Abstract
We generated from a single blood sample five independent human monoclonal antibodies that recognized the Sa antigenic site on the head of influenza HA and exhibited inhibitory activity against a broad panel of H1N1 strains. All five Abs used the VH3-7 and JH6 gene segments, but at least four independent clones were identified by junctional analysis. High throughput sequence analysis of circulating B cells revealed that each of the independent clones were members of complex phylogenetic lineages that had diversified widely using a pattern of progressive diversification through somatic mutation. Unexpectedly, B cells encoding multiple diverging lineages of these clones, including many containing very few mutations in the antibody genes, persisted in the circulation. Conversely, we noted frequent instances of amino acid sequence convergence in the antigen combining sites exhibited by members of independent clones, suggesting a strong selection for optimal binding sites. We suggest that maintenance in circulation of a wide diversity of somatic variants of dominant clones may facilitate recognition of drift variant virus epitopes that occur in rapidly mutating virus antigens, such as influenza HA. In fact, these Ab clones recognize an epitope that acquired three glycosylation sites mediating escape from previously isolated human antibodies.
Introduction
Induction and maintenance of a diversity of broadly neutralizing antibodies against viruses is desirable for immunity against reinfection, but the molecular features of human antibody repertoires specific for particular agents or epitopes is poorly understood. Isolation of limited panels of epitope-specific human monoclonal antibodies to viruses has suggested that the circulating human B cell response often is dominated by major clonal populations. In vivo selection in germinal centers of particular B cell clones using B cell receptors with high-affinity binding to virus epitopes likely leads to expansion of dominant clonal populations. The extent to which a dominant clone of B cells responding to a viral epitope represents a single B cell receptor with an optimal affinity for binding, versus a family of related somatic variants, has not been determined in the past because of the difficulty in generating large numbers of human antibodies.
The 2009 H1N1 influenza pandemic was the first influenza pandemic in over 40 years. Pediatric death rates were 10 times the rates for seasonal influenza in previous years (1). Elderly people had preexisting cross-reactive antibodies against this 2009 H1N1 virus (2–4). Preserved epitopes within H1N1 HA were the likely structural correlate for this cross-reactivity, particularly the Sa antigenic site on the globular head (5–7). We had shown previously that the Sa site-specific Ab 2D1 that was cloned from a survivor of the 1918 pandemic potently neutralized 2009 pandemic virus (5, 8). We elucidated the crystal structure of Ab2D1 in complex with 1918 HA (9). Ab2D1 uses the VH2-70 germline gene with a unique insertion close to CDRH2 that enhances the function of the antibody (8, 10).
Here, we describe a panel of H1N1-specific antibodies that was cloned from a 47 year old healthy woman after the pandemic. Like Ab2D1, Abs from this new panel bound the Sa site, but they shared VH3-7/JH6 germline gene usage and had HAI activity against a broader panel of H1N1 strains than 2D1, including viruses with glycosylation sites in the Sa site. These VH3-7/JH6 antibodies belonged to four different clones that arose independently, yet converged towards similar amino acid sequences. Ultra deep sequencing has been used previously to determine the combinatorial diversity of antibodies (11–14). We used this technology to elucidate the VH3 repertoire of this donor to find related antibody sequences using the VH3-7/JH6 H chain gene segments, to more fully define the molecular diversity of an epitope-specific human antibody repertoire. The data revealed unexpected features of the evolution of antibody repertoires and the persistence of corresponding B cells in the peripheral blood.
Materials and Methods
Hybridoma generation and recombinant antibody expression
Acquisition of human blood samples was approved by the Vanderbilt University Institutional Review Board. The animal studies were approved by the Institutional Review Boards of the CDC. PBMCs were isolated from a 47-year old healthy female donor with Histopaque-1077 (Sigma), EBV-transformed in 384 well plates (Nunc) in the presence of 2.5 μg/mL CpG ODN 2006 (Invivogen), 10 μM of Chk2 inhibitor II (Sigma C3742), and 1 μg/mL cyclosporine A (Sigma), essentially as described previously (10, 15). Supernatants from wells containing EBV-transformed lymphoblastoid cell lines were screened for binding activity by ELISA against a panel of recombinant soluble HA proteins. Positive wells were fused with HMMA2.5 myeloma cells and cloned molecularly using previously described primer sets (16) into pGEM-T Easy vector (Promega) and eventually into pEE12.4/pEE6.4 mammalian expression vectors (Lonza) from where they were expressed (9) and purified on a protein G column using an ÅKTA chromatography instrument (GE). All following studies were performed using recombinant Abs. We used Kabat numbering as determined using the Abnum server (17) for the antibodies and an H3 numbering scheme (18) for HA. Antibody clonality was defined strictly by shared VH gene, shared VDJ junction and a sequence of shared somatic mutations.
Generation and purification of recombinant soluble HA molecules
1918 or 2009 influenza HA constructs were ordered sequence-optimized for expression in human cells (GenScript) based on the extracellular domain of the respective HA, a thrombin recognition cleavage site, a fibritin trimerization domain, and a 6× His-tag (18). The constructs were cloned into pcDNA3.1 (+) (Invitrogen), expressed in 293F cells (Invitrogen), purified over nickel columns using an ÅKTA chromatography instrument (GE), and concentrated with Amicon Ultra centrifugal filters with a 30 kD molecular weight cut-off (Millipore).
ELISA
384-well clear plates (Nunc 242757) were coated with HA at 1 μg/mL in D-PBS overnight, blocked with 0.5% cow milk, 0.2% goat serum, and 0.05% TWEEN 20 (Sigma) in D-PBS. Five μL of hybridoma supernatant per well were transferred to 25 μL of blocking solution with a multi-channel pipette. Secondary goat anti-human IgG antibody (Meridian Life Science W99008A) was diluted 1:8000 in blocking solution and added after four automated washing steps. After another wash, phosphatase substrate (Sigma S0942) was dissolved in substrate buffer per the instructions of the manufacturer and dispensed onto the plates. The plates were read at 405 nm on a Power Wave HT (BioTek).
VLP expression and HAI assays
Expression plasmids encoding the parenteral or mutated 1918 HA were co-expressed with N1 neuraminidase to produce VLPs in 293T cells (10, 19). Two days post-transfection, supernatants were collected. HAI assays were performed as described (20) using VLPs (for 1918 H1N1 or H2) or live virus (all other influenza strains). Briefly, serially diluted antibodies were pre-incubated with eight hemagglutinating units of virus or VLP per well. Chicken red blood cells were added to a final concentration of 0.5%, and the plate was incubated on ice for 30 to 60 min.
In vivo antiviral effect of Ab4K8
Female 8-week-old BALB/c mice were inoculated intranasally with 5× LD50 in a 50 μL volume of the virulent reconstituted 1918 influenza virus. At 24 h after inoculation, mice were each administered 200, 20, or 2 μg (approximately 10, 1, or 0.1 mg/kg) of Ab4K8 or an equal volume of polyclonal human IgG (Sigma) by the i.p. route in groups of 10 mice. Mice were observed for weight loss for 14 days. Subsets of four animals treated with Abs were euthanized on day 4 after inoculation, and whole lungs were homogenized in 1 mL of sterile PBS. Virus titers in lung tissue homogenates were determined by plaque titration in Madin-Darby Canine Kidney cell monolayer cultures and expressed as log10 PFU/mL.
Isolation and characterization of antibody escape mutant viruses
We selected new antibody escape mutant viruses by incubating virus with neutralizing antibodies followed by inoculation in 10-day-old embryonated chicken eggs, essentially as described (21, 22). RNA was extracted from virus-infected allantoic fluid, then cDNA was generated by RT-PCR, cloned molecularly, and sequenced. The K166 escape mutations found in selected virus mutants were built back into 1918 VLPs for use in HAI assays to confirm that these alterations mediated escape from neutralization (10).
Pyrosequencing
Total RNA was extracted from 30 million PBMCs (RNeasy kit, Qiagen). A first round PCR was performed for 35 cycles using BIOMED-2 VH3 framework 1/JH primers (23) and the OneStep RT-PCR kit (Qiagen). 454-specific adapters (Roche) were added by 10 cycles of a second round PCR. The PCR products were sequenced on a GS Junior instrument (Roche). The primers were cartridge-purified (Invitrogen); the sequences were CCATCAGCTGGGGGGTCCCTGAGACTCTCCTG (forward) and CGCTCAGCTTACCTGAGGAGACGGTGACC (reverse) for the first round and CGTATCGCCTCCCTCGCGCCATCAGCTGGGGGGTCCCTGAGACTCTCCTG (forward) CTATGCGCCTTGCCAGCCCGCTCAGCTTACCTGAGGAGACGGTGACC (reverse) for the second round. The gene-specific elements are in bold; the key is underlined; Roche-specific primer A/B sequences are in italics. Sequence analysis was performed with IMGT/HighV-Quest (24). IMGT output was analyzed in Access 2010 (Microsoft).
Results
Hybridoma generation, antibody reactivity, animal studies
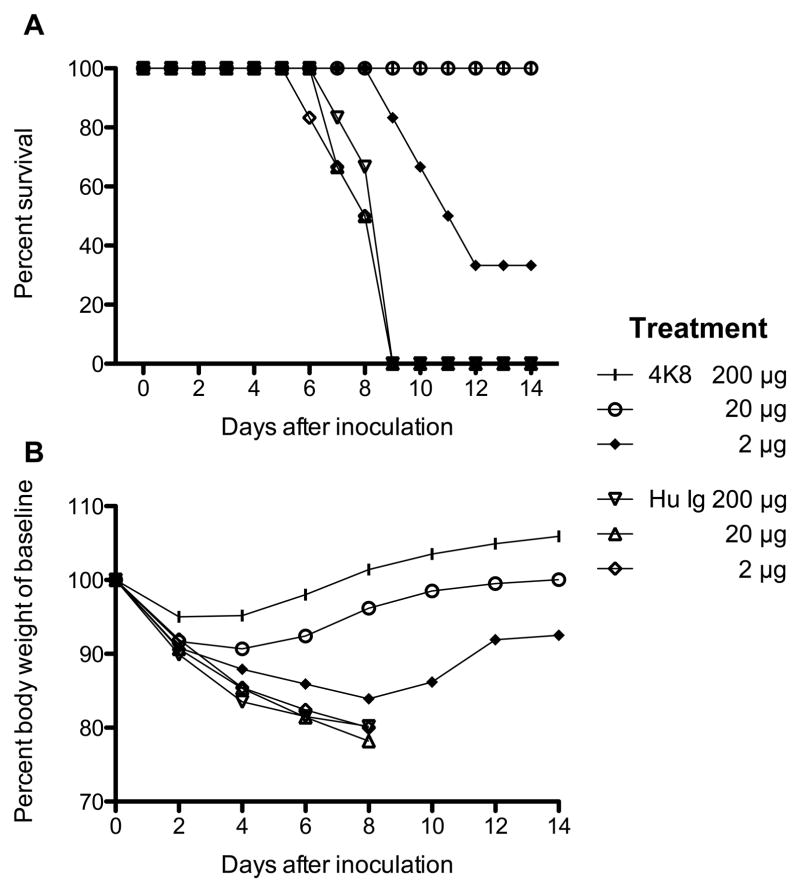
We isolated a panel of five human monoclonal antibodies named 4A10, 2O10, 4K8, 6D9, and 2K11 from a single blood sample of a human subject. All antibodies showed HAI activity against both 1918 and 2009 H1N1 pandemic viruses, the related swine influenza viruses from 1930 and 1976, and the H1N1 virus from 1977, but not against strains from the 1940s or H1N1 strains after 1977 (Table I). Ab 6D9 tested negative for functional activity against representative H2N2, H3N2, or H5N1 viruses by HAI (data not shown). Generally, Ab4K8 was the most potent antibody, with an HAI activity <0.01 μg/mL against pandemic H1N1 viruses. We selected this Ab for testing in a lethal mouse model of 1918 influenza virus infection (Fig. 1, Table II). Ab4K8 protected 6 out of 6 animals from death at both the highest and the intermediate dose levels and 2 out of 6 animals at the lowest dose(Fig. 1A). Ab4K8 reduced lung virus titers as compared to the human IgG control by 3.1 log10 PFU/mLat the 200 μg dose, 2.6 log10 PFU/mLat the 20 μg dose, and still 1.3 log10 PFU/mLat the 2 μg dose(Table II).
Table I.
Specific HAI activity of human antibodies against influenza viruses or VLPs
| Influenza A strain | HAI activity of indicated Ab [μg/mL] | ||||
| 4A10 | 2O10 | 4K8 | 6D9 | 2K11 | |
| A/South Carolina/1/1918 wt (VLP) | 0.08 | 0.04 | <0.01 | 0.2 | 0.16 |
| A/South Carolina/1/1918 K166E (VLP) | > | > | > | > | > |
| A/South Carolina/1/1918 K166N (VLP) | > | > | > | > | > |
| A/South Carolina/1/1918 K166Q (VLP) | > | > | > | > | > |
| A/South Carolina/1/1918 K166R (VLP) | 10 | > | > | 20 | > |
| A/swine/Iowa/15/1930 | 0.63 | 0.08 | <0.01 | 0.4 | 0.16 |
| A/Weiss/1943 | > | > | > | > | > |
| A/L3/47 | > | > | > | > | > |
| A/New Jersey/11/1976 | 2.5 | 2.5 | 0.16 | 1.3 | 1.3 |
| A/USSR/92/1977 | 0.32 | 2.5 | 0.32 | 2.5 | 0.32 |
| A/New Caledonia/20/1999 | > | > | > | > | > |
| A/Brisbane/59/2007 | > | > | > | > | > |
| A/California/04/2009 | 0.16 | 0.32 | <0.01 | 0.7 | 0.63 |
The > symbol indicates that activity was not detected at the highest concentration tested, 20 μg/mL.
FIGURE 1.
Therapeutic efficacy of Ab 4K8 against disease caused by the 1918 A (H1N1) virus in mice. Mice were inoculated on day 0 and treated on day 1 with the indicated antibody and dose. In each group, six mice were monitored every other day for survival (A) and weight (B). At all dose levels, the differences in survival distribution between the 4K8 and the human IgG control groups were significant by log-rank test (p = 0.001 for the high-dose level, p < 0.001 for the medium-dose level, p < 0.01 for the low-dose level).
Table II.
Therapeutic efficacy of Ab 4K8 against virus replication in mice inoculated with 1918 influenza A virus. Four mice were inoculated intranasally with 5× LD50 and administered 4K8 antibody or human IgG i.p. 24 h later. Mice were euthanized on day 4 after inoculation for the determination of lung titers
| Antibody | Dose [μg/mouse] | Mean lung virus titer* [log10 PFU/mL ± SD] |
|---|---|---|
| Ab 4K8 | 200 | 3.6 ± 0.2 |
| 20 | 4.3 ± 0.3 | |
| 2 | 5.3 ± 0.2 | |
|
| ||
| Human IgG control | 200 | 6.7 ± 0.1 |
| 20 | 6.9 ± 0.0 | |
| 2 | 6.6 ± 0.4 | |
At the α = 0.025 level controlling the overall type I error at 7.5%, the lung homogenates of the 4K8 groups had lower virus titers than the human IgG control groups for all dose levels by the Wilcoxon signed-rank test (p = 0.01429 for the each level).
Generation and validation of escape mutations in epitopes recognized by these five antibodies
Ab 4K8 selected for a K166R mutation in the A/USSR/97/1977 context and a K166E mutation in the A/California/04/2009 H1N1 context. To confirm that these mutations alone were sufficient to confer escape, we produced 1918 virus-like particles (VLPs) with mutant HA. The entire panel of antibodies was unable to inhibit hemagglutination of VLPs with K166E, K166N, or K166Q mutations (Table I). Abs 4A10 and 6D9 showed modest HAI activity against VLPs with the K166R mutation, but not 2O10, 4K8, or 2K11(Table I). These data suggested that all five Abs recognized a common epitope, possibly with minor differences in the mode of binding between those five antibodies.
Sequence analysis of the antibody variable gene sequences in the hybridomas
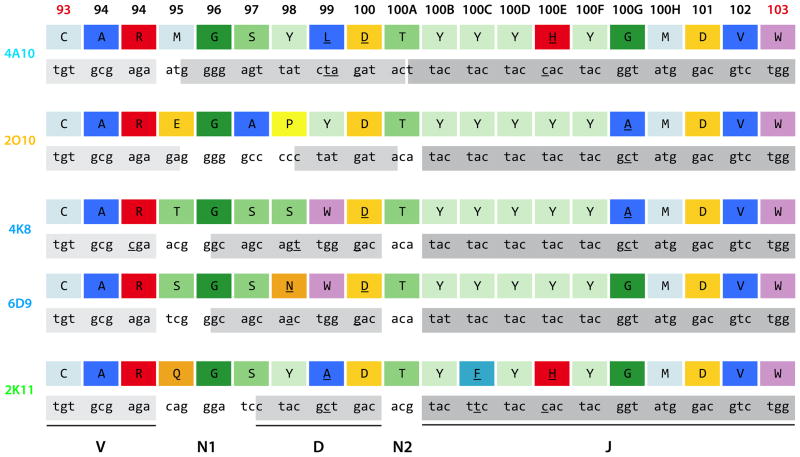
Next, we determined whether shared germline genes were the basis for the recognition of this epitope. Nucleotide sequence determination and sequence analysis of variable gene sequences (Table III, Fig. S1) using the international ImMunoGeneTics information system (IMGT) (24) revealed that all of the five antibodies shared usage of the VH3-7*01 gene and the JH6*02 gene; furthermore, 4A10, 2O10, and 2K11 used the same VL1-40*01 gene. Also, 4K8 and 6D9 used both VK3-20*01 and JK2. The D genes of these five antibodies were predicted to be of different origins except for 4K8 and 6D9, which shared the D6-13*01 gene (Table III). These data suggested that 4K8 and 6D9 might be derived from the same B cell ancestor clone. Careful review of the junctional sequences showed that indeed 4K8 and 6D9 were clonally related, while 4A10, 2O10, and 2K11 were derived independently of each other and of the 4K8/6D9 clone (Fig. 2). Despite four different clonal origins, the CDR H3s of these antibodies were remarkably similar (Fig. 2); for instance the CDR H3 length of 18 amino acids was identical across this panel. The amino acids in positions 93–95, 96, 100, 100A, 100B, 100D, 100F, and 100H-103 were fully conserved. Interestingly, somatic mutations were shared between the antibodies, for example the tyrosine to histidine mutation in position 100E of 2K11 and 4A10 or the glycineto alaninemutation in position 100G of 2O10 and 4K8 (Fig. 2). Also, several common mutated amino acid residues were found despite differing sequences in the inferred germlineorigin sequence: the glycine in position 96 was encoded entirely by the N1 segment (2K11, 2O10), by the D segment (4A10), or by both (4K8/6D9). The S97 was encoded by the D segment alone (4A10, 4K8/6D9) or by both N1 and D1 (2K11). An aspartic acid was found in position 100 of 2K11 and 2O10 because of their germline; clones 4A10 and 4K8/6D9 acquired this aspartic acid through a somatic mutation, suggesting that this panel converged towards a consensus sequence (Fig. 2). Position 100A was predicted by IMGT to be encoded by the N2 segment alone (2K11, 4K8/6D9), by the D chain and the N2 segment (2O10), or by the D and the J chain (4A10; Fig. 2). No matter the origin, a threonine was found in all five CDR H3s in this position (Fig. 2). Residues from 100B onward were encoded by the JH6 gene in all clones; four to six successive tyrosine residues are typically encoded by that germline gene segment(25), although somatic mutations were found in positions 100C (2K11) and 100E (4A10, 2K11).
Table III.
Genetic features of H1N1-specific human monoclonal antibodies
| Ab | Heavy chain genes | Light chain genes | ||||
|---|---|---|---|---|---|---|
| VH | D | JH | VK/VL | JK/JL | λ/κ | |
| 4A10 | 3–7*01 | 3–16*02 | 6*02 | 1–40*01 | 2*01 or 3*01 | λ |
| 2O10 | 3–7*01 | 3–22*01 | 6*02 | 1–40*01 | 1*01 | λ |
| 4K8 | 3–7*01 | 6–13*01 | 6*02 | 3–20*01 | 2 | κ |
| 6D9 | 3–7*01 | 6–13*01 | 6*02 | 3–20*01 | 2 | κ |
| 2K11 | 3–7*01 | 4–17*01 | 6*02 | 1–40*01 | 3*02 | λ |
FIGURE 2.
Comparison of antibody gene junctional sequences reveals four independent clones. The IgH gene segment junctions of the five VH3-7/JH6 antibodies 4A10, 2O10, 4K8, 6D9, and 2K11 are shown in amino acid and DNA sequence. Mutated amino acids and nucleotides are underlined. The amino acid residues are color-coded per standard IMGT color scheme based on chemical properties (43). Briefly, aliphatic (A, I, L, V) residues are dark blue, phenylalanine light blue, sulfur (C, M) residues cyan, glycine dark green, residues with hydroxyl groups (S, T) medium green, tryptophan pink, tyrosine light green, proline yellow, acidic (D, E) residues light orange, amide (N, Q) residues dark orange, and basic (H, K, R) residues red. Kabat numbering for amino acids is used instead of IMGT numbering, and is shown at the top level; the CDR H3 margins are denoted in red. The contributions of the VH, D, and JH genes are shown in light grey (VH), medium grey (D), and dark grey (JH).
We reviewed the variable gene encoded N-terminal part of heavy variable chains for common somatic mutations: 4A10 and 2K11 shared a threonine to serine mutation in position H28. All five antibodies shared a threonine instead of the germline serine in position H35 (Fig. 3). Abs 2O10 and 4K8 shared a lysineto asparaginemutation in position H52. Abs 2O10 and 6D9 mutated towards a threonine from an asparagine in position H76 (Fig. 3). Abs 2O10, 4K8, and 2K11 were found to have a valine instead of an alanine in position H84 (Fig. 3). An aspartic acid took the place of a glutamic acid in position H85 of Abs 4A10 and 4K8. Finally, both Abs 4A10 and 2K11 have a histidineto tyrosinemutation in position L34 of the λ chains in common (Fig. S1).
FIGURE 3.
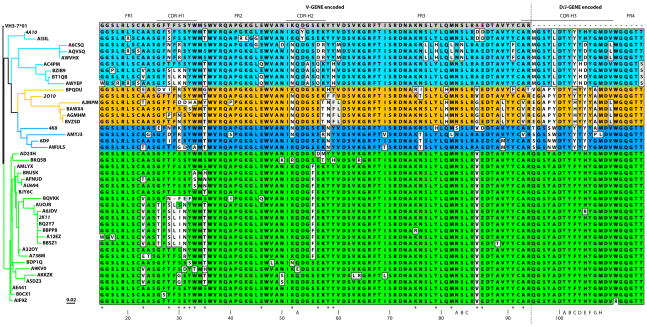
Phylogram and sequence alignment to the VH3-7*01 germline sequence of the heavy variable chain genes of Abs 4A10 (cyan), 2O10 (orange), 4K8/6D9 (medium blue), and 2K11 (green) from hybridoma technology and pyrosequencing. Five-letter alphanumeric labels denote sequences derived from pyrosequencing; hybridoma names are italicized. The location of the CDRs (based on IMGT analysis) is shown on top; Kabat numbering is shown at the bottom. Amino acids similar to the VH3-7*01 germline sequence (for the VH region) or to the consensus sequence (for the D/J regions) are in light gray, dissimilar amino acids in white. Variable gene segment (V-GENE) encoded sequences are separated from the diversity and joining gene segment (D/J-GENE) encoded sequences by a dashed line. Residues within the VH region with evidence of convergence are identified with an asterisk at the bottom. The phylogram was generated based on the protein sequences with MacVector software version 12 using neighbor joining, best tree, symmetric tie breaking, uncorrected (“p”) distance settings, and rooted to the deduced VH3-7*01 germ line protein sequence.
Deep sequencing of the VH3 gene encoded repertoire of this donor
The response of this donor towards 2009 H1N1 was dominated by antibodies encoded by the VH3-7/JH6 gene segments. We hypothesized that this limited panel of five clones might represent only a small portion of the circulating antigenic site Sa-specific repertoire in this individual. To test this idea, we extracted mRNA from total PBMCs from this donor six months after hybridoma generation and isolated antibody genes using an RT-PCR amplification specific for VH3. We used pyrosequencing to delineate the entire VH3 repertoire. We identified sister clones of the above antibodies based on shared V/J usage and identical VDJ junctions to more fully define the genetic diversity within these clones in circulating cells. We obtained 26.2 mega bases of data including 80,687 sequences that passed filter reads. The sequences had an average length of 325 base pairs after removal of primer sequences. Analysis with IMGT identified 60,484 of those sequences as productive; 60,447 belonged to the VH3 germline. VH3-23 and VH3-11 each accounted for 17% of productive VH3 sequences (the first IMGT assignment was used in case of ambiguities), VH3-30 for 13%, and VH3-7 for 9% (Fig. S2). A total of 1,917 VH3-7/JH6 IgH were identified, of which 203 shared a CDR H3 length of 18 amino acids with the five neutralizing influenza antibodies we had isolated. Of these 203 sequences, 138 were non-redundant on the protein level belonging to a total of 69clones based on review of the VDJ junctions. Eight of those sequences belonged to the 4A10 lineage (five to clone 2O10, two to clone 4K8/6D9, and 23 to clone 2K11) meaning that every single clone was still found in the peripheral blood of the individual six months after the initial blood draw for hybridoma generation. Since residues DTY at positions 100-100Bwere completely conserved across all antibodies, we screened the remaining clones for the presence of this motif, but none of them shared this DTY motif in CDR H3, suggesting that we likely had identified most of the clones of the VH3-7/JH6 gene segment-encoded Abs that recognized the influenza HA Sa site.
Intraclonal sequence divergence of the 2K11 clone
While we identified sequences highly related to those of Abs 4A10, 2O10, or 4K8/6D9, none of the sequences from the high throughput analysis was completely identical to those in the original clones. Remarkably, we were able to recover a sequence that was virtually identical to the 2K11 hybridoma cell IgH sequence in the high throughput sequence BQ2Y7. This occurrence may have been linked to the fact that sequences recovered that were related to Ab 2K11 represented by far the biggest clonal family identified and thus provided a more comprehensive snapshot of the evolution of this antibody. Within the panel related to Ab2K11, the AE441 and AIF9Z sequences embodied essentially a germline state with just a single non-silent somatic mutation in the variable gene encoded amino acid sequence, the valine in position 84. On the other hand, the Ab2K11 sequence itself and related sequences were highly mutated, particularly in the CDR H1, with up to five changes in amino acid sequence. While both germline states and highly mutated states were present simultaneously in the peripheral blood, sequences representing many of the intermediate predicted steps were not detected. For example, it was not clear in what order the mutations within CDR H1 of the hybridoma 2K11 occurred except that S31N probably occurred first because it was present in A32OY by itself. Also the S30I mutation probably occurred last within CDR H1 because AUOJR contained all of the other mutations except the aforementioned one. Since the other three clones had fewer representatives than the 2K11 clone, the order of mutations would be more difficult to establish for those clones. Nevertheless, sequence divergence from the germline sequence was readily apparent. Notably, insertion/deletion events seemed to play a minor role in this antibody panel with only sequence BQVKK within the 2K11 clone bearing evidence of a three base pair deletion leading to the loss of a single amino acid residue within CDR H1.
Interclonal sequence convergence
We defined convergence as the same altered amino acid introduced by somatic mutation present in two or more independent clones. Despite sequence divergence within the individual clones, sequence comparison revealed many further examples of interclonal sequence convergence not evident in the hybridoma sequences such as valine in H23 (members of clones 4A10/2K11) or threonine in the same position (2O10, 4K8/6D9, 2K11), leucine in H29 (4A10/2K11), asparagine in H31 (4A10/2K11), glutamine in H46 (4A10/2K11), asparagine in H58 (2O10/4K8/6D9), histidine in H59 (4A10, 2K11), and several others (Fig. 3). Overall, there were 20 positions within the VH protein sequence with evidence of convergence. Only eight of those 20 positions were found within CDR H1, H2, or the V-GENE encoded portion of H3, so 12 convergence positions were located in the framework regions. Remarkably, a sub clone each within two different clones converged towards a set of similar somatic mutations within the CDR H1 (amino acids SLIN in positions H28-31 in the 2K11 clones and amino acids SLKN in the same positions of the 4A10 lineage). An analysis of silent versus non-silent mutations by framework and CDR is presented in Supplemental Table 1.
Discussion
Glycosylation within site Sa does not always confer escape from neutralization
We describe a panel of four independent clones of human antibodies. Like Ab 2D1, they were Sa site antibodies based on the selection of escape mutations in position K166 and that showed potent HAI activity against pandemic H1N1 influenza (5, 9, 10). Unlike Ab2D1 however, this panel had HAI activity against USSR/77 H1N1 as well. This finding is striking because the USSR virus possesses three predicted N-linked glycosylation sites within its Sa antigenic site (9). Glycosylation at these sites is thought to shield HA from neutralization (7, 9). In support of this model, experimental introduction of glycosylationsites into the 1918 or 2009 HAs conferred resistance to neutralization by antiserum elicited by the unglycosylated HAs (7). The fact that neither the 1918 virus, nor the 2009 pandemic virus possess glycosylation sites within the Sa antigenic site, while human H1N1 viruses acquired N-linked glycosylation over the course of the 20th century, likely explains, at least in part, why elderly people had preexisting cross-reactive humoral immunity against the 2009 pandemic virus(7, 9). The fact that VH3-7/JH6 Abs inhibited USSR virus, despite the presence of the glycosylation sites, demonstrated that the presence of glycosylation sites within site Sa did not always confer escape from Sa-specific antibodies.
Redundancy of the immune response
Interestingly, this VH3-7/JH6 panel of four independent clones was derived from a single donor; her response to pandemic 2009 H1N1 may have been VH3-7/JH6 dominant and oligoclonalsince we were only able to generate another H1N1 globular head antibody, Ab 5J8, from this donor that selected for escape mutations along the receptor-binding pocket (unpublished data). The redundancy of this response may ensure adequate neutralization of a given pathogen such as influenza A virus. This redundancy may be quite common, but may only be detected more frequently through advances in antibody engineering and sequencing technology.
Same genes suggest a common epitope
Wrammert et al. described cross-reactive human Abs in subjects convalescing from infection with 2009 H1N1 pandemic virus (26); this work did not feature neutralizing VH3-7/JH6 antibodies—potentially due to a limited number of antibodies. While a similar genetic makeup (like VH3-7/JH6) suggests a common epitope, the Sa antigenic site can likely be reached from a variety of different germline configurations such as VH3-7/JH6 or the VH2-70 of the 2D1 antibody (5, 8, 10).
The VH3-7/JH6 antibodies described here showed strongest HAI activity against 1918 VLPs of a virus that circulated well before the birth of this donor and thus could not have served as the inciting agent. In fact, given that this donor was born between 1957 and 1977 when H1N1 did not circulate in humans, subtype H1N1 infection was unlikely to have been the cause of her first exposure to influenza or original antigenic sin (27). These antibodies could have been created in response to vaccination with the 1976 swine influenza virus (28) or vaccination or infection with a USSR/77-like virus or the 2009 H1N1 virus. Since these antibodies in general seemed to neutralize 2009 H1N1 best among those three candidates, we hypothesized that these antibodies were created in response to the 2009 virus and that the process of several antibodies independently hitting the same epitope with a similar genetic makeup was entirely stochastic.
Intraclonal sequence divergence and interclonal sequence convergence
The ultra deep sequencing in this study revealed that the VH3-7/JH6 clones represented large circulating phylogenies. Since pyrosequencing was performed on PBMCs isolated six months after the initial hybridoma generation, the phylogenies seemed to persist for at least several months in the peripheral blood. B-cells that are not stimulated because they do not express high-affinity antibody are supposed not to undergo further proliferation (29). Still, wide divergence of sequences from essentially unmutated germline states to extensively mutated sequences was found within clones in the peripheral blood of this donor. Persistent low-affinity memory populations may aid in the immunologic response towards a related HA antigen that the individual subsequently encounters (30, 31).
B cell development has been traced by molecular analysis of single cells picked from histological sections in human germinal centers (32, 33) and spleen (33, 34) showing the presence of both naïve and memory B cells. An alternative method to track the development of single memory-lineage B cells has been the use of a specific anti-idiotypic antibody E4, which recognizes a canonical V region (35). In the mouse study by Liu et al, this E4+ pool is derived from fewer than five canonical precursors. A lack of shared somatic mutations across clonally-related cells by day 13 indicates that the selective expansion of mutant sub clones typical of memory responses did not yet occur as it did in our panel of antibodies (35). The strength of our study was the combination of functional data from human hybridoma technology with ultra deep sequencing. A limitation of the deep sequencing was that we were not able to document the concurrent evolution of the corresponding light chains, although the hybridoma technology allowed us to find at least one hybridoma per clone with a matching heavy and light chain, adding further evidence that these hybridomas represented four distinct clones.
Despite different clonal origins within the VDJ junction, these four independent clones converged towards common amino acid residues within this junction and throughout the shared VH3-7 gene. This suggested a strong selection for optimal binding sites across clones. Since three clones also share the VL1-40 gene, but different J genes, CDRs H1, H2, H3, L1, and L2 would be expected to make contacts with the HA. Given the frequency of mutations within the CDR H1 segment and the conservation of the typically very variable CDR H3, we hypothesize that those two loops would be especially critical in the antibody-antigen interaction. The majority of the convergence positions are within framework regions; these residues can be at the surface, the core, or the heavy chain-light chain interface of the antibody. Core mutations partially determine stability (36) and may change the conformation of adjacent and distant CDR loops (8).
Sequence convergence has been found in a variety of proteins across individuals and species
Antibodies of the human VH1-69 germline from diverse phage display libraries, plasma blasts, and EBV-transformed B cells neutralize the stem of H1N1 and H5N1 influenza viruses (26, 37–40). Likewise, the phage-derived HIV antibodies Fab 8066 and D5 from entirely different antibody libraries and panning procedures show convergence in their sequences and also in the conformation of the CDR H2 loop (41). Indeed, pyrosequencing of zebrafish antibodies showed evidence of convergence, in which different individuals made the same antibody (12), but this was not correlated with functional data. Sequence convergence is not only seen in antibodies across individuals of the same species (or—as demonstrated here—within a single individual), but even in proteins that serve a similar purpose across distinct mammalian species (42). With continued progress in B cell technology, there will likely be further discoveries of converging sequence-related antibodies from one or multiple individuals that share common epitopes. Once common germline gene responses to defined epitopes of pathogens are better understood, it might be possible to diagnose an infection of an individual based on the immune repertoire documented by ultra deep sequencing.
Supplementary Material
Acknowledgments
The authors thank the anonymous blood donor; Jose A. Archuleta, Jr. and Cheryl Kinnard of the Vanderbilt Clinical Trials Center; M. Posner and L. Cavacini for the HMMA2.5 cell line; Kimberly S. Crimin for statistical help; Patricia Bethany Smith and the Vanderbilt Genome Sciences Resource (Kasey D. Lawrence, Robert Walker Woodhall) for technical assistance.
Abbreviations used in this article
- HA
hemagglutinin
- HAI
hemagglutination activation inhibition
- VLP
virus-like particle
Footnotes
J.E.C. is a Burroughs Well come Fund Clinical Scientist in Translational Research. This work was supported by NIH grants P01 AI058113, R21AI085306 and a pilot project from UL1RR029887 to C.F.B. and by DOD grant HDTRA1-08-10-BRCWMD-BAAand NIAID Contract HHSN272200900047C. The Genome Sciences Resource was supported by the Cancer Center Support Grant (CA068485), the Vanderbilt Digestive Disease Research Center (DK058404), and the Vanderbilt Vision Research Center (EY008126).
Antibody nucleotide sequences have been deposited in GenBank (accession numbers JF806284-JF806293).
Disclosures
The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency. Vanderbilt University submitted a patent covering the diagnostic and therapeutic use of the antibodies described in this paper. The authors have no financial conflict of interest.
References
- 1.Libster R, Bugna J, Coviello S, Hijano DR, Dunaiewsky M, Reynoso N, Cavalieri ML, Guglielmo MC, Areso MS, Gilligan T, Santucho F, Cabral G, Gregorio GL, Moreno R, Lutz MI, Panigasi AL, Saligari L, Caballero MT, Egues Almeida RM, Gutierrez Meyer ME, Neder MD, Davenport MC, Del Valle MP, Santidrian VS, Mosca G, Garcia Dominguez M, Alvarez L, Landa P, Pota A, Bolonati N, Dalamon R, Sanchez Mercol VI, Espinoza M, Peuchot JC, Karolinski A, Bruno M, Borsa A, Ferrero F, Bonina A, Ramonet M, Albano LC, Luedicke N, Alterman E, Savy V, Baumeister E, Chappell JD, Edwards KM, Melendi GA, Polack FP. Pediatric hospitalizations associated with 2009 pandemic influenza A (H1N1) in Argentina. N Engl J Med. 2010;362:45–55. doi: 10.1056/NEJMoa0907673. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep. 2009;58:521–524. [PubMed] [Google Scholar]
- 3.Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 4.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, Ogasawara K, Kawaoka Y. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krause JC, Tumpey TM, Huffman CJ, McGraw PA, Pearce MB, Tsibane T, Hai R, Basler CF, Crowe JE., Jr Naturally occurring human monoclonal antibodies neutralize both 1918 and 2009 pandemic influenza A (H1N1) viruses. J Virol. 2010;84:3127–3130. doi: 10.1128/JVI.02184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manicassamy B, Medina RA, Hai R, Tsibane T, Stertz S, Nistal-Villan E, Palese P, Basler CF, Garcia-Sastre A. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 2010;6:e1000745. doi: 10.1371/journal.ppat.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei CJ, Boyington JC, Dai K, Houser KV, Pearce MB, Kong WP, Yang ZY, Tumpey TM, Nabel GJ. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med. 2010;2:24ra21. doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause JC, Ekiert DC, Tumpey TM, Smith PB, Wilson IA, Crowe JE., Jr An insertion mutation that distorts antibody binding site architecture enhances function of a human antibody. MBio. 2011:2. doi: 10.1128/mBio.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, Stevens J, Wilson IA, Aguilar PV, Altschuler EL, Basler CF, Crowe JE., Jr Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, Ni I, Mei L, Sundar PD, Day GM, Cox D, Rajpal A, Pons J. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A. 2009;106:20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Fire AZ. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Collins AM. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, McGraw PA, House FS, Crowe JE., Jr An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods. 2008;336:142–151. doi: 10.1016/j.jim.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, Wilson PC. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abhinandan KR, Martin AC. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol Immunol. 2008;45:3832–3839. doi: 10.1016/j.molimm.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 19.Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization Collaborating Centers for Reference and Research on Influenza. Concepts and Procedures for Laboratory-Based Influenza Surveillance. Centers for Disease Control and Prevention; Atlanta, GA: [Google Scholar]
- 21.Caton A, Brownlee G, Yewdell J, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 22.Yewdell JW, Webster RG, Gerhard WU. Antigenic variation in three distinct determinants of an influenza type A haemagglutinin molecule. Nature. 1979;279:246–248. doi: 10.1038/279246a0. [DOI] [PubMed] [Google Scholar]
- 23.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 24.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and VDJ sequence analysis. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Jr, Kirkham PM. Expressed murine and human CDR H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O’Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davenport FM, Hennessy AV, Francis T., Jr Epidemiologic and immunologic significance of agedistribution of antibody to antigenic variants of influenza virus. J Exp Med. 1953;98:641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause R. The swine flu episode and the fog of epidemics. Emerg Infect Dis. 2006;12:40–43. doi: 10.3201/eid1201.051132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.French DL, Laskov R, Scharff MD. The role of somatic hypermutation in the generation of antibody diversity. Science. 1989;244:1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- 30.Herzenberg LA, Black SJ, Tokuhisa T. Memory B cells at successive stages of differentiation. Affinity maturation and the role of IgD receptors. J Exp Med. 1980;151:1071–1087. doi: 10.1084/jem.151.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fish S, Zenowich E, Fleming M, Manser T. Molecular analysis of original antigenic sin. I. Clonal selection, somatic mutation, and isotype switching during a memory B cell response. J Exp Med. 1989;170:1191–1209. doi: 10.1084/jem.170.4.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuppers R, Zhao M, Hansmann ML, Rajewsky K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J. 1993;12:4955–4967. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tierens A, Delabie J, Michiels L, Vandenberghe P, De Wolf-Peeters C. Marginal-zone B cells in the human lymph node and spleen show somatic hypermutations and display clonal expansion. Blood. 1999;93:226–234. [PubMed] [Google Scholar]
- 34.Dunn-Walters DK, Isaacson PG, Spencer J. Analysis of mutations in immunoglobulin heavy chain variable region genes of microdissected marginal zone (MGZ) B cells suggests that the MGZ of human spleen is a reservoir of memory B cells. J Exp Med. 1995;182:559–566. doi: 10.1084/jem.182.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu AH, Jena PK, Wysocki LJ. Tracing the development of single memory-lineage B cells in a highly defined immune response. J Exp Med. 1996;183:2053–2063. doi: 10.1084/jem.183.5.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark LA, Ganesan S, Papp S, van Vlijmen HW. Trends in antibody sequence changes during the somatic hypermutation process. J Immunol. 2006;177:333–340. doi: 10.4049/jimmunol.177.1.333. [DOI] [PubMed] [Google Scholar]
- 37.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corti D, Suguitan AL, Jr, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, Weiss RA, Sallusto F, Subbarao K, Lanzavecchia A. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gustchina E, Li M, Louis JM, Anderson DE, Lloyd J, Frisch C, Bewley CA, Gustchina A, Wlodawer A, Clore GM. Structural basis of HIV-1 neutralization by affinity matured Fabs directed against the internal trimeric coiled- coil of gp41. PLoS Pathog. 2010;6:e1001182. doi: 10.1371/journal.ppat.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Cotton JA, Shen B, Han X, Rossiter SJ, Zhang S. Convergent sequence evolution between echolocating bats and dolphins. Curr Biol. 2010;20:R53–54. doi: 10.1016/j.cub.2009.11.058. [DOI] [PubMed] [Google Scholar]
- 43.Pommie C, Levadoux S, Sabatier R, Lefranc G, Lefranc MP. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J Mol Recognit. 2004;17:17–32. doi: 10.1002/jmr.647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.