Abstract
Variations in gene expression are essential for the evolution of novel phenotypes and for speciation. Studying allelic specific gene expression (ASGE) within interspecies hybrids provides a unique opportunity to reveal underlying mechanisms of genetic variation. Using Xiphophorus interspecies hybrid fishes and high-throughput next generation sequencing technology, we were able to assess variations between two closely related vertebrate species, X. maculatus and X. couchianus, and their F1 interspecies hybrids. We constructed transcriptome-wide SNP polymorphism sets between two highly inbred X. maculatus lines (JP 163 A and B), and between X. maculatus and a second species, X. couchianus. The X. maculatus JP 163 A and B parental lines have been separated in the laboratory for ≈ 70 years and we were able to identify SNPs at a resolution of 1 SNP per 49 kb of transcriptome. In contrast, SNP polymorphisms between X. couchianus and X. maculatus species, which diverged ≈ 5–10 million years ago, were identified about every 700 bp. Using 6,524 transcripts with identified SNPs between the two parental species (X. maculatus and X. couchianus), we mapped RNA-seq reads to determine ASGE within F1 interspecies hybrids. We developed an in silico X. couchianus transcriptome by replacing 90,788 SNP bases for X. maculatus transcriptome with the consensus X. couchianus SNP bases and provide evidence that this procedure overcomes read mapping biases. Employment of the insilico reference transcriptome and tolerating 5 mismatches during read mapping allow direct assessment of ASGE in the F1 interspecies hybrids. Overall, these results show that Xiphophorus is a tractable vertebrate experimental model to investigate how genetic variations that occur during speciation may affect gene interactions and the regulation of gene expression.
Keywords: Interspecies hybrids, Allele Specific Gene Expression, SOLiD, Next Generation Sequencing, SNP detection, Xiphophorus
1 Introduction
The genus Xiphophorus has at least 27 species of live-bearing fishes found from northern Mexico south into Belize and Guatemala (Kallman and Kazianis, 2006). The Xiphophorus genus couples extreme genetic variability among Xiphophorus species with the capability of producing fertile interspecies hybrids that have allowed chromosomal inheritance of complex traits to be followed into individual F1 and backcross hybrid progeny. Induction of cancer in select Xiphophorus interspecies hybrids was an early hallmark of genetic dysregulation within interspecies hybrid genetic background (for reviews see, Walter and Kazianis, 2001; Meierjohann and Schartl,2006). Xiphophorus interspecies hybrids exhibit genetic control of tumor susceptibility for a variety of spontaneous and induced neoplasms, including several melanoma models and less well-studied tumors such as neuroblastomas, neurofibromas, and fibrosarcomas (Schwab et al., 1978a; Schwab et al., 1978b; Schwab et al., 1979; Kazianis et al., 2001a). Studies using various Xiphophorus interspecies crosses have shown reduced DNA repair capabilities in interspecies backcross (BC1) hybrids and severely dysregulated DNA repair activities in some hybrid tissues relative to either parent (Mitchell et al., 1993; Walter et al., 2001; David et al., 2004). Further, in one case, expression of a specific enzyme in the base excision repair pathway was shown to be compromised in the interspecies hybrid genetic background (Heater et al., 2007). Thus, for over 70 years Xiphophorus interspecies hybrids have been used as a valuable model to study the genetics underlying various phenotypes (Walter et al., 2005; Walter et al., 2006a). The use of Xiphophorus in genetic studies led to the early establishment of the Xiphophorus Genetic Stock Center and a pedigreed breeding program for many Xiphophorus lines that stretches over 80 years (Walter et al., 2005; Walter et al., 2006a). This breeding program has produced lines that represent over 100 generations of inbreeding and serve as an unparalleled vertebrate genetic resource.
Despite many reports regarding expression of specific genes within the Xiphophorus interspecies hybrid genetic background, thus far no attempt has been made to assess modulations in gene regulatory patterns on a global transcriptomicscale. Since hybrid gene expression is under the influence of two different and divergent genomes, gene expression levels in hybrids may reveal information about interactions of two parental alleles and their impacts. Allele-specific gene expression (ASGE) within interspecies hybrids has been employed in insect and plant systems to illuminate the direct impact of parental influences on gene regulation (Yan et al., 2002; Pastinen and Hudson, 2004; Main et al., 2009; Tirosh et al., 2009a; Pickrell et al., 2010). Pioneering interspecies hybrid studies between two Drosophila species employed allele specific primer and quantitative RT-PCR to assess gene expression and ASGE levels for 29genes (Wittkopp et al., 2004). More recently specially designed micro array chips capable of differentiating different alleles have been employed to study hybrid gene expression in maize crosses and in yeast interspecies hybrids. Use of these special micro arrays to survey ASGE at a whole transcriptome level suggests that cis-element variations (eg. promoter or enhancer polymorphisms) appeared to be the major contributor to expression differences within the hybrid genetic background (Stupar and Springer, 2006; Tirosh et al., 2009b). However, despite being sensitive and high throughput, microarray based ASGE methods require a deep characterization of genome wide polymorphisms between two species and specially designed probes to assess the abundance of each allele independently. Therefore, it is difficult to study ASGE in less well-characterized species and/or genetic models that possess little information of known polymorphisms. With rapid progress in next generation sequencing technologies (NGS), RNA-Seq technology has now been shown to provide both a single-base resolution and quantitative information of thousands of genes simultaneously (Pastinen, 2010). Notably, this approach does not rely on any previous knowledge of known variations and can be used for both identifying polymorphisms and quantifying ASGE.
Very little is known regarding gene expression and gene interactions within vertebrate interspecies hybrids. We recently developed deep transcriptome reference assembly for the X. maculatus Jp 163 A line and we were able (1) to identify transcriptome-wide SNPs between two highly inbred lines of X. maculatus (Jp 163 A and B) and between these X. maculatus lines and an inbred line of a second species, X. couchianus; (2) to develop reference in silico transcriptomes for each species to eliminate normal read mapping bias, and (3) to directly assess ASGE in the F1interspecies hybrid genetic background. These results indicate the Xiphophorus may serve as a tractable vertebrate experimental model to investigate allele specific gene regulatory effects of interspecies hybridization.
2 Material and methods
2.1 Fish and RNA isolation
All fishes were supplied by the Xiphophorus Genetic Stock Center (seehttp://www.Xiphophorus.txstate.edu/). The X. maculatus Jp 163 B parental line in its100th generation of inbreeding (0.73 g) and one male and one female fishes were sacrificed for RNA when they were sexually dimorphic but immature at 1.5 months post birth. X. couchianus in its 77th generation of inbreeding produced a brood of 10 unsexed fry(0.88 g) that were utilized for RNA isolation at 1.5 month post birth. The parents crossed to produce F1 interspecies hybrids were from 99th and 75th generation of X. maculatus and X. couchianus parental lines, respectively. The X. maculatus Jp 163 B(x) X. couchianus F1 interspecies hybrid RNA was isolated a brood of 10 unsexed fishes (0.84 g) at1 month post birth.
Total RNA was isolated after maceration of whole fish frozen in liquid nitrogen using a pestle followed by resuspension in Trizol (Invitrogen, Carlsbad, CA). RNA was further purified using RNaeasy mini RNA isolation kit (Qiagen, Valencia, CA). Anyresidual DNA was eliminated by performing column DNase digestion at 37 °C (30 min). The integrity of RNA was determined by gel electrophoresis and its concentration was determined using a Nanodrop spectrophotometer.
2.2 RNA-Seq reads: sequencing and Mapping
At least 20 μg total RNA were used for library construction and subjected to ABISOLiD3sequencing (Cofactor Genomics LLC, St. Louis, Mo www.cofactorgenomics.com). Two barcodes of 50 bp single endsequences were run for each sample (X. maculatus, X. couchianus, and the F1 interspecies hybrid, Table 1). Bowtie (Langmead et al., 2009) was used to map raw reads to X. maculatus Jp 163 A reference transcriptome (X. maculatus Jp 163 A, version 3) assembled using total Illumina GAII RNA sequences derived from 10 distinct stages of development or isolated organs from Xiphophorus (cumulatively about13E+08 reads). This reference assembly contained 40,140 contigs with an N50 of2,707 bp and average length of 1,586 bp (Catchen et al., unpublished). Using ABISOLiD data from the three samples (two parental and the F1 hybrid), about four million reads were mapped to X. maculatus reference transcriptome when we set parameters to allow 2 mismatches. This corresponds to about 15 to 18% of all raw reads (Table 1). The mapping percentage is lower compared with other RNA-seq projects (≈50%, Pickrell et al., 2010). This low percentage might be due to the fact that the reference transcriptome rather than genome is used in our case. There are less false-positive mapping since reads will not be mapped to intergenic or other non-related regions. Another possibility is that the current reference transcriptome assembly might not cover all transcribed mRNAs. RNA-seq reads from genes with low expression level or rare alternative splicing isoforms will be less likely to be mapped to current transcriptome assembly.
Table 1.
Deep sequencing and mapping of two Xiphophorus fishes and their F1 hybrid using RNA-Seq
| Fish type | Total # of reads | # of mapped readsa | % of mapped reads | Reads coverageb |
|---|---|---|---|---|
| X. maculatus Jp 163 B | 26,156,645 | 4,816,169 | 18.41 | 3.78 |
| X. couchianus | 31,204,498 | 4,586,358 | 14.70 | 3.60 |
| F1 hybrid | 28,335,078 | 4,348,411 | 15.35 | 3.45 |
reads are mapped to the reference transcriptome of X. maculatus Jp 163 A
average number of reads per base in the reference transcriptome. Total size of transcriptome is 63.6 Mb.
2.3 SNP base calling
Using the Samtools’ pileup program (Li and Durbin, 2009), we examined results of X. couchianus reads mapped to the X. maculatus Jp 163 A reference transcriptome to identify specific SNPs between the two species. A SNP was called when all X. couchianus reads produced a consensus base that was different from that in the X. maculatus reference. Reads from X. maculatus were further tested to rule out possible sequencing or assembly errors that may have led to an incorrect SNP base call in the reference transcriptome. In total, 90,788 SNP locations between X. couchianus and X. maculatus Jp 163 A reference were initially identified using Samtools. Further criteria were used to establish a usable SNP database; at least three reads were needed to support each SNP and all reads had to agree on same consensus base calls at each SNP coordinate – note that this an advantage of using highly inbred parental lines. SNPs that passed this the initial round of quality control were examined and the number of reads covering these SNP positions counted individually in each of the three SOLiD transcriptomes.
2.4 Data normalization and allele-specific expression analysis
Because the X. maculatus Jp 163 A transcriptome served as a reference, X. couchianus allele usage in the F1 interspecies hybrid may be underestimated due to an error that favors mapping for reads from the X. maculatus Jp 163 B allele in orthologous regions of the X. maculatus Jp 163 A reference transcriptome where the X. couchianus read may differ by more than the preset 2 nt mismatch criterion. Thus, using our transcriptome SNP data, we created an in silico X. couchianus reference transcriptome by replacing all bases in the X. maculatus Jp 163 A reference with the consensus SNP base calls identified from the X. couchianus reads. We then changed read mapping parameters to tolerate more mismatches (five) to increase the likelihood of mapping X. couchianus reads. For the X. couchianus parental allele, and in the F1 hybrid, read mapping and counting used this in silico X. couchianus reference transcriptome while the X. maculatus Jp 163 B alleles in the hybrid were mapped and counted based on the X. maculatus Jp 163 A reference transcriptome. A custom written Perl script was utilized to count reads carrying species-specific alleles separately for each SNP. We then combined read counts from all SNPs mapping to each gene and normalized them based on total number of mapped reads in each sample to eliminate the differences in number of reads obtained from sequencer in the first place. For F1 hybrid read counting, genes showing less than 20 SNP supporting reads were discarded to increase the power of allele-specific expression data at the expense of losing rare transcript representation.
3 Results
3.1 Deep sequencing and mapping of RNA-Seq reads
RNA-Seq technology has proven to be an efficient way to uncover structures of transcriptomes and to assess allele-specific gene expression (Pastinen, 2010). To measure ASGE patterns in a Xiphophorus interspecies model, RNA-Seq was employed to amplify and sequence cDNA fragments from two highly inbred fish lines: X. maculatus (Jp 163 B) X. couchianus and their F1 interspecies hybrids. Two barcodes of ABI-SOLiD 3 sequencing were run for each RNA sample derived from of age-matched(≈1.5 month old) fry. These sequencing runs produced from 26 to 31 million 50 bp single end reads for these samples (Table 1).
RNA-seq reads were mapped to a reference Xiphophorus transcriptome de novo assembled from 10 life stages or organs of X. maculatus Jp 163 A that had been produced using the Illumina NGS platform (120 million RNA-seq reads of 36 and 60 nt, a mix of single and paired-end reads; Catchen et al, unpublished). This reference transcriptome contains 40,140 transcripts with an average length of 984 bp. Upon mapping the RNA-seq reads to the assembled reference transcriptome we determined 18.4% of the X. maculatus Jp 163 B, and 14.7% of the X. couchianus, reads were able to produce at least one alignment to the transcriptome. Among X. maculatus Jp 163 B, X. couchianus, and their F1 interspecies hybrids, it is not surprising that reads from X. maculatus Jp 163 B had higher read mapping percentage than X. couchianus due to the expected divergence between these fishes and the Jp 163 A reference transcriptome. RNA-seq reads from the F1 hybrid exhibited an intermediate mapping percentage(15.4%) compared with the two parental species. For every base in the reference transcriptome, there are on average 3.5 reads mapped from each of the fish types (Table 1), and this provides a foundation for consensus base-calling.
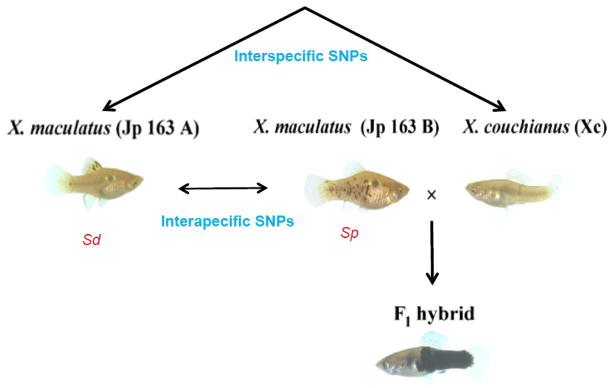
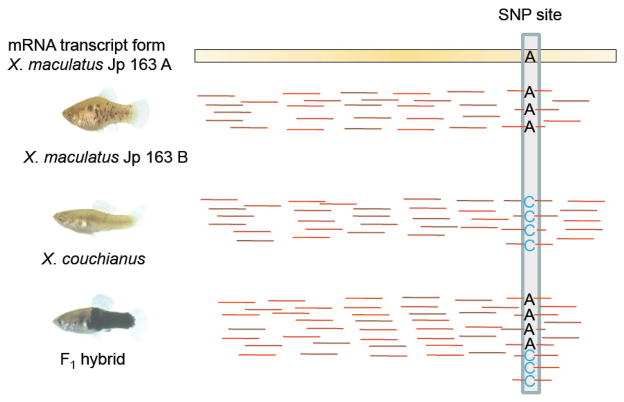
3.2 Identification of intraspecific and interspecific SNPs in Xiphophorus
The Xiphophorus Genetic Stock Center (XGSC, see http://www.xiphophorus.txstate.edu/) currently maintains fourteen X. maculatus lines. The X. maculatus Jp 163 A and Jp 163 B lines represent highly inbred stocks often used in tumor induction studies. These two lines are derived from a single female collected in 1939 that upon reaching the laboratory produced a brood of progeny carrying two distinct macromelanophore spotting patterns (Figure 1) termed, spot dorsal(Sd) and spot side (Sp)(Kallman, 1970). Based on this difference in phenotype, the two X. maculatus lines (Jp 163 A and B) were split around 1944 and have been maintained separately in the XGSC for 106 and 100 generations, respectively, prior to use in these studies. Despite several phenotypic differences between the two lines and documented distinct behavior with regard to tumor induction among interspecies hybrids (Walter and Kazianis, 2001; Meierjohann and Schartl, 2006), little is known about the precise genetic variations separating these two X. maculatus lines. To characterize SNP polymorphisms between two X. maculatus lines, we mapped RNA-seq reads generated from Jp 163 B onto the Jp 163 A reference transcriptome assembly and identified variations between the two transcriptomes (Figure 2). Reads were “piled-up” based on coordinates corresponding to the transcriptome assembly and a SNP site was “called” when all reads from Jp 163 B were found to be different from the reference transcriptome at that particular coordinate. To further improve the quality of SNP base calling, we used criteria requiring that at least three RNA-seq reads be present to map each coordinate in order to call the SNP. In toto, 1,294 SNPs were identified (“called”) between the two X. maculatus lines, Jp 163 A and Jp 163 B. The density of intraspecific SNPs is about 1 SNP per 49 kb of transcriptome.
Figure 1.
Fishes used in this study. X. maculatus Jp 163 A carrying the Sd pigmentpattern is the species utilized for deep transcriptome development and eventual assembly of the reference transcriptome. F1 interspecies hybrids utilized in these studies were produced by crossing the X. maculatus Jp 163 B (Sp pigment pattern) and X. couchianus parental species. RNA-seq reads analyzed in this study were sequenced from X. maculatus Jp 163 B, X. couchianus and their F1 interspecies hybrids respectively.
Figure 2.
A diagrammatic example of identification of SNPs and measurement of ASGE in F1 interspecies hybrids. Red bars represent RNA-Seq reads mapped to the reference transcriptome. Most reads from X. maculatus Jp 163 B match perfectly to the Jp 163 A reference transcriptome. RNA-seq reads from X. couchianus were also mapped to X. maculatus Jp 163 A reference transcriptome and SNPs sites were identified by comparing consensus bases of RNA-seq reads (C in this case) to the corresponding base in the reference transcriptome (A in this case). In the hybrid, reads mapped to SNPs sites are classified by the bases they carry and counted separately as the measurement of ASGE. In this SNP, 4 X. maculatus allele reads and 3 X. couchianus allele reads were counted in the hybrid.
In addition to intraspecific SNPs, we characterized polymorphisms between two Xiphophorus species. To do this, X. couchianus reads were mapped onto the X. maculatus Jp 163 A reference transcriptome and SNPs were identified using similar methods and criteria as described above (Figure 2). SNPs were called when a consensus base in a mapped X. couchianus RNA-seq read was clearly different from the X. maculatus base in that position. Overall, we were able to identify 90,788 SNPs between X. maculatus Jp 163 A and X. couchianus species, with an overall density of 1SNP per 700 bp of transcriptome.
3.3 Unbiased mapping of reads from Xiphophorus F1 interspecies hybrid
Xiphophorus fishes have been well-documented to produce fertile interspecies hybrid progeny upon crossing two parental species lines (Walter and Kazianis, 2001). This makes them quite different from most vertebrates where interspecies hybrids are difficult to produce or are infertile. Use of Xiphophorus interspecies hybrids in tumor induction protocols and other studies has indicated that novel phenotypes may arise from the interaction of two genomes. However, the genetic mechanisms involved in producing novel phenotypes within interspecies hybrids are not well understood.
We wished to ascertain ASGE between X. maculatus Jp 163 B, X. couchianus and the F1 hybrid produced from these two species. Melanoma induction phenotype in this hybrid is well documented where backcross fishes of this particular cross develop melanoma after MNU or UVB treatment (Kazianis et al., 2001a). Thus, we first determined that the90,788 SNPs, identified between X. maculatus Jp 163 A and X. couchianus (see above)were also polymorphic between X. maculatus Jp 163 B and X. couchianus. To further improve the accuracy of ASGE analysis in the hybrid, we scored only genes that exhibited greater than 20 SNP supporting reads. These quality control criteria resulted in our focus on 38,746 SNPs between X. maculatus Jp 163 B and X. couchianus that could be clearly assigned to one or the other parental alleles. These 38,746 SNPs were unambiguously mapped to 6,524 Xiphophorus contigs (i.e., transcripts) in the reference transcriptome. On average, there are ≈75 + species-specific RNA-Seq reads pertranscript in the hybrid RNA-seq data allowing a reliable estimation of the relative abundances for each parental allele in the F1 hybrid transcriptome.
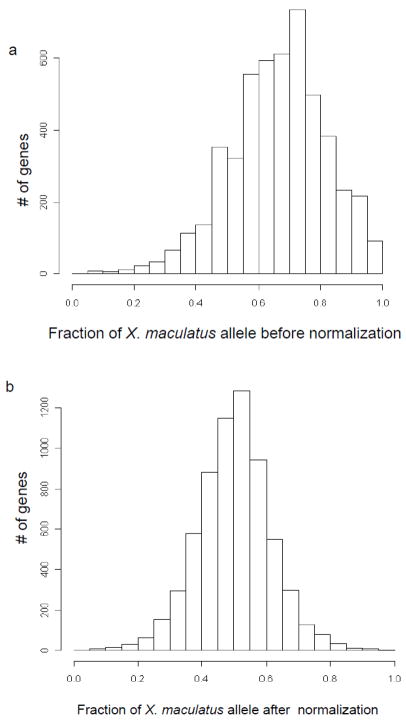
To assess ASGE, RNA-Seq reads derived from each parental allele were mapped to transcripts based on allele-specific SNPs previously identified and the frequency of reads for each allele was determined for each transcript. To test if this method can accurately measure allele expression in hybrids, we plotted the distribution of the ratio of X. maculatus Jp 163 B SNP reads to all mapped reads for each transcript(Figure 3a). Transcripts that exhibit a fraction larger than 0.5 are those that mapped more X. maculatus Jp 163 B reads than X. couchianus ones in the hybrid genetic background. Those, with ratios of less than 0.5 indicate genes where the X. couchianus alleles were expressed at a higher level than the X. maculatus Jp 163 B allele. As shown in Figure 3a, we found over 84% of genes in the transcriptome were biased toward over-representation of the X. maculatus allele (fraction > 0.5). Because our reference transcriptome was developed from X. maculatus Jp 163 A, it was not surprising the RNA-seq reads from the X. maculatus Jp 163 B parental alleles mapped with better efficiency than those from X. couchinaus (using Bowtie default parameters) in the F1 hybrid. Bowtie only allow two mismatches between reads and reference sequences, reads carrying X. couchianus allele have natural disadvantages in mapping efficiency since they carry an extra mismatch (SNP) compared with X. maculatus reads. To derive a better representation of ASGE in F1 hybrids, we took several steps to eliminate this bias.
Figure 3.
Allele distribution in F1 hybrid background. A: A histogram shows the distribution of F1 transcripts carrying different parental alleles before normalization. X axis is the fraction of reads carrying X. maculatus allele. 0.5 means in that gene, half of F1 hybrid reads can be identified from X. couchianus and another half are from X. maculatus. 1.0 and 0.0 means reads exclusively carrying X. maculatus and X. couchianus alleles, respectively. B: Fraction of X. maculatus in hybrid background after normalization. We masked X. maculatus reference with consensus bases from X. couchianus and allowing five mapping mismatches.
To eliminate the read count bias created from a single reference transcriptome source (i.e., X. maculatus Jp 163 A), and thus perform head-to-head comparison in allele specific expression, we created an in silico X. couchianus reference transcriptome by constructing a dataset wherein each of the 90,788 SNP bases in the X. maculatus Jp163 A reference were replaced with the consensus X. couchianus SNP bases we had previously identified. The induction of X. couchianus reference transcriptome allows reads with X. couchianus allele have comparable chances of being counted in ASGE study. We also adjusted our read mapping parameters within the Bowtie program to tolerate more mismatches (5 mismatches) to increase the likelihood of mapping X. couchianus reads onto the X. couchianus in silico transcriptome reference. We recounted allele frequencies in the F1 hybrid and plotted the distribution of alleleratios as above (Figure 3b). As shown, using the in silico corrected reference transcriptome allowed both X. maculatus and X. couchianus alleles to exhibit balanced expression patterns in the hybrid genetic background. In this case, 51% of genes are biased toward over representation of X. maculatus allele (fraction > 0.5), compared to84% bias towards X. maculatus allele using the X. maculatus reference transcriptome (Figure 3a).
3.4 Assessment of ASGE in Xiphophorus F1 interspecies hybrid
Having established steps to eliminate ASGE mapping bias in the hybrid reads, the number of X. maculatus and X. couchianus RNA-seq reads corresponding to each species allele appear to be fairly symmetrical in the F1 hybrids (51% to 49%, Figure 3b). Detailed analyses of the distribution of ASGE in F1 hybrids indicate that most genes(5,980 of 6,524 genes or 92%) exhibit relatively balanced allele expression in the hybrid genetic background (<70% of preference of one allele, those between 0.3 and 0.7 in Figure 3b). However, 544 genes showed strong preference for expression of a single parental allele in the F1 hybrid genetic background (>70% preference of one allele, fraction <0.3 or >0.7, Figure 3b).
To further investigate genes showing strong allele expression bias in the F1 hybrids, we analyzed 27 genes exhibiting expression almost exclusively from one parental allele (over 90% from one parent, Table 2). Among these 27 allele-specific expressed genes, 15 predominantly expressed the X. maculatus allele while the other12 expressed primarily the X. couchianus allele. Blast annotation of these X. maculatus Jp 163 A transcripts indicated that 12 of them are homologous to known and functionally characterized genes. Genes having kinase activity or involving metabolic pathways such as dehydrogenases or hydrolases are common in this list. In addition, two genes (unc-13 homolog b and perforin 1), thought to be involved in apoptosis are divergently regulated in F1 hybrids.
Table 2.
27 genes show bias towards the usage of one parental allele in F1 hybrid
| Gene IDa | Length(bp) | # of XMb reads in F1 | # of XCb reads in F1 | Blast best hit |
|---|---|---|---|---|
| G012369 | 743 | 22 | 0 | ---NA--- |
| G024659 | 2424 | 86 | 3 | ubiquitin-conjugating enzyme e2g 2 |
| G023261 | 11830 | 28 | 1 | pr gag-pro-pol |
| G004299 | 776 | 105 | 5 | unnamed protein product [Tetraodon nigroviridis] |
| G008510 | 5192 | 20 | 1 | protein tyrosine phosphatase-like a domain containing 1 |
| G030907 | 1127 | 783 | 43 | ---NA--- |
| G013530 | 579 | 33 | 2 | ---NA--- |
| G011143 | 1834 | 28 | 2 | ---NA--- |
| G021904 | 469 | 82 | 7 | ---NA--- |
| G000614 | 1458 | 23 | 2 | ---NA--- |
| G013483 | 949 | 21 | 2 | dual specificity phosphatase 14 |
| G007003 | 418 | 223 | 22 | inter-alpha inhibitor h3 |
| G005248 | 3969 | 20 | 2 | unc-13 homolog b ( elegans) |
| G010297 | 3224 | 20 | 2 | novel kruppel-like factor |
| G003097 | 238 | 177 | 19 | ---NA--- |
| G021727 | 1707 | 2 | 19 | ---NA--- |
| G039744 | 2614 | 3 | 29 | novel protein vertebrate acyl- thioesterase |
| G033133 | 504 | 2 | 20 | ---NA--- |
| G037369 | 471 | 3 | 30 | nadh dehydrogenase 1 beta subcomplex subunit 1 |
| G012123 | 339 | 2 | 21 | ---NA--- |
| G001545 | 714 | 6 | 64 | perforin 1 |
| G027192 | 608 | 4 | 43 | ---NA--- |
| G018381 | 487 | 2 | 22 | serine (or cysteine) proteinase inhibitor |
| G009372 | 680 | 2 | 25 | ---NA--- |
| G000402 | 2523 | 7 | 99 | methylmalonate-semialdehyde dehydrogenase |
| G037005 | 1055 | 2 | 49 | ---NA--- |
| G003510 | 339 | 0 | 71 | ---NA--- |
Gene ID as in X. maculatus transcript assembly (Catchen et al., unpublished).
abbreviation: XM X. maculatus, XC X. couchianus
Nucleotide sequences were searched against GenBank nr (All non-redundant protein) database using blastx with a minimum e-value of 1.0E-3. No hit found means no homologous sequence is found that meets the minimum e-value cutoff.
4 Discussion
The study of ASGE in interspecies hybrids can take either of two general approaches (Pastinen, 2010). The first, a polymorphism-directed approach, utilizes known genome variants and can achieve highly specific ASGE results (Wittkopp et al.,2004; Stupar and Springer, 2006). A second approach employs specially designed SNParrays to allow concurrent examination of tens of thousands of ASGE sites (Tirosh et al., 2009b; Zhang and Borevitz, 2009). While informative, both of these approaches require previous knowledge of precise polymorphisms between two species and usually involve species with fully sequenced genomes, and thus cannot be translated to studies using species that may not have extensive genomic resources available. We present a different general approach to provide an unbiased view of gene regulation between two species and their interspecies hybrid that has been developed using deep transcriptome sequencing of each parental species (Babak et al., 2008; Hillier et al., 2008; Serre et al.,2008; Main et al., 2009; Emerson et al., 2010; Heap et al., 2010). This approach simultaneously provides single-base resolution for identifying polymorphisms, quantitative information regarding the expression of thousands of genes, and does not rely on previous knowledge of known genetic variation. Using both 454 and Illumina sequencing platforms respectively, allelic expression imbalances have been assessed in Drosophila hybrids and in humans (Serre et al., 2008; Daelemans et al., 2010; Fontanillas et al., 2010). Here we present results employing ABI-SOLiD (Sequencing by Oligonucleotide Ligation and Detection) reads to identify SNP polymorphisms between two Xiphophorus species and to quantify ASGE within F1 interspecies hybrids. Compared with Illumina sequencing technology, the bicolor space character of ABISOLiD (Sequencing by Oligonucleotide Ligation and Detection) platform and its comparatively low error rate [0.1%~0.2%; 1–2% in Illumina reads (McKernan et al.,2009)] make SOLiD data extremely well suited for identification of SNPs and measurement of ASGE.
The extreme genetic variability among Xiphophorus species and ability to produce fertile interspecies hybrids make this a good candidate system for exploration of evolutionary divergence in gene expression and to study complex gene interactions. Herein, we identified a density of SNPs between X. maculatus Jp 163 A and Jp 163 B fish lines of 1 per 49 kb of transcriptome. These two Jp 163 lines are derived from the progeny of a single collection of several dozen platyfish from the Rio Jamapa, Veracruz, Mexico in 1939 (Kallman, 2001). Once in the laboratory two types of macromelanophore pigment patterns; “spot dorsal” (Sd) and “spot side” (Sp) were observed. The Sd pattern results in black spots (macromelanophores) on the dorsal fin while Sp compartmentalizes black spots onto the flanks of the animals (Figure 1). A single cross of an Sp female with an Sd male gave rise to the pedigree Jp 163 (Jp for the RioJamapa and 163, as the cross number; Gordon, 1947). After nine generations of closed colony random sibling mating, progeny from the Jp 163 pedigree were split into two distinct lines based on the macromelanophore Sd (Jp 163 A) and Sp (Jp 163 B) pigment patterns. These two Jp 163 lines have been and inbred by brother-sister mating eversince (Kalman, 2001 Kalman, 1965). The Sd and Sp traits are considered alleles and both are linked to a “melanoma determining locus” (mdl) on the X. maculatus X chromosome(Walter et al., 2006b; Meierjohann, S., Schartl, M., 2006). Thus, RNA-Seq reads we recollected from Jp 163 A and Jp 163 B fish lines that had been separated in the laboratory for about 91 generations spanning 67 years. Assuming most polymorphisms in the two parents became homogenous during the 9 generations of colony breeding, and than fixed when the Jp 613 A and B lines were established by inbreeding, the SNPsidentified here may be expected to have derived from variations occurring after separation of the two lines. If so, we may estimate a mutation rate in the Xiphophoru stranscriptome to be about 2.24 ×10−7per base per generation or 3.09 ×10−7per base per year. This number is comparable to the estimated human genome mutation rate, which is similarly estimated to be ~1.1×10−8per base per generation (Roach et al.,2010). The mutation rate calculated in the Xiphophorus transcriptome is the first such estimate in laboratory bred fishes. With the current trancriptomes for each line as a fixed time point, we can reassess variation within the Jp 163 A and B fish lines periodically and assess both mutation rate and distribution in the X. maculatus transcriptome as time moves forward.
In this paper, we characterized transcriptome-wide polymorphisms between two Xiphophorus species and assessed the ASGE pattern in their F1 hybrid. This work paves the way for further understanding of variations in select gene sets that have occurred as the parental species diverged. For example, each of the 544 genes showing aberrant regulation in the F1 interspecies hybrid, relative to either parent, may have their own divergent pattern that results in altered gene regulation. It will be interesting to see if sets of these genes share common modalities leading to similar regulatory dysfunction in the hybrid genetic background.
After two species diverge from a common ancestor, changes in gene expression within each new species occur in cis-regulatory and/or trans-regulatory elements(Wittkopp et al., 2004). Alteration of cis-elements may affect the strength of promoters, action of enhancers, transcript stability, etc., whereas trans-element changes may involve structure, binding affinities, or intercellular levels of factors that influence transcription. If variations happen in cis-elements, the ratio of expression levels of two parental alleles will be the same as the ratios of two parental alleles in the hybrids. On the other hand, if trans-element changes occur in the parents, two alleles should show no difference in expression within the F1 hybrid because both alleles are exposed to the same subcellular environment. Therefore, comparison of expression levels for alleles derived from two diverged species within an F1 interspecies hybrid allows functional assessment of cis- and trans-regulatory variations that have occurred in the two parents.
In this study, only young (1.5 mo) intact fish were used for the RNA source and thus tissues or life stage specific effects on parental allele expression may have been muted. How age, tissue specificity, stress induction, different interspecies crosses, or cross directions may affect allele specific gene expression patterns within the many interspecies crosses possible among the 27 Xiphophorus species leaves much to be determined. The advent of next generation sequencing and the power of RNA-seq methodologies plays to many of the strengths of the Xiphophorus model for utility in exploration of gene regulatory and gene interaction phenomena upon interspecies hybridization.
Acknowledgments
The authors would like to thank Markita Savage and the other employees of the Xiphophorus Genetic Stock Center, Texas State University, for maintaining the pedigreed fish lines, performing interspecies crosses, and caring for the hybrid animals used in this study. This work was supported by Texas State University and the National Institutes of Health, National Center for Research Resources grant #R24-RR024790 including an American Recovery and Reinvestment act supplement to this award.
Footnotes
This paper is based on a presentation given at the 5th Aquatic Annual Models of Human Disease conference: hosted by Oregon State University and Texas State University-San Marcos, and convened at Corvallis, OR, USA September 20-22, 2010.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babak T, DeVeale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP. Global Survey of Genomic Imprinting by Transcriptome Sequencing. Current Biol. 2008;18:1735–1741. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- Daelemans C, Ritchie ME, Smits G, Abu-Amero S, Sudbery IM, Forrest MS, Campino S, Clark TG, Stanier P, Kwiatkowski D, Deloukas P, Dermitzakis ET, Tavare S, Moore GE, Dunham I. High-throughput analysis of candidate imprinted genes and allele-specific gene expression in the human term placenta. BMC Genet. 2010;11:25. doi: 10.1186/1471-2156-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David WM, Mitchell DL, Walter RB. DNA repair in hybrid fish of the genus Xiphophorus. Comp Biochem Physiol C Toxicol Pharmacol. 2004;138:301–309. doi: 10.1016/j.cca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Hsieh LC, Sung HM, Wang TY, Huang CJ, Lu HH, Lu MY, Wu SH, Li WH. Natural selection on cis and trans regulation in yeasts. Genome Res. 2010;20:826–836. doi: 10.1101/gr.101576.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanillas P, Landry CR, Wittkopp PJ, Russ C, Gruber JD, Nusbaum C, Hartl DL. Key considerations for measuring allelic expression on a genomic scale using highthroughput sequencing. Mol Ecol. 2010;19(Suppl 1):212–227. doi: 10.1111/j.1365-294X.2010.04472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. Genetics of Platypoecilus mauclatus, IV; the sex determining mechanosm in two wild populaions on Mexican platyfish. Genetics. 1947;32:8–17. doi: 10.1093/genetics/32.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap GA, Yang JH, Downes K, Healy BC, Hunt KA, Bockett N, Franke L, Dubois PC, Mein CA, Dobson RJ, Albert TJ, Rodesch MJ, Clayton DG, Todd JA, van Heel DA, Plagnol V. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum Mol Genet. 2010;19:122–134. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heater SJ, Rains JD, Wells MC, Guerrero PA, Walter RB. Perturbation of DNA repair gene expression due to interspecies hybridization. Comp Biochem Physiol C Toxicol Pharmacol. 2007;145:156–163. doi: 10.1016/j.cbpc.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, Magrini VJ, Richt RJ, Sander SN, Stewart DA, Stromberg M, Tsung EF, Wylie T, Schedl T, Wilson RK, Mardis ER. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- Kallamn KD. Different genetic basis of identica pigment patterns in two populations of platyfish, Xiphophorus maculatus. Copeia. 1970;3:472–487. [Google Scholar]
- Kallman KD. How the Xiphophorus problelm arrived in San Marcos, Texas. Mar Biotechnol (NY) 2001;3:S6–16. doi: 10.1007/s10126-001-0022-5. [DOI] [PubMed] [Google Scholar]
- Kallman KD, Kazianis S. The genus Xiphophorus in Mexico and central america. Zebrafish. 2006;3:271–285. doi: 10.1089/zeb.2006.3.271. [DOI] [PubMed] [Google Scholar]
- Kazianis S, Gimenez-Conti I, Setlow RB, Woodhead AD, Harshbarger JC, Trono D, Ledesma M, Nairn RS, Walter RB. MNU induction of neoplasia in a platyfish model. Lab Invest. 2001a;81:1191–1198. doi: 10.1038/labinvest.3780333. [DOI] [PubMed] [Google Scholar]
- Kazianis S, Gimenez-Conti I, Trono D, Pedroza A, Chovanec LB, Morizot DC, Nairn RS, Walter RB. Genetic analysis of neoplasia induced by N-nitroso-Nmethylurea in Xiphophorus hybrid fish. Mar Biotechnol (NY) 2001b;3:S37–43. doi: 10.1007/s10126001-0025-2. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main BJ, Bickel RD, McIntyre LM, Graze RM, Calabrese PP, Nuzhdin SV. Allele-specific expression assays using Solexa. BMC Genomics. 2009;10:422. doi: 10.1186/1471-2164-10-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, Zhang Z, Ranade SS, Dimalanta ET, Hyland FC, Sokolsky TD, Zhang L, Sheridan A, Fu H, Hendrickson CL, Li B, Kotler L, Stuart JR, Malek JA, Manning JM, Antipova AA, Perez DS, Moore MP, Hayashibara KC, Lyons MR, Beaudoin RE, Coleman BE, Laptewicz MW, Sannicandro AE, Rhodes MD, Gottimukkala RK, Yang S, Bafna V, Bashir A, MacBride A, Alkan C, Kidd JM, Eichler EE, Reese MG, De La Vega FM, Blanchard AP. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierjohann S, Schartl M. From Mendelian to molecular genetics: the Xiphophorus melanoma model. Trends Genet. 2006;22:654–661. doi: 10.1016/j.tig.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Mitchell DL, Scoggins JT, Morizot DC. DNA repair in the variable platyfish (Xiphophorus variatus) irradiated in vivo with ultraviolet B light. Photochem Photobiol. 1993;58:455–459. doi: 10.1111/j.1751-1097.1993.tb09590.x. [DOI] [PubMed] [Google Scholar]
- Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet. 2010 doi: 10.1038/nrg2815. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Abdo S, Ahuja MR, Kollinger G, Anders A, Anders F, Frese K. Genetics of susceptibility in the platyfish/swordtail tumor system to develop fibrosarcoma and rhabdomyosarcoma following treatment with N-methyl-N-nitrosourea (MNU) Z Krebsforsch Klin Onkol Cancer Res Clin Oncol. 1978a;91:301–315. doi: 10.1007/BF00312292. [DOI] [PubMed] [Google Scholar]
- Schwab M, Haas J, Abdo S, Ahuja MR, Kollinger G, Anders A, Anders F. Genetic basis of susceptibility for development of neoplasms following treatment with Nmethyl- N-nitrosourea (MNU) or x-rays in the platyfish/swordtail system. Experientia. 1978b;34:780–782. doi: 10.1007/BF01947324. [DOI] [PubMed] [Google Scholar]
- Schwab M, Kollinger G, Haas J, Ahuja MR, Abdo S, Anders A, Anders F. Genetic basis of susceptibility for neuroblastoma following treatment with N-methyl-Nnitrosourea and X-rays in Xiphophorus. Cancer Res. 1979;39:519–526. [PubMed] [Google Scholar]
- Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, Bibikova M, Chudin E, Barker DL, Dickinson T, Fan JB, Hudson TJ. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Springer NM. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics. 2006;173:2199–2210. doi: 10.1534/genetics.106.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N, Verstrepen KJ. Promoter architecture and the evolvability of gene expression. J Biol. 2009a;8:95. doi: 10.1186/jbiol204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A Yeast Hybrid Provides Insight into the Evolution of Gene Expression Regulation. Science. 2009b;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- Walter RB, Ju Z, Martinez A, Amemiya C, Samollow PB. Genomic resources for Xiphophorus research. Zebrafish. 2006a;3:11–22. doi: 10.1089/zeb.2006.3.11. [DOI] [PubMed] [Google Scholar]
- Walter RB, Hazlewood L, Kazianis S. In: The Xiphophorus Genetic Stock Center Manual. 1. Kallman Klaus D, Schartl Manfred., editors. Texas State University; 2006b. p. 113. [Google Scholar]
- Walter RB, Kazianis S, Hazlewood L, Johnston DJK. The Xiphophorus Genetic Stock Center. In: Grier MCUaH., editor. Viviparous Fishes. New Life Publications; Homestead, FI: 2005. pp. 343–350. [Google Scholar]
- Walter RB, Sung HM, Obermoeller RD, Mitchell DL, Intano GW, Walter CA. Relative base excision repair in Xiphophorus fish tissue extracts. Mar Biotechnol (NY) 2001;3:S50–60. doi: 10.1007/s10126-001-0027-0. [DOI] [PubMed] [Google Scholar]
- Walter RB, Kazianis S. Xiphophorus interspecies hybrids as genetic models of induced neoplasia. ILAR J. 2001;42:299–321. doi: 10.1093/ilar.42.4.299. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Zhang X, Borevitz JO. Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics. 2009;182:943–954. doi: 10.1534/genetics.109.103499. [DOI] [PMC free article] [PubMed] [Google Scholar]