Abstract
Cancer immunoediting describes the process whereby highly immunogenic tumor cells are removed, or edited, from the primary tumor repertoire by the immune system. In immune deficient mice, the editing process is hampered, and “unedited” tumor cells can be recovered and studied. In this study we compared unedited and edited tumors for their expression of NKG2D ligands, a family of surface proteins expressed on tumor cells that can activate natural killer (NK) cell cytotoxic activity. We found that the expression of the NKG2D ligand H60a was more heterogeneous in groups of unedited compared to edited 3’methylcholanthrene (MCA) sarcoma cell lines, i.e., some unedited cell lines expressed very high levels of H60a, while other unedited and edited cell lines expressed very low levels. We also found that some highly immunogenic cell lines displayed a bimodal distribution consisting of H60a-hi and H60a-lo cells. In one of these cell lines, the H60a-hi cells could be removed by passaging the cells through recombinase activating gene-2 deficient (RAG2−/−) mice, resulting in edited cell lines that were poor targets for NK cells and displayed progressive tumor growth. This editing of H60a-hi cells required NK cells and NKG2D. Our studies show that the expression of H60a on tumors cells can be actively modulated by the immune system, thereby implicating this NKG2D ligand in tumor immunosurveillance.
Keywords: Cancer immunoediting, NKG2D ligands, H60a, tumor surveillance
Introduction
Significant interactions between immune cells and tumor cells occur during tumor development and can lead to tumor promotion or tumor destruction (1). These interactions have been codified into a cancer immunoediting process that results in the generation of a tumor cell repertoire capable of surviving in immune competent hosts (2–4). The substrate for the editing process is thought to be a primary repertoire of nascently transformed immunogenic cells, so called “unedited” cells. In the most complex form of cancer immunoediting, the immune system acts on the unedited tumor cell repertoire by eliminating immunogenic tumor cells, leading to an equilibrium process (5) that eventually culminates in the escape and outgrowth of an edited repertoire of poorly immunogenic tumor cells.
We previously found that adaptive lymphocytes and the IFN system are required for effective tumor surveillance (2–4, 6, 7). When MCA is given to mice that lack adaptive immunity (RAG2−/−) or responsiveness to type I interferon (IFNAR−/−), the tumor incidence is higher compared to syngeneic WT mice. Given the requirement for RAG2 and IFNAR in tumor surveillance, we reasoned that tumors that arise in these mice would be unedited. Indeed, we found that tumors from RAG2−/− or IFNAR−/− mice displayed increased immunogenicity (3, 6). In these studies, the immunogenicity of a cell line was measured by transplanting it into wild-type (WT) and RAG2−/− mice and measuring its growth. Cell lines that did not grow in WT but grew in RAG2−/− mice were termed regressor cell lines (3, 8) and were considered highly immunogenic. Progressor cell lines grew in both WT and RAG2−/− mice and are thought to be poorly immunogenic. We found that certain unedited tumor cell lines derived from immune deficient RAG2−/− or IFNAR−/− mice displayed a regressor growth phenotype while others displayed a progressor growth phenotype (3, 6). In contrast, all edited tumor cell lines derived from WT mice displayed progressor growth phenotypes. It is important to note that this assay of immunogenicity, involving tumor cell transplantation, is a functional assay that does not necessarily define a molecular target for cancer immunoediting. To date, a molecular assay has not been established that can reliably identify unedited versus edited tumor cell groups.
In this study, we compared the edited and unedited tumor cell repertoire by examining MCA sarcoma cell lines generated in carcinogen-treated syngeneic WT and immunodeficient mice (3, 6). We focused on evaluating the expression of ligands for the activating receptor Natural Killer Group 2D (NKG2D) (9) within the unedited tumor repertoire. NKG2D is a receptor expressed on NK cells, CD8+ T cells, γδ-T cells, and NK-T cells that mediates the detection of stressed cells that are infected by viruses or undergoing transformation (10–14). It binds a ligand family that is generally not expressed at functional levels in normal tissues, but can be up-regulated by certain stimuli, including DNA damage and virus infection (10, 15). In mice deficient in NKG2D, there is increased development of prostate tumors and lymphomas compared to control mice, but the development of MCA sarcomas was not changed (12). In contrast, mice treated with blocking antibody to NKG2D developed more MCA sarcomas than control-treated mice (13), although it is not clear whether NK cell function was affected throughout the course of this study.
We have described that the NKG2D ligand H60a (16–19), but not other NKG2D ligands, is down-regulated by IFNγ in MCA sarcomas (19). Considering the important role of IFNγ and NKG2D in preventing tumor formation (6, 20–22), we wished to determine whether the IFNγ-regulated NKG2D ligand H60a could participate in the surveillance of developing tumors. Whereas H60a can mediate tumor rejection when expressed at very high levels (23), its endogenous role in tumor immunosurveillance is not known. Furthermore, the above-mentioned studies with NKG2D-deficient mice (12, 13) were performed on the C57BL/6 background, which does not express H60a, and therefore does not address the role of H60a in tumor editing. Interestingly, H60a can be induced within days of carcinogen exposure in mouse skin (24), but its expression in the unedited tumor cell repertoire has not been studied.
In this study, we examined the expression of NKG2D ligands in MCA sarcoma cell lines derived from mice with varying levels of immune activity. We show that the heterogeneity of NKG2D ligand levels is inversely correlated with the degree of cancer immunoediting. This heterogeneity can be detected by the level of H60a expression in groups of tumor cell lines or can manifest in a single tumor cell line via bimodal distribution of H60a expression. When a cell line expressing bimodal levels of H60a was passaged through a RAG2−/− mouse and subjected to innate immune pressure, the H60a expression was reduced and it showed a progressive growth phenotype in WT mice, indicating that H60a had been edited. When NK cells were depleted or NKG2D function was blocked during passaging, H60a levels were not reduced and these cell lines showed a rejection phenotype in WT mice, showing that NK cell recognition of H60a high cells through NKG2D drives this editing process. Additionally, overexpression of H60a into passaged tumor cells lines that displayed a progressive growth phenotype was sufficient to cause rejection in WT mice. We propose that cancer immunoediting acts in part by limiting the heterogeneity of a diverse primary repertoire of tumor cells. H60a may be a suitable substrate or a surrogate marker for the editing process.
Materials and Methods
All experiments involving mice were conducted under animal protocols approved by the Washington University Animal Studies Committee and the University of California, San Diego Institutional Animal Care and Use Committee (IACUC protocol #S06201) and were in accordance with their ethical guidelines.
Cell lines and mice
MCA sarcoma cell lines were generated as described (3) by injecting 129/Sv mice with 1–400 µg of 3’methylcholanthrene (Sigma, St. Louis) dissolved in peanut oil, harvesting the tumor mass after it had reached >10 mm in average diameter, dissociating the mass via collagenase treatment, and culturing the cells in vitro. All experiments were done with cells passaged between 4 and 12 cycles. 129/Sv, RAG2−/−, RAG2−/−xSTAT1−/− (RkSk), and STAT1−/− mice used were described (3, 6). Cell lines were maintained in RPMI 1640 supplemented with 10% FCS, L-glutamine, NEAA, sodium pyruvate, sodium bicarbonate, pen/strep, and β-mercaptoethanol. The IFNAR−/− MCA sarcoma cell line d100 was passaged through RAG2−/− mice and cell lines were generated from harvested tumor masses. The passaged cell lines which were now progressors were transduced with a retrovirus expressing either green fluorescent protein (GFP), or GFP and H60a, and sorted as a >95% GFP+ population using a FACS Aria cell sorter.
Antibodies and FACS analysis
All cell stains were done with 50–80% confluent cells and were repeated at least twice. Cells were harvest with dPBS or HBSS supplemented with 2.5 mM EDTA. Trypsin was not used since it decreased NKG2D tetramer staining, presumably by cleaving the ligands. NKG2D tetramers were generated as described (25). Monoclonal antibodies to H60a, pan-RAE-1, and MULT1 were obtained from R&D (Minneapolis, MN). Secondary antibodies were obtained from Biolegend (San Diego, CA). Staining was conducted for 15–30 minutes at 4° in FACS tubes containing 0.5–2 million total cells, 0.5–1 µl of antibody, and 100 µl of FACs buffer (PBS+1% FCS+0.09% NaN3, Sigma). All analyses were done on live cells identified by forward and side scatter properties. Significance of variance was determined between groups by using the F-test. The Levene statistic was calculated to determine homogeneity within all groups. The Welch’s t test, which accommodates unequal variances, was used to determine differences between the means of two groups. The median fluorescence intensity was used to quantitate antibody binding.
Tumor transplantation
Subconfluent tumor cell lines were harvested by trypsinization, washed 3x with HBSS+Ca/Mg, and injected subcutaneously into syngeneic recipient WT or RAG−/− mice as described (6, 19, 26). RAG−/− mice were injected i.p. with either PBS, anti-ASGM1, or anti-NKG2D on days −2 and 0 and every four days thereafter until tumor harvest. NK cell depletion was verified by FACS analysis of both spleen and tumor cell suspensions at the time of harvest. Mice were monitored for tumor growth by measurement of mean tumor diameter, defined as the average of the 2 maximum dimensions of the tumor mass. On various days post-transplantation, tumors were excised from mice, minced, and treated with 1 mg/mL type IA collagenase (Sigma) as described (26). Cells were vigorously resuspended, washed in FACS buffer and filtered before staining. Antibodies to CD45, CD122, DX5, NK1.1, CD69, MAC1, Kb, Kd, I-A/I-E, and Ly6G were from eBiosciences (San Diego, CA).
Chromium release assay
Splenocytes from RAG2−/− mice were activated by culturing in media with 1000 U/ml human IL-2 (Chiron, Emeryville, CA). Day 7 IL-2-activated NK effector cells were used in a 4-hour 51Cr release assay using tumor target cells labeled with 51Cr as described (19) with either control IgG or anti-NKG2D. Bars depict standard error of triplicates. All experiments were done at least twice. ANOVA was used to assess statistical significance between parent and passaged cell lines.
Results
Unedited tumors display a wide range of NKG2D ligand expression
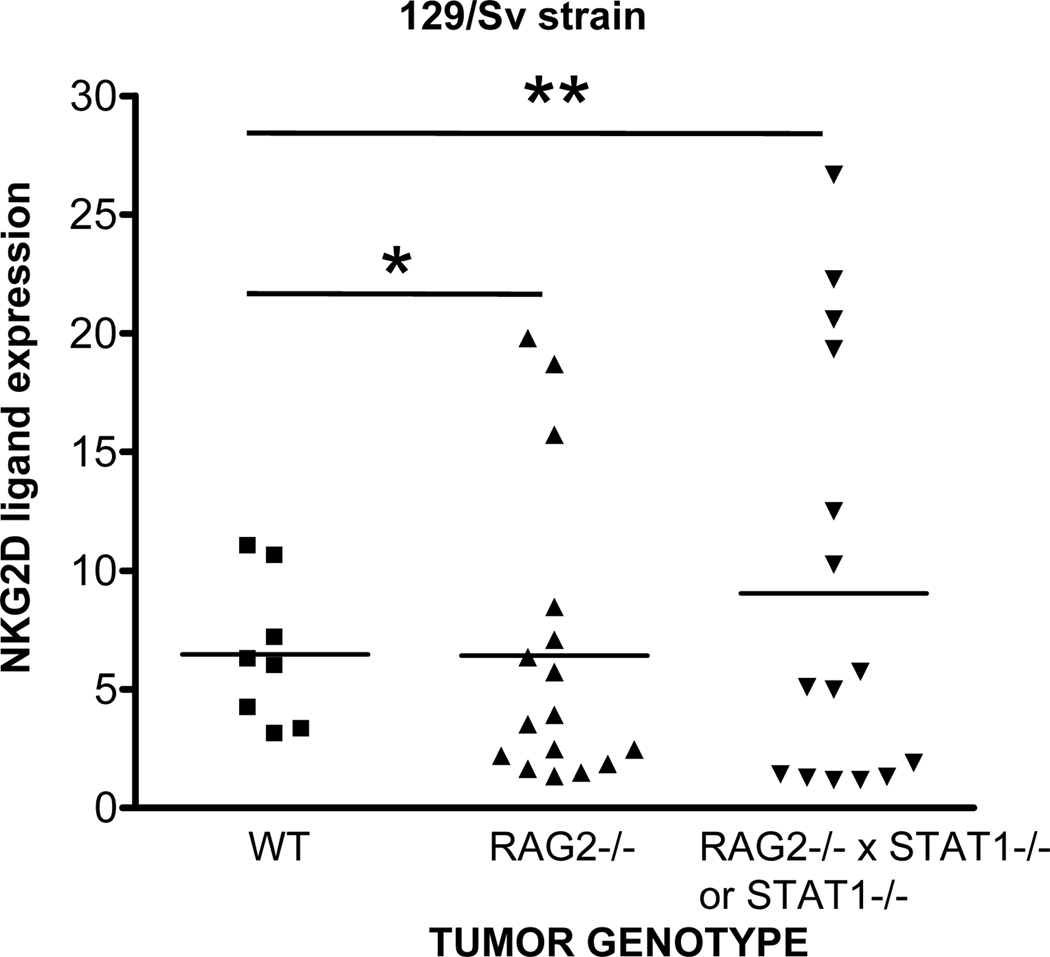
Previous carcinogenesis studies comparing primary tumor formation in WT versus NKG2D-deficient mice found no significant difference in MCA sarcoma formation (12). However, these experiments were performed using mice on a C57BL/6 background, where H60a is not expressed (18, 27). Therefore, we measured NKG2D ligand expression in MCA sarcoma cell lines generated in WT mice and mice with varying levels of immune deficiency (3, 6), all on a 129/Sv background, which display an intact H60a gene (28). Figure 1 shows NKG2D tetramer staining of 39 129/Sv-strain MCA sarcoma cell lines. We found that the range of NKG2D ligand expression was highest in the MCA sarcomas that developed in the most immune deficient mice (RAG2−/−xSTAT1−/−, denoted as RkSk, or STAT1−/−). MCA sarcomas from RAG2−/− mice displayed moderate heterogeneity of NKG2D ligand expression, whereas edited MCA sarcomas that developed in immunocompetent WT mice had tightly grouped levels of NKG2D ligand expression. Although the mean level of NKG2D ligand expression was similar between the groups, using the F test to compare variances, we found that there was significant difference in the variances of NKG2D ligand expression between WT and RkSk/STAT1−/− groups (p=0.0079), WT and RAG2−/− groups (p=0.043), but not between RAG2−/− and RkSk/STAT1−/− groups (p=0.166). We also calculated the Levene statistic to test the homogeneity of variances between all groups, and found significant heterogeneity in NKG2D ligand expression (p=0.007).
Figure 1. NKG2D ligand expression displays heterogeneous expression in unedited tumors.
MCA sarcoma cell lines were generated in WT or immune deficient mice. Cell lines were stained with NKG2D tetramer to measure all NKG2D ligands. NKG2D ligand expression was calculated by dividing the median fluorescences of NKG2D tetramer staining and SA-PE control. Similar results were obtained when the median channel shift was calculated by subtracting the median fluorescences. Each symbol represents a different individual cell line. The F-test was used to measure significant differences in variances between groups. (* Indicates p value < 0.05, ** Indicates p value < 0.01).
Regressor cell lines display a higher level of heterogeneous NKG2D ligand expression than matched progressor cell lines
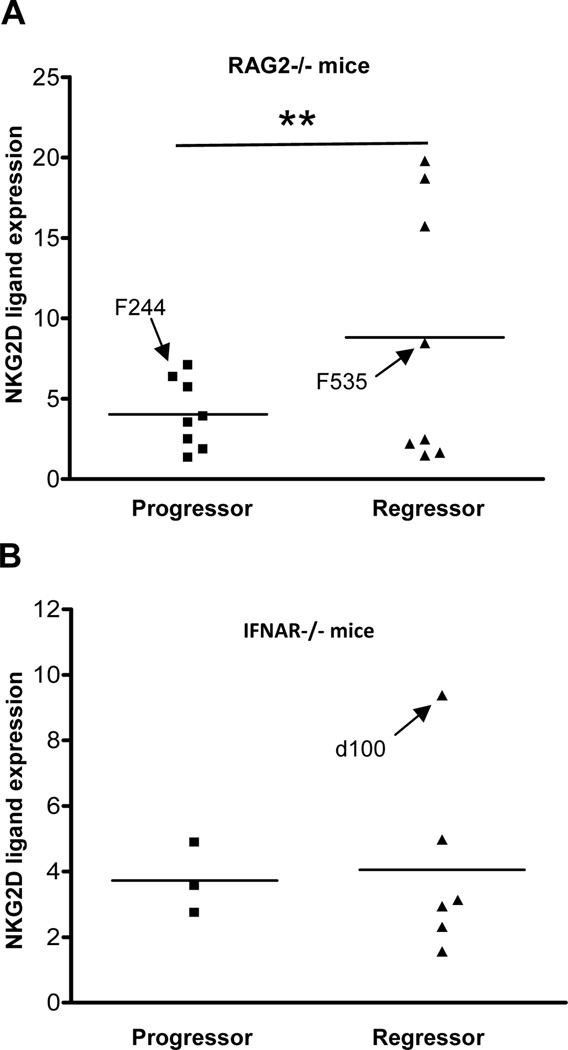
Previously, we found that approximately 40% of unedited MCA sarcoma cell lines tested were rejected when transplanted into syngeneic, naïve WT mice, whereas 60% grew progressively (3, 6). The regressor and progressor phenotypes of these cell lines were reproducible and were only observed in tumor cell lines derived from immunodeficient mice (either RAG2−/− or IFNAR−/−). Edited tumor cell lines from WT mice all displayed progressor phenotypes. We determined whether the tumor growth phenotype was correlated with the expression of NKG2D ligands, by examining unedited MCA sarcomas from RAG2−/− or IFNAR−/− mice that had already been categorized into regressor and progressor phenotypes (3, 6). We found that the heterogeneity in NKG2D ligand levels was associated with the regressor phenotype of unedited cells (Figs. 2A–B). This difference in heterogeneity was significant when the variances of the populations were compared in the group of tumors (F test, p=0.0024; Levene statistic p=0.001) from RAG2−/− but not IFNAR−/− mice.
Figure 2. NKG2D ligand expression displays heterogeneous expression in regressor compared to progressor cell lines.
MCA sarcoma cell lines derived from (A) 129/Sv RAG2−/− and (B) 129/Sv IFNAR−/− mice were categorized into progressor and regressor phenotypes, and NKG2D ligand expression was plotted similar to Figure 1. The F-test was used to measure significant differences in variances between groups. (** Indicates p value < 0.01). Several cell lines (F244, F535, d100) were chosen for later studies, and their data points are indicated in the bar graph for comparison.
H60a expression correlates with the heterogeneity in NKG2D tetramer staining seen in unedited tumors
We next examined whether the heterogeneous NKG2D tetramer staining could be explained by heterogenous expression of a specific NKG2D ligand. We found that our MCA sarcoma cell lines did not express MULT1 (19), so we focused on examining the expression of H60a and RAE1 using monoclonal antibodies. We found that H60a expression was highly variable in the groups of unedited tumors (Fig. 3A). This mirrored our results with NKG2D tetramer staining. Interestingly, RAE1 expression, while lower than H60a expression, also displayed heterogeneity (Fig. 3B). For both H60a and RAE1, there were significant differences in variance between the RAG progressor and RkSk/STAT1−/− groups (p=0.0079 for H60a, p=0.0014 for RAE1) and between RAG progressor and RAG regressor groups (p=0.0087 for H60a, p=0.001 for RAE1). The Levene statistic indicated that RAE had a slightly higher level of heterogeneity than H60a (p=0.048 for RAE variance; p=0.075 for H60a variance). Although the heterogeneity in expression of H60a and RAE1 was found in similar groups of tumors, most regressor tumors expressed H60a while few expressed RAE1. Therefore, we focused on H60a.
Figure 3. Heterogeneity in NKG2D ligand expression in 129/Sv strain tumors is seen in H60a but not RAE1 expression.
H60a and RAE1 expression was determined in the indicated cell lines by flow cytometry using monoclonal antibodies. NKG2D ligand expression was calculated by dividing the median fluorescences of anti-H60a or anti-RAE-1 staining by isotype control. The F-test was used to measure significant differences in variances between groups. (** Indicates p value < 0.01, *** Indicates p value < 0.001).
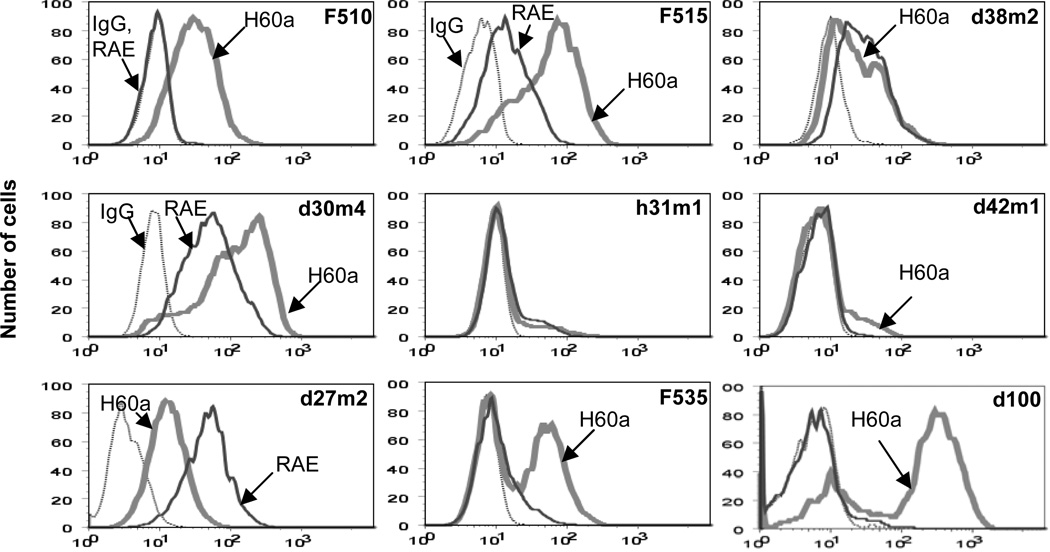
H60a staining was bimodal in many cell lines
In examining the expression of H60a in our cell lines, we noticed that the staining of H60a in some MCA sarcoma cell lines did not display unimodal staining, as would be expected of a homogeneous population of cells. We have chosen nine representative regressor cell lines (Fig. 4A) to demonstrate that the pattern of H60a staining ranged from unimodal (d27m2, F510, h31m1), broad staining (F515, d30m4), to bimodal staining, with equal distribution (F535, d100) between the two cell populations or uneven, “shoulder”-type distribution (d38m2, d42m1), suggesting that within a cell line, there were populations of cells that expressed differing levels of H60a. This pattern of staining did not change even after three weeks of in vitro passaging for the bimodal d100 cell line (Suppl Fig. 1), indicating that it was a relatively stable characteristic of this cell line. RAE1 did not display this pattern of staining, even though in some regressors (d30m4, d27m2), it was highly expressed.
Figure 4. Heterogeneity in H60a expression can be seen even within a single cell line.
Nine regressor cell lines were stained with control isotype (dotted line), anti-RAE (solid line), or anti-H60a (thick line).
Cell lines with bimodal expression of H60a lose H60a expression after in vivo passage
We were struck by the fact that bimodal H60a, but not RAE1 expression could be seen in some cell lines (F535, d100) and wondered whether H60a high cells could be edited by components of the immune system in vivo. Since d100 and F535 are both regressor cell lines that reject in WT mice, we transplanted them into immune deficient mice in order to test if partial immune function could eliminate H60a-hi cells in vivo. We transplanted d100 (derived from an IFNAR−/− mouse) into RAG2−/− mice and F535 (derived from a RAG2−/− mouse) into IFNAR−/− mice. After in vivo growth for 20–30 days, we harvested the tumor mass and generated cell lines. Figure 5 shows that the passaged cell lines indeed had lower expression of H60a compared to the parental cell lines. Whereas 70% of parental d100 cells had high expression of H60a (as measured by NKG2D-tet), this was significantly reduced to 35% in the passaged cell lines (Fig. 5A and C, p<0.001). Similarly, two of the three passaged F535 cell lines completely lost the H60a-hi population of cells and became unimodally low in H60a expression, while the third passaged cell line had decreased levels of H60a-hi cells (Fig. 5B and D). The loss of H60a-hi cells seen after in vivo passage was not seen in the same cell line cultured in vitro for a similar period of time (Suppl Fig. 1), and therefore it is unlikely due to inherent differences in proliferation or cell cycle between H60a-hi and H60a-lo cells.
Figure 5. Editing of H60a-hi cells after in vivo passage.
(A) The IFNAR−/−-derived regressor tumor d100 was transplanted into three RAG2−/− mice, and daughter cell lines were generated, designated with the suffix “Ragp” to indicated their passage through RAG2−/− mice. Cell lines d100.Ragp.a, d100.Ragp.b, and d100.Ragp.c were obtained from three different RAG2−/− mice, and H60a expression was detected by NKG2D-tetramer. (B) The RAG2−/−-derived regressor tumor f535 was transplanted into three IFNAR−/− mice, and daughter cell lines were generated, designated with the suffix IFNARp. Cell lines f535.IFNARp.a, f535.IFNARp.b, and f535.IFNARp.c were obtained from three different IFNAR−/− mice. H60a expression was detected by flow cytometry. (*** Indicates p value < 0.001).
The regressor tumor d100 shows delayed growth in RAG2−/− mice and increased NK cell infiltration
Next, we examined the cell types that could mediate the editing process that removed H60a-hi cells. We hypothesized NK cells would infiltrate and selectively destroy H60a-hi cells. Therefore, we transplanted d100 and a control progressor cell line F244 that expresses moderate levels of H60a (Suppl Fig. 2) into WT and RAG2−/− mice. As shown previously (3, 29), d100 was rejected, but F244 grew progressively in WT mice (Fig. 6A). Interestingly, in RAG2−/− mice, the progressor F244 developed a tumor mass greater than 5 mm (mean diameter) within 10 days of transplant (Fig. 6B) while the d100 tumor remained ≤3 mm for more than 2 weeks after tumor transplant. This delayed growth was associated with a preponderance of infiltrating NK cells in the tumor mass. When both tumors were harvested at the same size (roughly 13 mm), 10% of the infiltrating CD45+ cells in the regressor d100 tumor were NK cells (DX5+CD122+) compared to 3% in the progressor F244 tumor (p<0.001) (Fig. 6C).
Figure 6. d100, a regressor tumor, shows delayed growth in RAG2−/− mice, which is associated with NK cell infiltration.
The d100 and F244 cell lines were injected into (A) WT and (B) RAG2−/− mice, and tumor size was measured over time. (C) Tumor masses growing in RAG2−/− mice were harvested at day 25, disaggregated, and analyzed as a single cell suspension by FACS analysis. Infiltrating NK cells were identified as the DX5+CD122+ population from CD45+ events. Results were reproduced in an independent experiment. (*** Indicates p value < 0.001).
Passage of the d100 regressor tumor through RAG2−/− mice leads to reduced NK cell recognition
Next, we examined whether the NK cell infiltration and delayed growth of d100 in RAG2−/− mice led to a sculpted tumor repertoire that was resistant to NK cell cytotoxicity. We performed a standard chromium release assay using IL-2 activated NK cells as effectors and the parental or RAG2−/−-passaged cell lines as targets. As predicted, we found that the passaged cell lines were two-fold less susceptible to NK cell killing compared to the parental cell line (Fig. 7, p = 0.0216).
Figure 7. Passage of d100 in RAG2−/− mice leads to decrease in NK recognition.
The parental unpassaged d100 or RAG2−/−-passaged cell lines were used as targets in a conventional cytotoxicity assay using IL-2 activated NK cells as effectors. Results were reproduced in an independent experiment. (* Indicates p value < 0.05)
Passage of the d100 regressor tumor through RAG2−/− mice converts it into a progressor tumor through NK cell NKG2D editing of H60a-hi cells
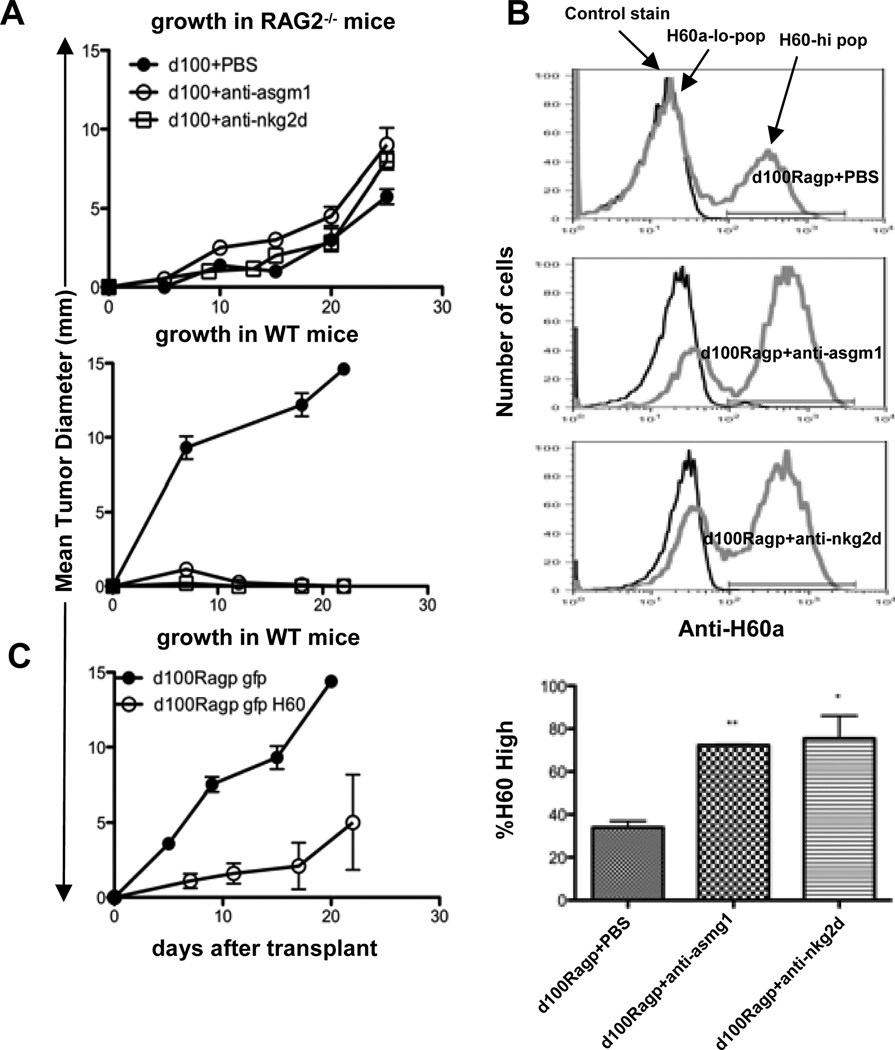
Having shown that NK recognition was decreased in vitro in RAG2−/−-passaged cell lines, we next determined whether RAG2−/−-passaging of d100 in the presence or absence of NK cells or NKG2D activity had functional consequences on tumor growth in vivo. We transplanted d100 into RAG2−/− mice that had received either i.p. injections of PBS, anti-ASGM1 (to deplete NK cells), or a blocking anti-NKG2D monoclonal antibody and monitored tumor growth in vivo. Depletion of NK cells with anti-ASGM1 showed a slight increase in tumor growth in RAG2−/− mice compared to control PBS treatment, while there was no change in growth when NKG2D function was blocked with anti-NKG2D (Fig 8A top panel). When cell lines were generated from the tumors, we found that cells passaged in the presence of functional NK cells (PBS) showed a progressive growth phenotype in WT mice, while those passaged without NKG2D function or NK cells still retained their regressor phenotype (Fig 8A bottom panel). Moreover, daughter cell lines generated in mice treated with anti-ASGM1 or anti-NKG2D still maintained ~70% H60a-hi cells (Fig 8B, p = 0.006, 0.03, respectively, when compared to H60a expression in cell lines passaged through mice treated with PBS), suggesting that NK cell recognition of H60a-hi cells through NKG2D leads to their elimination during RAG2−/−-passaging.
Figure 8. H60a is functionally edited by NK cell recognition through NKG2D.
The IFNAR−/−-derived regressor tumor d100 was injected into (A) Top-RAG2−/− mice that received i.p injections of either PBS, anti-ASGM1, or anti-NKG2D, and tumor size was measured over time. Bottom-Tumor masses growing in RAG2−/− mice were harvested at day 25, disaggregated, and daughter cell lines were generated and injected into WT mice. (B) H60a expression of daughter cell lines was detected by flow cytometry. (C) An edited, passaged, progressor d100 cell line was transduced either with retrovirus-expressing green fluorescent protein (GFP), or both GFP and H60a and injected into WT mice. (** Indicates p value < 0.01, * Indicates p value < 0.05).
We tested NKG2D-dependent recognition by a standard chromium release assay using IL-2 activated NK cells as effectors and the parental d100 cell line as a target incubated with IgG or anti-NKG2D antibody. Targets incubated with anti-NKG2D antibody were lysed 5 times less than control IgG, indicating that NK cell recognition through NKG2D resulted in higher killing of the d100 parental cell line (Suppl. Fig. 3, p = 0.0102).
Editing of H60a by NK cells and NKG2D is required for conversion to a progressor phenotype
It is possible that H60a serves as a surrogate marker of an editing process and is not required for tumor rejection. To test this, we restored H60a expression in RAG2−/−-passaged d100 cell lines that were now progressor tumors by transducing with a retrovirus expressing either green fluorescent protein (GFP), or both GFP and H60a. We restored H60a expression to the level that was originally on the parent cell line (Suppl. Fig. 4). Figure 8C shows that control GFP cell lines grew progressively, while those overexpressing H60a displayed delayed growth or a rejection phenotype in vivo, confirming that the reduction of H60a by NK cells and/or NKG2D is required for the conversion of a regressor into a progressor.
Discussion
Tumor heterogeneity has been appreciated as an inherent characteristic of genetically unstable neoplasms (30, 31). Various studies have shown that among the cancerous cells in a tumor mass, or even within a tumor cell line, there is heterogeneity in the phenotype (32, 33), capacity to metastasize (34, 35), response to chemotherapy (36), ability to initiate new tumors (32, 35, 37), and signalling through surface receptors (33, 38). Indeed, the cancer stem cell hypothesis (32, 37) is supported by the observation that a tumor mass is heterogeneous, and therefore made of cells that have different capacities to initiate new tumors after transplantation. In this study, we have applied the idea of tumor heterogeneity to the measurement of immunogenicity. We have hypothesized that the tumor cell repertoire is immunologically heterogeneous, i.e., it consists of cells that have varying levels of immunogenicity due to diversity in antigens or innate ligands. We further hypothesized that cancer immunoediting acts by limiting the immunologic heterogeneity of the tumor cell population by removing cells of increased immunogenicity. In support of our hypotheses, we have found that the expression of H60a, as a potential surrogate marker of immunogenicity, is more heterogeneous in unedited compared to edited tumor cells. This heterogeneity in H60a expression is found in groups of cell lines (unedited versus edited) and even within single unedited cell lines. These data suggest that tumor heterogeneity may be detected and eliminated by the immune system via a cancer immunoediting process, and NKG2D ligands may serve as markers for this process.
We conclude that H60a is an edited molecule, and this editing process requires NK cells and NKG2D activity. A previous study (39) using NKG2D-deficient mice found that NKG2D is not involved in the surveillance of MCA sarcomas, whereas a study from another group showed increased MCA tumor formation in mice treated with blocking antibodies to NKG2D (13). Both of these studies were performed in C57BL/6 mice, which lack H60a, and therefore do not shed light on the mechanism of H60a editing in our model system. The finding that some unedited cell lines from mice deficient in STAT1 or γc display even higher levels of H60a points to NK cells as an effector of the editing process, since they are functionally defective in the absence of STAT1 (40) and are completely absent in mice lacking γc (41). We found increased levels of activated NK cells in the d100 regressor tumor as it is undergoing editing when transplanted into RAG2−/− mice compared to a progressor tumor control. In addition, daughter cell lines generated in the presence of functional NK cells showed a progressive growth phenotype in WT mice, while those generated without NKG2D function or NK cells still retained their rejection phenotype, suggesting that NK cell killing through NKG2D is required for the editing of these tumors in vivo. Overexpression of H60a into edited tumors causes their rejection in vivo, showing that H60a can be a functional marker of immunoediting for certain tumors.
We found that H60a can be expressed at higher levels in progressor than regressor tumor cell lines. These results suggest that H60a is not sufficient or necessary for tumor rejection. We envision that NKG2D ligands are only one component of the immunologic landscape of tumor cells, and tumors with very high antigenicity or innate ligands might not need NKG2D ligand expression to be rejected. On the other hand, tumor cells with moderate NKG2D ligand expression can escape immune detection if they decrease their antigenicity or increase inhibitory ligands or cytokines. In our model system using d100, the decrease in H60a expression is required for the conversion to a progressor. In contrast, although H60a is edited in F535 after passage through IFNAR−/− mice, the IFNAR−/−-passaged F535 daughter cell lines remain regressors, indicating that other immune targets can be sufficient to mediate rejection. It will be interesting to examine the expression of inhibitory molecules such as CD47 (42) and B7-H1 (43) on progressors with moderate H60a expression and innate ligands such as HMGB1 and advanced glycation end-products (44) on regressors with low levels of H60a, such as IFNAR−/−-passaged F535 cells (Fig. 5B). It is also possible that F535 has very strong tumor antigens that mediate rejection independent of NKG2D ligand expression or NK cell priming of adaptive responses (23, 45).
We found that some regressor cell lines displayed bimodal H60a expression. This pattern was fairly stable over time, and therefore is unlike the bimodal H60c expression that is induced via “culture shock” in primary keratinocytes (46). Nevertheless, we cannot rule out that cultured cell lines induce H60a expression via a response to the stress of in vitro growth. We consider this possibility unlikely, since we have shown previously that H60a expression on tumor cells harvested ex vivo is comparable to levels of the same tumor cell grown in vitro (19). In addition, other groups have found that primary tumor cells also express high levels of NKG2D ligands (14, 47). Regardless of whether in vitro culture induces NKG2D ligand expression, we advocate that the level of expression is an inherent characteristic of the cell line and can reflect the level of editing that the cell line has undergone in vivo.
It should be noted that regressor cell lines that are bimodal in H60a expression (d100, F535) are completely rejected in WT mice, rather than undergoing an equilibrium phase that promotes the escape of H60a-lo clones. We speculate that even though the H60a expression is bimodal, the T cell antigens may be shared among the H60a-hi and H60a-lo cell clones. Since it is known that NK cell activation and killing can lead to T cell priming (23, 45), we envision that NK/NKG2D-dependent killing of H60a-hi cells releases antigens that are targeted in an adaptive immune response to H60a-lo cells, leading to complete tumor rejection. Future studies will address whether the H60a-hi cells can immunize against edited H60a-lo cells.
It is not known what regulatory mechanism allows for very high and very low NKG2D ligand levels in similarly treated cells. The fact that H60a expression is also heterogeneous within cell lines indicates that H60a expression could be controlled at the level of a single cell via intrinsic mechanisms. We envision that during carcinogenesis, some tumor cells suffer lesions that induce H60a expression, while others do not. These lesions could be quantitative in nature, and may loosely correlate with H60a expression. We believe these lesions could be in the cis elements that regulate H60a and also throughout the genome, to be sensed by trans factors that regulate H60a. Although DNA damage has been shown to regulate Rae-1, Mult-1, and MICA, we have not found similar regulation of H60a (JDB, unpublished observations). We have found that the transcript levels for H60a also displayed heterogeneity and is not always correlated with H60a protein levels, implying that post-transcriptional mechanisms of H60a regulation may exist. Indeed, we and others have found that microRNAs can regulate the human NKG2D ligand MICA (48, 49) and H60a (28).
Our cell lines are not cloned, and therefore, the heterogeneous expression of H60a is likely due to multiple cell clones expressing different levels of H60a. Interestingly, when we sorted H60a-lo cells from the bimodal parental d100 cell line, the H60a expression was unstable, and the sorted H60a-lo cells gave rise to H60a-hi cells after several days of culture (data not shown). The sorted H60a-lo cells, when transplanted into WT mice as a bimodal population due to H60a instability, were still regressor cells. We could not obtain cell clones that stably expressed H60a at low levels, even after multiple limiting dilution cloning procedures. Thus, we still do not know whether the parent d100 is a mixture of cell lines or is a single “unstable” cell line that then gives rises to different daughter cells. Nevertheless, our observations are consistent with the concept that the substrate for editing is a mixture of various cell clones, each with differing immunogenicities. Tumors that are not edited would therefore be more heterogeneous in their expression of immunogenic molecules such as NKG2D ligands.
An alternative scenario could be that H60a expression is associated with intrinsic cellular physiology, and could be related to the proliferation state of the cell, as has been shown for MICA (50). Although cell starvation is not sufficient to affect H60a levels (19), cell cycle signals could collaborate with other signals to regulate H60a. It should be noted that the Rae-1 levels in our cell lines are fairly homogeneous, i.e., displays a unimodal, narrow peak in flow cytometric analysis, and thus, the mechanisms that underlie the observed heterogeneous expression of H60a must be specific for individual NKG2D ligands.
Supplementary Material
Acknowledgments
We would like to thank Jennifer Ngolab, Michael White, and Cora Arthur for help with generating and maintaining the MCA sarcoma cell lines. We would like to thank Ravi Uppaluri and Deepak Yadav for critical discussion and comments on the manuscript.
Footnotes
This work was supported by grants to J.D.B. from the Cancer Research Institute, the American Cancer Society (ACS-IRG #70-002), the Cancer Research Coordinating Committee (6-444951-34384), the V Foundation Scholar Award, the Concern Foundation, and the NIH (CA128893); and grants to R.D.S. from the NIH (CA43059), the Cancer Research Institute, and the Ludwig Institute for Cancer Research
References
- 1.Bui JD, Schreiber RD. Cancer immunosurveillance, immunoediting and inflammation: independent or interdependent processes? Curr Opin Immunol. 2007;19:203–208. doi: 10.1016/j.coi.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nature immunology. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 3.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 4.Smyth MJ, Dunn GP, Schreiber RD. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Advances in immunology. 2006;90:1–50. doi: 10.1016/S0065-2776(06)90001-7. [DOI] [PubMed] [Google Scholar]
- 5.Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 6.Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, Bui JD, Diamond MS, Koebel CM, Arthur C, White JM, Schreiber RD. A critical function for type I interferons in cancer immunoediting. Nature immunology. 2005 doi: 10.1038/ni1213. [DOI] [PubMed] [Google Scholar]
- 7.Crowe NY, Smyth MJ, Godfrey DI. A critical role for natural killer T cells in immunosurveillance of methylcholanthrene-induced sarcomas. J Exp Med. 2002;196:119–127. doi: 10.1084/jem.20020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flood PM, Schreiber H, Ron Y. Protective immunity to progressive tumors can be induced by antigen presented on regressor tumors. J Immunol. 1987;138:3573–3579. [PubMed] [Google Scholar]
- 9.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 10.Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001;1:41–49. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- 11.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 12.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, Knoblaugh S, Cado D, Greenberg NR, Raulet DH. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. The Journal of experimental medicine. 2005;202:583–588. doi: 10.1084/jem.20050994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 15.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, Lanier LL. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 17.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nature immunology. 2000;1:119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 18.Malarkannan S, Shih PP, Eden PA, Horng T, Zuberi AR, Christianson G, Roopenian D, Shastri N. The molecular and functional characterization of a dominant minor H antigen, H60. J Immunol. 1998;161:3501–3509. [PubMed] [Google Scholar]
- 19.Bui JD, Carayannopoulos LN, Lanier LL, Yokoyama WM, Schreiber RD. IFN-dependent down-regulation of the NKG2D ligand H60 on tumors. J Immunol. 2006;176:905–913. doi: 10.4049/jimmunol.176.2.905. [DOI] [PubMed] [Google Scholar]
- 20.Burnet FM. The concept of immunological surveillance. Prog Exp Tumor Res. 1970;13:1–27. doi: 10.1159/000386035. [DOI] [PubMed] [Google Scholar]
- 21.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nature reviews. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 22.Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nature reviews. 2009;9:568–580. doi: 10.1038/nri2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, Hobby P, Sutton B, Tigelaar RE, Hayday AC. Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294:605–609. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- 25.Carayannopoulos LN, Naidenko OV, Kinder J, Ho EL, Fremont DH, Yokoyama WM. Ligands for murine NKG2D display heterogeneous binding behavior. European journal of immunology. 2002;32:597–605. doi: 10.1002/1521-4141(200203)32:3<597::aid-immu597>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Bui JD, Uppaluri R, Hsieh CS, Schreiber RD. Comparative analysis of regulatory and effector T cells in progressively growing versus rejecting tumors of similar origins. Cancer Res. 2006;66:7301–7309. doi: 10.1158/0008-5472.CAN-06-0556. [DOI] [PubMed] [Google Scholar]
- 27.Samarakoon A, Chu H, Malarkannan S. Murine NKG2D ligands: "double, double toil and trouble". Molecular immunology. 2009;46:1011–1019. doi: 10.1016/j.molimm.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Hardamon C, Sagoe B, Ngolab J, Bui JD. Studies of the H60a locus in C57BL/6 and 129/Sv mouse strains identify the H60a 3'UTR as a regulator of H60a expression. Molecular immunology. 2011;48:539–545. doi: 10.1016/j.molimm.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn GP, Ikeda H, Bruce AT, Koebel C, Uppaluri R, Bui J, Chan R, Diamond M, White JM, Sheehan KC, Schreiber RD. Interferon-gamma and cancer immunoediting. Immunologic research. 2005;32:231–245. doi: 10.1385/ir:32:1-3:231. [DOI] [PubMed] [Google Scholar]
- 30.Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978;38:2651–2660. [PubMed] [Google Scholar]
- 31.Weinberg RA. The Biology of Cancer. New York: Garland Science, Taylor & Francis Group, LLC; 2006. [Google Scholar]
- 32.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 33.Kuwai T, Nakamura T, Kim SJ, Sasaki T, Kitadai Y, Langley RR, Fan D, Hamilton SR, Fidler IJ. Intratumoral heterogeneity for expression of tyrosine kinase growth factor receptors in human colon cancer surgical specimens and orthotopic tumors. Am J Pathol. 2008;172:358–366. doi: 10.2353/ajpath.2008.070625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dexter DL, Leith JT. Tumor heterogeneity and drug resistance. J Clin Oncol. 1986;4:244–257. doi: 10.1200/JCO.1986.4.2.244. [DOI] [PubMed] [Google Scholar]
- 37.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 38.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, Knoblaugh S, Cado D, Greenberg NM, Raulet DH. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen KB, Salazar-Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY, Caligiuri MA, Durbin JE, Biron CA. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol. 2002;169:4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- 41.Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 42.Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, Weissman IL. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 44.Logsdon CD, Fuentes MK, Huang EH, Arumugam T. RAGE and RAGE ligands in cancer. Curr Mol Med. 2007;7:777–789. doi: 10.2174/156652407783220697. [DOI] [PubMed] [Google Scholar]
- 45.Kelly JM, Darcy PK, Markby JL, Godfrey DI, Takeda K, Yagita H, Smyth MJ. Induction of tumor-specific T cell memory by NK cell-mediated tumor rejection. Nat Immunol. 2002;3:83–90. doi: 10.1038/ni746. [DOI] [PubMed] [Google Scholar]
- 46.Whang MI, Guerra N, Raulet DH. Costimulation of dendritic epidermal gammadelta T cells by a new NKG2D ligand expressed specifically in the skin. J Immunol. 2009;182:4557–4564. doi: 10.4049/jimmunol.0802439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groh V, Smythe K, Dai Z, Spies T. Fas-ligand-mediated paracrine T cell regulation by the receptor NKG2D in tumor immunity. Nat Immunol. 2006;7:755–762. doi: 10.1038/ni1350. [DOI] [PubMed] [Google Scholar]
- 48.Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- 49.Yadav D, Ngolab J, Lim RS, Krishnamurthy S, Bui JD. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-gamma-induced microRNA. J Immunol. 2009;182:39–43. doi: 10.4049/jimmunol.182.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkataraman GM, Suciu D, Groh V, Boss JM, Spies T. Promoter region architecture and transcriptional regulation of the genes for the MHC class I-related chain A and B ligands of NKG2D. J Immunol. 2007;178:961–969. doi: 10.4049/jimmunol.178.2.961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.