical, 2010]. Logic dictates that D-loop intermediates formed via the homologous pairing and strand exchange activities of presynaptic filaments must also be channeled into the downstream steps of HDR. To complete dHJ, SDSA, BIR or error-free lesion bypass, a D-loop intermediate must be converted into a DNA replication fork so that D-loop extension can occur. Therefore the D-loop is handed off to DNA helicase and/or translocase activities that sequentially dissolve the post-synaptic filament of recombinase bound to heteroduplex DNA, assemble the leading strand replication machinery, facilitate unwinding of the duplex template, and (in the case of BIR) coordinate the assembly of lagging strand replication machinery. It is tempting to speculate that there is specific coupling between the clamp loading process (e.g. PCNA/RFC) in the replicative component of HDR and turnover of the post-synaptic filament (e.g. Rad54/Rad51) in the recombinational component. This could involve sequential handoffs of the invading-strand primer-template junction between the three AAA+ ATPase components Rad51, Rad54, and RFC that serve to channel D-loop structures into HDR. Further downstream in dHJ and SDSA pathways, there is clear evidence for the function of recombinase and recombination mediators in second end capture that leads to a second round of replication dependent on leading strand synthesis machinery. It is reasonable to postulate that the extension of both ends is coordinated. Orchestrating this coordination could be SMC proteins such as Rad50, which is proposed to tether the two broken ends of a DSB [Connelly and Leach, 2002], and which along with Mre11 and Xrs2 (Nbs1) forms the MRX (MRN) complex that plays a central role in DSB resection, thus linking the late stages of error-free DSB repair to the beginning.
These pieces of the recombination and replication machineries undoubtedly interface with regulatory elements involved in crossover control and the DNA damage checkpoint. Understanding the complexities of these interactions is the next challenge in the quest to understand genome stability mechanisms in all organisms, and how to exploit them to cure human disease.
Acknowledgements
Funded by NIH grant nos. GM48847 and CA98993/P3 to S.W.M. R.L.M. was supported by American Cancer Society postdoctoral fellowship no. PF-09-254-01-DMC. A.M.B. was supported by NIH training grant no. ES07122.
Abbreviations
- HR
homologous recombination
- HDR
homology-directed repair
- RDR
recombination-dependent replication
- DSB
DNA double-strand break
- DSBR
double-strand break repair
- dHJ
double Holliday Junction
- SDSA
synthesis-dependent strand annealing
- BIR
break-induced replication
- SDR
stable DNA replication, a RecA-dependent, DnaA-independent replication pathway in Escherichia coli
References
- Beernink HT, Morrical SW. RMPs: recombination/replication mediator proteins. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu Rev Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuit JS, Xu H, Ma Y, Wang T, Liu J, Morrical SW. Mediator proteins orchestrate enzyme-ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proc Natl Acad Sci U S A. 2001;98:8298–8305. doi: 10.1073/pnas.131007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira A, Kowalczykowski SC. BRCA2: Shining light on the regulation of DNA-binding selectivity by RAD51. Cell Cycle. 2009;8:3445–3447. doi: 10.4161/cc.8.21.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavito S, Prakash R, Sung P. Promotion and regulation of homologous recombination by DNA helicases. Methods. 2010;51:329–335. doi: 10.1016/j.ymeth.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Leach DR. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem Sci. 2002;27:410–418. doi: 10.1016/s0968-0004(02)02144-8. [DOI] [PubMed] [Google Scholar]
- Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. Erratum in 2007 Nat Struct Mol Biol 14:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas KC, Kreuzer KN. Bacteriophage T4 helicase loader protein gp59 functions as gatekeeper in origin-dependent replication in vivo. J Biol Chem. 2005;280:21561–21569. doi: 10.1074/jbc.M502351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ede C, Rudolph CJ, Lehmann S, Schurer KA, Kramer W. Budding yeast Mph1 promotes sister chromatid interactions by a mechanism involving strand invasion. DNA Repair (Amst) 2011;10:45–55. doi: 10.1016/j.dnarep.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T, Spies M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010;38:2917–2930. doi: 10.1093/nar/gkp1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. DNA recombination: the replication connection. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CE, Mueser TC, Dudas KC, Kreuzer KN, Nossal NG. Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: a versatile couple with roles in replication and recombination. Proc Natl Acad Sci U S A. 2001;98:8312–8318. doi: 10.1073/pnas.121009398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell. 2002;10:359–371. doi: 10.1016/s1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer KN. Recombination-dependent DNA replication in phage T4. Trends Biochem Sci. 2000;25:165–173. doi: 10.1016/s0968-0004(00)01559-0. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD. RAD54 controls access to the invading 3'-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:638–646. doi: 10.1093/nar/gkn980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol Cell. 2009;36:704–713. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ehmsen KT, Heyer WD, Morrical SW. Presynaptic filament dynamics in homologous recombination and DNA repair. CRC Crit Rev Biochem Mol Biol. 2011;46:240–270. doi: 10.3109/10409238.2011.576007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Marians KJ. PriA-directed assembly of a primosome on D loop DNA. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- Liu J, Morrical SW. Assembly and dynamics of the bacteriophage T4 homologous recombination machinery. Virology J. 2010;7:357. doi: 10.1186/1743-422X-7-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Morrical SW. Dynamics of protein-ssDNA interactions in the bacteriophage T4 homologous recombination system. In: Williams MC, Maher LJ III, editors. Biophysics of Protein-DNA Interactions: From Single Molecules to Biological Systems. New York: Springer; 2011. pp. 213–240. [Google Scholar]
- Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, Haber JE. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Asai T, Kubota Y, Arai K, Kogoma T. Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J. 1994;13:5338–5345. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwraith MJ, West SC. DNA repair synthesis facilitates RAD52-mediated second-end capture during DSB repair. Mol Cell. 2008;29:510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Morrical SW, Hempstead K, Morrical MD. The gene 59 protein of bacteriophage T4 modulates the intrinsic and single-stranded DNA-stimulated ATPase activities of gene 41 protein, the T4 replicative DNA helicase. J Biol Chem. 1994;269:33069–33081. [PubMed] [Google Scholar]
- Nimonkar AV, Sica RA, Kowalczykowski SC. Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc Natl Acad Sci U S A. 2009;106:3077–3082. doi: 10.1073/pnas.0813247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panico ER, Ede C, Schildmann M, Sch√°rer KA, Kramer W. Genetic evidence for a role of Saccharomyces cerevisiae Mph1 in recombinational DNA repair under replicative stress. Yeast. 2010;27:11–27. doi: 10.1002/yea.1727. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Ploquin M, Bransi A, Paquet ER, Stasiak AZ, Stasiak A, Yu X, Cieslinska AM, Egelman EH, Moineau S, Masson JY. Functional and structural basis for a bacteriophage homolog of human RAD52. Curr Biol. 2008;18:1142–1146. doi: 10.1016/j.cub.2008.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranatunga W, Jackson D, Lloyd JA, Forg AL, Knight KL, Borgstahl GE. Human RAD52 exhibits two modes of self-association. J Biol Chem. 2001;276:15876–15880. doi: 10.1074/jbc.M011747200. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking BRCA2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- Shivji MK, Davies OR, Savill JM, Bates DL, Pellegrini L, Venkitaraman AR. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Peterson CL. A Rad51 presynaptic filament is sufficient to capture nucleosomal homology during recombinational repair of a DNA double-strand break. Mol Cell. 2008;30:803–810. doi: 10.1016/j.molcel.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;3:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Taniyama C, Arai K, Masai H. ATPase/helicase motif mutants of Escherichia coli PriA protein essential for recombination-dependent DNA replication. Genes Cells. 2003;8:251–261. doi: 10.1046/j.1365-2443.2003.00630.x. [DOI] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ira G, Tercero JA, Holmes AM, Diffley JF, Haber JE. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6891–6899. doi: 10.1128/MCB.24.16.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1996;378:789–792. doi: 10.1038/378789a0. Erratum in: 1996. Nature 379:749. [DOI] [PubMed] [Google Scholar]
- Wu Y, Sugiyama T, Kowalczykowski SC. DNA annealing mediated by Rad52 and Rad59 proteins. J Biol Chem. 2006;281:15441–15449. doi: 10.1074/jbc.M601827200. [DOI] [PubMed] [Google Scholar]
- Xi J, Zhuang Z, Zhang Z, Selzer T, Spiering MM, Hammes GG, Benkovic SJ. Interaction between the T4 helicase-loading protein (gp59) and the DNA polymerase (gp43): a locking mechanism to delay replication during replisome assembly. Biochemistry. 2005;44:2305–2318. doi: 10.1021/bi0479508. Errata in: 2005. Biochemistry 44:4600. 2005. Biochemistry 44:12264. [DOI] [PubMed] [Google Scholar]
- Xu H, Beernink HT, Morrical SW. DNA-binding properties of T4 UvsY recombination mediator protein: polynucleotide wrapping promotes high-affinity binding to single-stranded DNA. Nucleic Acids Res. 2010;38:4821–4833. doi: 10.1093/nar/gkq219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]


