Abstract
The streptococcal antigen I/II (AgI/II)-family polypeptides are cell wall-anchored adhesins expressed by most indigenous oral streptococci. Proteins sharing 30–40% overall amino acid sequence similarities with AgI/II-family proteins are also expressed by Streptococcus pyogenes. The S. pyogenes M28_Spy1325 polypeptide (designated AspA) displays an AgI/II primary structure, with alanine-rich (A) and proline-rich (P) repeats flanking a V region that is projected distal from the cell. In this study it is shown that AspA from serotype M28 S. pyogenes, when expressed on surrogate host Lactococcus lactis, confers binding to immobilized salivary agglutinin gp-340. This binding was blocked by antibodies to the AspA-VP region. In contrast, the N-terminal region of AspA was deficient in binding fluid-phase gp-340, and L. lactis cells expressing AspA were not agglutinated by gp-340. Deletion of the aspA gene from two different M28 strains of S. pyogenes abrogated their abilities to form biofilms on saliva-coated surfaces. In each mutant strain, biofilm formation was restored by trans complementation of the aspA deletion. In addition, expression of AspA protein on the surface of L. lactis conferred biofilm-forming ability. Taken collectively, the results provide evidence that AspA is a biofilm-associated adhesin that may function in host colonization by S. pyogenes.
Introduction
Streptococcus pyogenes (group A Streptococcus; GAS) is a causative agent of tonsillitis, pharyngitis, otitis media, impetigo, scarlet fever, cellulitis and necrotizing fasciitis. The species is part of a phenotypically heterogeneous group of bacteria that readily colonize humans and animals, ordinarily forming part of the commensal microbiota (Nobbs et al., 2009). Under appropriate conditions, S. pyogenes can cause opportunistic invasive infections with high (10–35%) mortality rates (NCIRD, 2008). In order to colonize, proliferate and persist, S. pyogenes must in the first instance adhere to host tissues. Numerous cell wall-anchored proteins have been identified on the surface of S. pyogenes (Nobbs et al., 2009) that mediate adherence to host tissue proteins such as fibronectin (Cue et al., 2001), fibrinogen (Hryniewicz et al., 1972; Courtney et al., 2002), albumin (Schmidt et al., 1993), immunoglobulins (Fagan et al., 2001; Okamoto et al., 2008) and mucin (Ryan et al., 2001). However, the mechanisms by which S. pyogenes colonizes oral or nasopharyngeal surfaces are not fully understood.
Streptococcus pyogenes has the potential to form biofilms in the oral cavity and nasopharynx (Doern et al., 2009), and on skin and wounds. Cases of pharyngitis from which biofilm-forming S. pyogenes have been isolated have been associated with multiple episodes of disease and 30% treatment failure (Lembke et al., 2006). It is clear that the ability of S. pyogenes to form biofilms varies considerably with respect to serotype and strain (Lembke et al., 2006; Courtney et al., 2009). One of the mechanisms proposed for initial adherence and accumulation of M1 serotype S. pyogenes involves the formation of complexes between cell surface lipoteichoic acid (LTA) and members of the M protein family (Courtney et al., 2009). Another more recent finding is that pilus structures (Mora et al., 2005) are involved in development of biofilm aggregates (Manetti et al., 2007; Koller et al., 2009). Clearly GAS utilizes different complements of surface proteins to adhere to host tissues and form biofilms.
In oral viridans group streptococci, the antigen I/II (AgI/II) family of cell surface proteins plays many roles in colonization of the host. The AgI/II-family polypeptides are cell wall-anchored and comprise between 1310 and 1653 amino acid (aa) residues (Brady et al., 2010). The SpaP (AgI/II-family) protein in Streptococcus mutans mediates adherence to salivary glycoproteins within the tooth enamel pellicle (Bowen et al., 1991). Antibodies to SpaP block adherence of S. mutans (Munro et al., 1993), and a peptide vaccine derived from SpaP has been shown to protect rodents against dental decay caused by S. mutans (Takahashi et al., 1991). Other indigenous streptococci, for example Streptococcus gordonii, express two AgI/II proteins, designated SspA and SspB, from duplicated genes (Jakubovics et al., 2005). The SpaP, SspA and SspB proteins have a common host receptor in gp-340, a member of the scavenger receptor cysteine-rich (SRCR) protein family (Madsen et al., 2010). Gp-340 is a mucin-like protein found in saliva, tears, and at mucosal surfaces. It functions in innate immune defence, complexing with other mucosal components such as mucins, collectins (Ligtenberg et al., 2007) and secretory immunoglobulin A (S-IgA) (Ligtenberg et al., 2004), and trapping microbes for removal from the body (Ligtenberg et al., 2007). However, when gp-340 becomes adsorbed to a surface, such as within salivary pellicle, gp-340 acts as a receptor for microbial adherence (Loimaranta et al., 2005). Additionally, AgI/II-family proteins have been shown to bind fibronectin (Nobbs et al., 2007), collagen (Soell et al., 2010), the oral bacteria Actinomyces naeslundii (Jakubovics et al., 2005) and Porphyromonas gingivalis (Daep et al., 2008), and the yeast Candida albicans (Silverman et al., 2010). These proteins therefore play potentially multiple roles in Streptococcus adherence, colonization and microbial community development.
Epidemiological studies of GAS associated with puerperal sepsis, a major cause of death of young women in the past, have identified serotype M28 strains as being largely responsible. The genome sequences of a series of M28 invasive strains revealed that they had all acquired a 37.4 kb element, shared with group B Streptococcus (GBS), designated region of difference 2 (RD2). This region contained genes encoding prophage virulence factors, R-28 surface protein antigen (Johnson, 1975; Stålhammar-Carlemalm et al., 1999) and six other inferred secreted proteins (Green et al., 2005). One of these genes encoded a protein (designated M28_Spy1325) with structural and sequence similarities to oral streptococcal AgI/II-family proteins (Fig. 1) (Jenkinson and Demuth, 1997). Preliminary characterization of the S. pyogenes AgI/II protein M28_Spy1325 (predicted molecular mass 148.36 kDa) indicated that it interacted with gp-340 in a manner consistent with that of other AgI/II-family proteins (Zhang et al., 2006). In addition, bioinformatic analyses revealed common ancestral characteristics of M28_Spy1325 (denoted AspA for group AStreptococcussurface protein A) that placed it firmly within the AgI/II-family of streptococcal proteins. Loss of sections of the N-terminal A and V regions has resulted in AspA being slightly smaller than many of the other oral streptococcal AgI/II proteins (Fig. S1) (Brady et al., 2010).
Fig. 1.
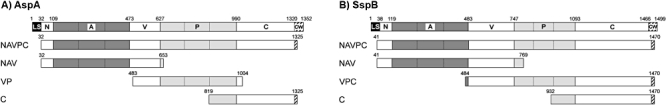
Pictorial representation of AgI/II proteins of (A) S. pyogenes serotype M28 (strain MGAS6180) and (B) S. gordonii DL1, showing the recombinant fragments generated in this study. Included are the aa residue numbers demarcating each of the protein fragments. Structural features are as follows: LS, leader sequence; N, N-terminal domain; A, alanine-rich repeats; V, variable region; P, proline-rich repeats; C, C-terminal domain; and CW, cell wall anchor.
Although AspA binds gp-340 (Zhang et al., 2006), the primary structural differences between AspA and SspB suggested a differential role for AspA may have evolved in pathogenic GAS. To test this hypothesis we investigated the structural and functional properties of AspA in more detail, particularly in respect to binding gp-340 and in biofilm community development. We show that AspA expressed on the surrogate host Lactococcus lactis binds immobilized gp-340, but the N-terminal region is defective in interaction with fluid-phase gp-340. In addition, expression of AspA appears to be essential for biofilm formation by two independently isolated M28 serotype strains of S. pyogenes. It is suggested that expression of AspA may confer on GAS the ability to successfully compete with indigenous commensal streptococci in colonizing host surfaces while avoiding, at least in part, host innate immune pressures.
Results
Primary sequence differences between AspA and SspB
The AgI/II-family polypeptides have a common NAVPC region structure (Fig. 1), as described above, with highest conservation of aa sequences within the C-terminal region (Jenkinson and Demuth, 1997). The Ala-rich repeat (A) and Pro-rich repeat (P) regions also show various degrees of sequence conservation across the family, related to maintaining the N-terminal α-helix and C-terminal polyproline helix structures, which together generate a stalk projecting the V region distal from the cell surface-anchorage point (Brady et al., 2010; Larson et al., 2010). The GAS AgI/II polypeptide A and P regions show approximately 28% and 30% aa sequence identities, respectively, to the A and P regions of SspB from S. gordonii (see Fig. S1 for aa sequence alignments). The C regions of AspA and SspB have 38% aa residue identity, while the V region sequences appear to be unrelated (< 10% identical aa residues) (Fig. S1).
Antigenic differences between AspA and SspB
Previous studies have investigated the binding properties of regions of AgI/II-family proteins by expressing recombinant fragments in Escherichia coli (Crowley et al., 1993; Hajishengallis et al., 1994). These studies have been paramount in deducing the 3D structure of AgI/II proteins (Forsgren et al., 2009; 2010; Brady et al., 2010; Larson et al., 2010). We have previously produced recombinant fragments of S. gordonii SspB and shown that these purified fragments have various binding activities with gp-340 (Nobbs et al., 2007). To compare therefore the binding properties of AspA regions with corresponding regions of SspB, we generated two sets of recombinant polypeptides (Fig. 1). The AspA polypeptides generated (Fig. 1A) were, as far as possible, analogous to those that had already been derived from SspB (Fig. 1B), except that we were unable to express a stable VPC fragment from AspA (see Fig. 1A).
Since the aa sequence of the V region of AspA was unique, we utilized fragment VP-AspA (Fig. 1A) to generate AspA-specific polyclonal antibodies. The reactivity of these antibodies with AspA or SspB protein fragments was compared with the reactivity of a polyclonal antibody raised to SpaP from S. mutans (Jakubovics et al., 2005). Anti-AspA-VP antibodies reacted as anticipated with the homologous antigen, recombinant VP (rVP-AspA) polypeptide (Fig. 2), but not with rNAV-SspB polypeptide (Fig. 2) or with rNAVPC-SspB (not shown). The anti-SpaP antibodies reacted with rNAV-SspB (Fig. 2) and with all of the recombinant SspB polypeptides i.e. rNAVPC, rVPC and rC (not shown). These observations were consistent with the relatively well-conserved sequences across the entire lengths of the SpaP and SspB proteins, with the exception of the V regions (Brady et al., 2010). Conversely, the SpaP antibodies did not react with any of the AspA recombinant polypeptides, reflecting the lower aa sequence identities between AspA and SspB, and suggesting major antigenic distinctions between the two polypeptides.
Fig. 2.
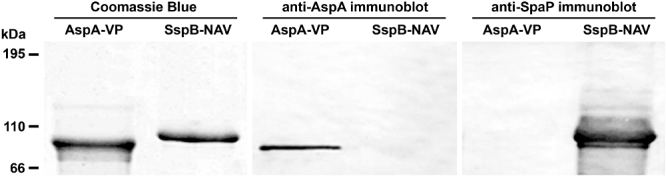
Western blots of recombinant protein fragments VP-AspA and NAV-SspB probed using anti-VP-AspA or anti-SpaP antibodies. Equivalent amounts of protein (100 ng) as determined by Bradford assay were applied to each lane. Proteins were also stained with Coomassie blue as a protein loading control.
Metal ion binding associated with V region
Within the V region aa sequence of AspA there is a His/Asp motif, HDDWVHDTH (608–616 aa), which is not annotated in any protein databases, and that is found only in the AgI/II-family proteins identified in GAS and GBS. It is known that clustered His residues form a region of high electron density to which positively charged metal ions may bind. Accordingly, inductively coupled plasma optical emission spectrometry (ICP-OES) was utilized to determine trace metal ion binding to the rNAV polypeptides. The analyses found that Zn2+ ions bound to rNAV-AspA at a metal ion : protein ratio of approximately 1:1 (Fig. 3). However, ICP-OES analysis of rNAV-SspB, which does not contain the HD cluster, suggested that this region did not specifically bind Zn2+ ions over and above similar amounts of residual Fe2+ and Ni2+ associated with the protein (Fig. 3). In addition, Mg2+ (or Ca2+) was found to be associated with both AspA and SspB polypeptides at metal ion : protein ratios of approximately 2:1 and 1:1 respectively (Fig. 3). Under the ICP-OES conditions used, it was not possible to distinguish between Mg2+ and Ca2+. Zinc (Zn2+) has not previously been identified as being associated with the AgI/II-family protein V regions. The residual amounts of Ni2+ present in both protein preparations (Fig. 3) may have arisen from the Ni2+ affinity column utilized to purify the His-tagged polypeptides.
Fig. 3.
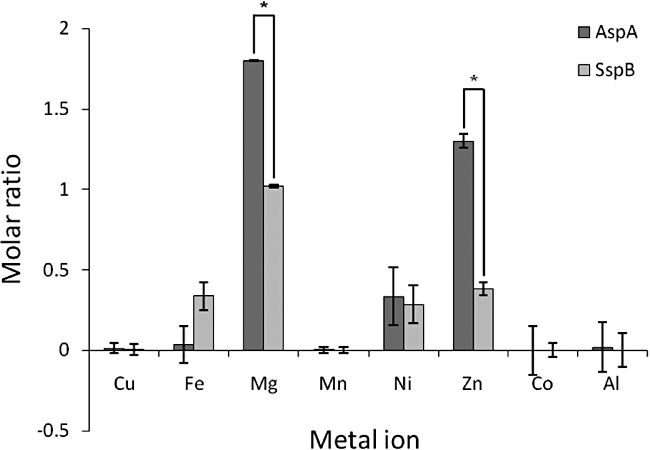
Metal ion analysis of recombinant protein fragments NAV-AspA (dark grey bars) and NAV-SspB (light grey bars) by ICP-OES using ICP multi-element standard solution IV (Merck) and 100 ng µl−1 protein. Values are metal ion : protein ratios comparative to a buffer standard containing no protein. Error bars represent ± SD from three independent experiments. *P < 0.001 between samples as indicated.
Interactions of fluid-phase gp-340 with AspA fragments
By utilizing the recombinant polypeptides described in Fig. 1, we then determined in far-Western blot overlays with gp-340 which of the various recombinant polypeptides were able to interact with gp-340. In these experiments, binding of gp-340 to the blotted polypeptides was determined using monoclonal antibody to gp-340 polypeptide backbone and anti-mouse HRP-conjugated secondary antibody. All of the SspB fragments, i.e. rNAV, rVPC and rC, and full-length rSspB, were found to bind gp-340 (Fig. 4). Purified full-length rSspB was subject to degradation and tended to bind gp-340 rather weakly. In contrast, only the full-length rAspA and rC-AspA polypeptides bound fluid-phase gp-340, while the rNAV-AspA and rVP-AspA polypeptides did not (Fig. 4). There were some apparent discrepancies between predicted and observed molecular masses on SDS-PAGE for some of these polypeptides. Anomalous migration on SDS-PAGE is common for polypeptides that are largely α-helical, e.g. A region, or that contain regions of random coils, e.g. P region. The C regions, on the other hand, resolved with predicted molecular mass of approximately 55 kDa (Fig. 4).
Fig. 4.
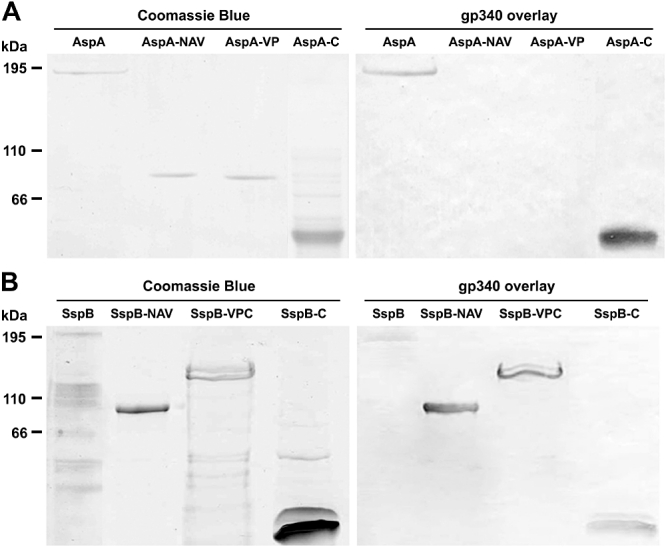
Binding of fluid-phase gp-340 by recombinant AspA or SspB protein fragments. SDS-PAGE patterns of proteins stained with Coomassie blue (left-side panels) and corresponding far-Western blots of (A) AspA-recombinant proteins AspA (NAVPC), NAV, VP and C, and (B) SspB-recombinant proteins SspB (NAVPC), NAV, VPC and C. Recombinant proteins (100–200 ng) were resolved by SDS-PAGE, blotted onto nitrocellulose and overlayered with 100 ng gp-340 ml−1. Binding of gp-340 to the bands was detected by probing with mouse monoclonal gp-340 antibody followed by HRP-conjugated goat anti-mouse antibody.
Binding of L. lactis expressing AspA to immobilized gp-340
The structurally conserved A, P and C regions of AgI/II-family proteins are present in AspA and SspB, and sequences within these three regions in SspB have all been shown to interact with gp-340 (Brady et al., 2010). To determine the ability of AspA to bind immobilized gp-340, we cloned the aspA gene, and separately the sspB gene, into the expression vector pKS80 (Hartford et al., 2001) that replicates in L. lactis. The abilities of L. lactis MG1363 strains expressing AspA (pKS80 aspA+) or SspB (pKS80 sspB+) to adhere to gp-340 were compared with control L. lactis MG1363 containing empty vector, utilizing a crystal violet spectrophotometric assay. Binding levels of L. lactis expressing AspA to immobilized gp-340 were found to be similar to those of L. lactis expressing SspB (Fig. 5A), while the control cells showed < 5% binding of the protein-expressing strains.
Fig. 5.
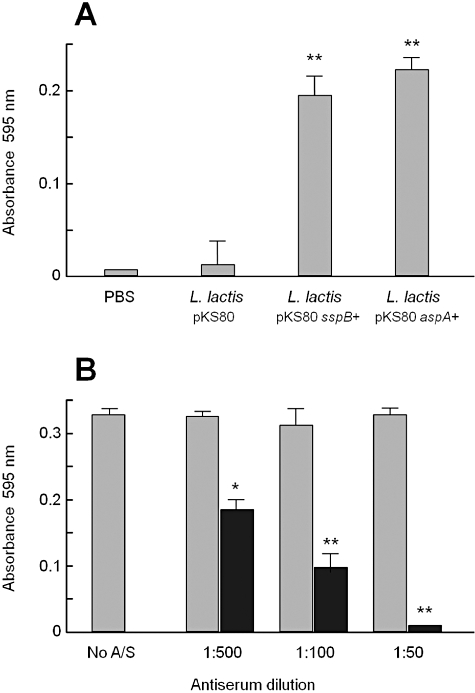
Interactions of L. lactis strains expressing AspA or SspB with immobilized gp-340 (50 ng). A. Binding of L. lactis MG1363 (pKS80) control cells, L. lactis (pKS80 sspB+) and L. lactis (pKS80 aspA+). Data represent the average ± SD of three independent experiments with triplicate samples. **P < 0.005 relative to L. lactis pKS80. B. Effect of dilutions of SpaP antiserum (grey fill) or rVP-AspA antiserum (black fill) on adherence of L. lactis (pKS80 aspA+) cells to gp-340. Control indicates no antiserum added. Input cell numbers in all experiments were 5 × 107 cells well−1. Error bars are ± SD of two independent experiments with duplicate samples. **P < 0.005, *P < 0.05, relative to no antiserum control.
Although only the C-terminal fragment of AspA bound fluid-phase gp-340 on far-Western blot overlays, it is known that fluid-phase gp-340 presents different receptor conformations compared with immobilized gp-340 (Loimaranta et al., 2005). Accordingly, to determine if the VP region of AspA carried sequences recognizing immobilized gp-340, we investigated the effects of rVP-AspA antibodies on adherence of L. lactis (pKS80 aspA+) to immobilized gp-340. It was found that antiserum raised to rVP-AspA inhibited adherence of L. lactis (pKS80 aspA+) cells to gp-340 in a dose-dependent manner (Fig. 5B), whereas corresponding dilutions of antiserum raised to S. mutans SpaP, which does not react with AspA (Fig. 2), had no inhibitory effects (Fig. 5B). Antiserum against rVP-AspA had no significant inhibitory effect on adherence of L. lactis (pKS80 sspB+) to immobilized gp-340 (data not shown).
Aggregation properties of L. lactis cells expressing AspA
To then test the ability of AspA to mediate aggregation of L. lactis by gp-340, aggregation of lactococcal suspensions mixed with gp-340 was determined. Assays were also performed with a peptide, designated SRCR Peptide 2 (SRCRP2), in place of gp-340. This peptide comprises a 16-aa-residue sequence found within the SRCR domains of the backbone of gp-340 (Leito et al., 2008). The peptide is reported to be active in aggregating a wide range of bacteria and it also interacts with AgI/II-family proteins (Jakubovics et al., 2005). In aggregation assays with gp-340, it was found that L. lactis cells expressing SspB were strongly aggregated in the presence of gp-340 while L. lactis cells expressing AspA were only weakly aggregated (Fig. 6). On the other hand, L. lactis cells expressing AspA or SspB were aggregated equally well in the presence of SRCRP2 (Fig. 6). These results indicate that the overall mechanism involved in cell aggregation as mediated by SRCRP2 is different from the mechanism associated with gp-340-mediated cell aggregation. Moreover, these gp-340-mediated aggregation data are in keeping with the above experimental results showing that the N-terminal region of AspA did not bind fluid-phase gp-340 (Fig. 4). Although the C regions of AspA and SspB were both shown to bind fluid-phase gp-340 (Fig. 4), clearly this interaction alone is not sufficient to promote aggregation of L. lactis (pKS80 aspA+) cells by gp-340.
Fig. 6.
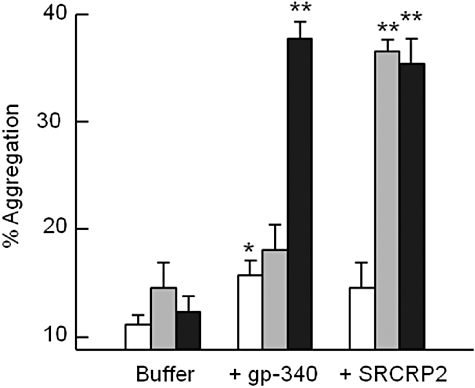
Aggregation of L. lactis strains expressing AspA or SspB in the presence of gp-340 or SRCRP2 peptide. Cells (108 cells ml−1) of each strain were incubated with 100 ng ml−1 gp-340 or 100 ng ml−1 SRCRP2 peptide, or in PBS alone, at 37°C for up to 5 h. Optical density (600 nm) was measured at intervals over 5 h and % aggregation was calculated from change in OD600, corrected for OD600 decrease in controls. Columns are L. lactis MG1363 (open), L. lactis (pKS80 aspA+) (grey fill) and L. lactis (pKS80 sspB+) (black fill). The data represent the average ± SD of three independent experiments with triplicate samples. **P < 0.005, *P < 0.05, relative to respective buffer only controls.
An additional property of SspB polypeptide is that it mediates co-aggregation between S. gordonii and A. naeslundii (Jakubovics et al., 2005). When expressed on the surface of L. lactis, SspB confers the ability on L. lactis to co-aggregate with A. naeslundii T14V (renamed A. oris T14V). In a similar manner, SspA from S. gordonii expressed on L. lactis confers co-aggregation with A. naeslundii PK606 but not with strain T14V (Jakubovics et al., 2005). L. lactis expressing AspA showed no co-aggregation ability with six different Actinomyces strains, including T14V and PK606 (data not shown). It is concluded that AspA does not interact with A. naeslundii, a major bacterial component of early dental plaque and of other oral biofilm communities (Palmer et al., 2003).
Role of AspA in GAS biofilm formation
Since various AgI/II-family adhesins have been implicated in mediating adherence and biofilm formation (Pecharki et al., 2005; Ahn et al., 2008), we investigated the abilities of two serotype M28 strains of S. pyogenes, MGAS6180 and H360, to form biofilms on salivary pellicle. Knockout mutants of these strains were generated as described in Experimental procedures and were then complemented in trans (see Fig. S2A). The allelic replacement resulted in deletion of the complete coding sequence, leaving in place the native promoter and terminator, as confirmed by sequencing. Moreover, sequencing determined that the deduced aa sequence of AspA from strain H360 was identical to that from MGAS6180.
To confirm that the aspA gene was inactivated, cell wall proteins were extracted from strains MGAS6180 and H360 following incubation with mutanolysin/lysozyme, separated by SDS-PAGE, and Western blots of proteins were incubated with rVP-AspA antibodies. The AspA protein in wild-type strain extracts appeared as a single immunoreactive band (Fig. S2B), and this band was absent from extracts of strains MGAS6180 ΔaspA and H360 ΔaspA. Cell wall extracts of complemented strains MGAS6180 ΔaspA (pKS80 aspA+) and H360 ΔaspA (pKS80 aspA+) both showed presence of many-fold higher levels of AspA protein than respective wild-type strains (Fig. S2B).
The ΔaspA mutant strains of MGAS6180 and H360 were identical in colony morphology to the respective parent strains, and had similar growth rates in minimal C medium to the parent strains. In adherence assays to immobilized gp-340, the mutants did not show any significant differences in adherence levels compared with respective wild-type (results not shown). This phenotype was not unexpected since at least two other proteins on the surface of GAS are known to interact with gp-340 (Edwards et al., 2008; Loimaranta et al., 2009).
Confocal laser scanning microscopy (CLSM) was employed to investigate differences in the development and structure of S. pyogenes biofilms as a consequence of deleting aspA. Wild-type strain MGAS6180 formed a biofilm after 24 h that was about 25 µm thick, consisting of densely packed cells with very little of the underlying salivary pellicle substratum visible (Fig. 7A). The corresponding knockout mutant, MGAS6180 ΔaspA, showed a much reduced biofilm thickness, with areas of the pellicle substratum clearly visible, and an approximately 60% reduction in biomass. Biofilms produced by strain H360 after 24 h were also approximately 25 µm thick with an ordered structure comprised of densely packed cells (Fig. 7B). The H360 ΔaspA knockout mutant was severely deficient in biofilm formation, with only small clumps of cells adhering to the pellicle substratum and a > 80% reduction in biomass (Fig. 7B). The complemented mutants were fully restored in biofilm formation, in each case reaching similar levels of biomass to the wild-type strains (Fig. 7). However, the architectures of the complemented strain biofilms were more disorganized than wild-type strain biofilms and the cells appeared to be more loosely packed (Fig. 7). These observations strongly suggest that AspA production is essential for biofilm formation by these two M28 serotype strains of GAS.
Fig. 7.
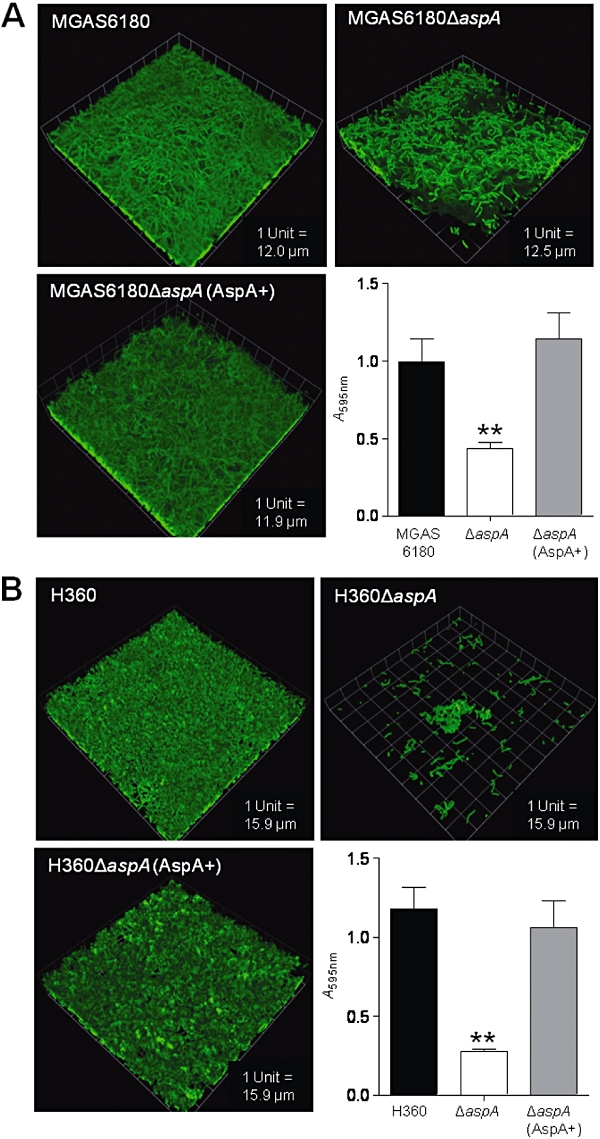
Confocal laser scanning microscopy images and biomass measurements of 24 h biofilms formed on salivary pellicle-coated coverslips, comparing biofilm structures of (A) S. pyogenes MGAS6180 and (B) strain H360, and corresponding isogenic ΔaspA mutants and complemented strains ΔaspA (aspA+). Biomass data were obtained by crystal violet assay (A595) of corresponding biofilms formed on coverslips. One unit is equivalent to the grid square side length. Data are averages ± SD of three independent experiments. **P < 0.005 relative to MGAS6180 (A) or H360 (B) wild-type strains.
To further define the specificity of AspA in mediating biofilm formation, the abilities of ΔaspA and complemented mutants to form biofilms on gp-340, saliva or polystyrene compared with wild-type strains were investigated. Total biomass measurements for each wild-type strain on the three substrata were not significantly different (Fig. 8). However, ΔaspA mutants of strains MGAS6180 and H360 were significantly impaired in biofilm formation on gp-340 compared with wild-type, with 60% reduced biomass (Fig. 8A and B). Likewise, ΔaspA mutant biofilms formed on 10% saliva-coated polystyrene showed reduced biomass by approximately 50%. In contrast, there were no significant differences in biofilm production on polystyrene between wild-type strains and their respective ΔaspA mutants (Fig. 8A and B). The complemented strains exhibited similar levels of biofilm formation (biomass) to wild-type strains on all three substrata tested (Fig. 8). These results suggest AspA specificity for salivary glycoprotein substratum.
Fig. 8.
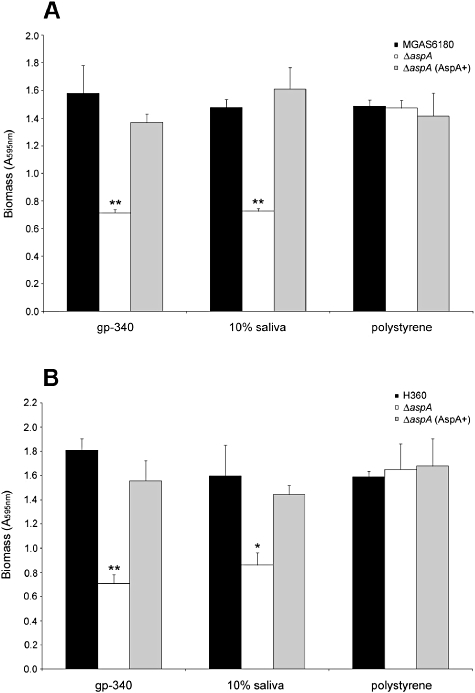
Biofilm formation on gp-340, saliva and polystyrene by (A) S. pyogenes MGAS6180 and (B) strain H360 (black fill), and corresponding isogenic ΔaspA mutants (no fill) and complemented strains ΔaspA (AspA+) (grey fill). Biomass data were obtained by crystal violet (A595) assay. Values given represent mean ± SD of three biological replicates from an experiment repeated twice. **P < 0.01, *P < 0.05, relative to respective MGAS6180 or H360 wild-type strains.
AspA directly enhances biofilm development
To confirm that AspA plays a direct function in promoting adherence and biofilm formation, the effects of expressing AspA in L. lactis on biofilm formation were investigated. Wild-type strain MG1363 cells produced a very sparse biofilm on salivary pellicle after 24 h incubation (Fig. 9A). However, L. lactis (pKS80 aspA+) expressing AspA protein produced a biofilm approximately 26 µm thick of densely packed cells (Fig. 9B), with more than threefold higher biomass (Fig. 9D). We also compared the ability of SspB protein to promote biofilm formation in L. lactis. Expression of SspB polypeptide led to a slight increase in coverage of the substratum by L. lactis (pKS80 sspB+) compared with wild-type L. lactis (Fig. 9C and D), but not to the same extent as expression of AspA did (Fig. 9B and D). This result may support the notion that AgI/II proteins may be less intimately involved in biofilm formation by S. gordonii (Zhang et al., 2005). Collectively, the results provide strong evidence that AspA is directly involved in GAS biofilm development upon salivary glycoprotein substrata. In addition, while AspA appears to differ functionally compared with some oral AgI/II-family polypeptides, it is suggested that AspA may constitute a novel colonization factor in GAS.
Fig. 9.
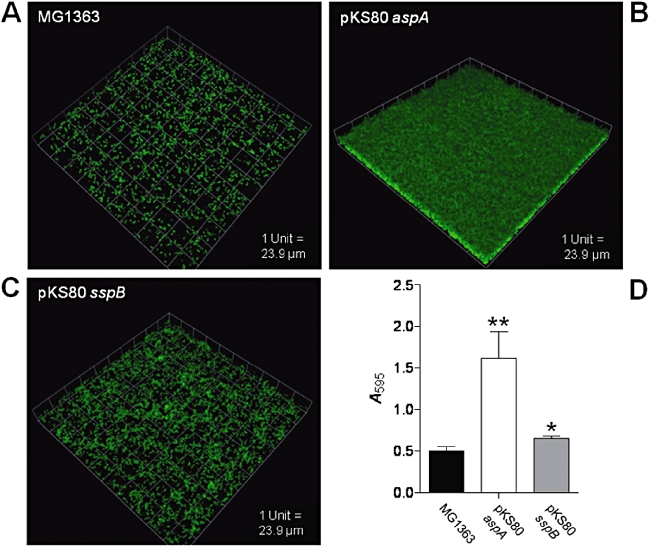
Confocal laser scanning microscopy images and corresponding biomass measurements of 24 h biofilms formed on salivary pellicle-coated coverslips of (A) wild-type L. lactis MG1363; (B) L. lactis (pKS80 aspA+); and (C) L. lactis (pKS80 sspB+). One unit is equivalent to the grid square side length. Biomass results (D) were obtained by crystal violet assay (A595) of corresponding biofilms formed on coverslips. The biomass data are averages ± SD of three independent experiments. **P < 0.005, *P < 0.05, relative to L. lactis MG1363 wild-type.
Discussion
The AgI/II proteins have been a focus in studies of the abilities of oral viridans streptococci to colonize oral cavity and nasopharyngeal surfaces, and to cause tooth decay. A primary host factor shown to be recognized by the AgI/II-family proteins is the mucin-like glycoprotein designated salivary agglutinin (or gp-340). The significance of this interaction is believed to be at least double-edged: gp-340 acts in concert with other innate factors, such as mucins, collectins and defensins, to restrict the growth of commensal bacteria, while at the same time providing a potential substratum for adhesion of bacteria to oral cavity surfaces. Some oral streptococci, e.g. S. mutans, depend strongly upon AgI/II protein for adherence, evidenced by analyses of AgI/II gene knockout mutants that are rendered deficient in adherence to salivary glycoprotein pellicle (Koga et al., 1990). Other oral streptococci, such as S. gordonii, appear to have evolved a wider repertoire of salivary pellicle adhesins (Zhang et al., 2005), and so AgI/II-deficient mutants of these organisms may not be noticeably affected in adherence (Jenkinson et al., 1993). While the structural features and biological properties of these proteins are to various degrees conserved, their biological functions in respect of microbial physiology and ecology may be quite different depending upon the complement of adhesins that are expressed.
The overall structure of the AgI/II-family proteins may be envisaged in terms of a head (V) region (Troffer-Charlier et al., 2002), projected by a semi-flexible stalk (formed by interactions of the A and P regions) linked to a wall-proximal basal C-region (Brady et al., 2010). In the SspB protein there is evidence that the head (V) and stalk (A–P) regions provide gp-340-binding activity, and that the NAV region functions in SspB-mediated co-aggregation with A. naeslundii (Jakubovics et al., 2005). The C-terminal region of SspB also has gp-340-binding activity, and carries a sequence (designated BAR) that is recognized by the periodontal disease-associated bacterium P. gingivalis (Daep et al., 2008). The results in this article suggest that the AspA N-terminal region does not interact in the same way with fluid-phase gp-340, although the C-terminal region retains gp-340-binding sequences. These results are somewhat different from those previously published (Zhang et al., 2006), where it was shown that N-terminally derived fragments of M28_Spy1325 bound fluid-phase gp-340. One reason for this discrepancy could be related to the different gp-340 preparations being employed in the two studies. In our study, gp-340 was purified from parotid saliva, which contains no mucins, by adsorption to S. mutans cells expressing AgI/II adhesin. However, Zhang et al. (2006) utilized whole saliva (from several glands) and adsorption to S. pyogenes cells expressing a range of salivary glycoprotein adhesins. Secretory IgA is known to be present in some gp-340 preparations and could bind, complexed with gp-340, to AspA (Ligtenberg et al., 2004). However, in the case of our gp-340 preparations, S-IgA is not present at the level of chemical detection (Loimaranta et al., 2005). Regardless of the reasons for the discrepancy, the present data suggest a C-terminally located AspA binding site for fluid-phase gp-340, while Zhang et al. (2006) indicate there may be multiple gp-340 binding sites in AspA.
The observation that the N-terminal region of AspA does not apparently bind fluid-phase gp-340 would be in keeping with some of the features of the AspA primary sequence that differ from those of SspB. The N-terminal region of SpaP (and of SspB) has been shown to contain a series of linear aa sequences that interact with salivary glycoproteins (SGPs) (Brady et al., 2010). These sequences are depicted as SGP1 and SGP2 in Fig. S1 and comprise SGP1 (TYEAALKQYEADL), repeated four times with some conserved aa residue changes (Okahashi et al., 1993), and SGP2 (TELARVQKANADAKAAY), repeated three times with various conserved aa residue changes (Moisset et al., 1994). In AspA, these SGP-binding sequences are poorly conserved. The corresponding SGP1 sequences in AspA contain only between two and five identical aa residues respectively out of 13 within SspB (15–38%), while the SGP2 sequences in AspA contain between two and five identical aa residues out of 17 aa residues within SspB (12–29%). Although L. lactis cells expressing AspA were not enhanced in gp-340-mediated aggregation, they were agglutinated by SRCRP2 peptide (Bikker et al., 2002), confirming that the two aggregation reactions occur by different mechanisms.
On the other hand, the ADH1 and ADH2 sequences identified by Kelly et al. (1995) within the C-terminal region of SpaP that bind gp-340 are much better conserved in AspA (Fig. S1). These sequences in AspA and SspB have 47–50% identical aa residues. Potentially therefore the gp-340-binding regions within the C-terminus have been relatively well conserved in AspA, and this is supported by the far-Western blot data showing that the C-terminal region of AspA binds gp-340.
The expression of aspA within the surrogate host L. lactis showed clearly that the expressed protein mediated adherence to immobilized gp-340, in a similar manner to SspB. This appeared to be dependent upon the VP region, since rVP-specific antibodies blocked adherence to gp-340. Antibodies to the V region of SpaP have also been shown to block adherence of S. mutans to gp-340 (Brady et al., 1992). This finding indicates that the V region of S. pyogenes AspA might be a target when devising new means to inhibit colonization of mucosal surfaces by S. pyogenes.
The SspA and SspB proteins in S. gordonii interact with other bacteria and promote the formation of mixed species communities. The N-terminal domains have been shown to mediate co-aggregation of S. gordonii with A. naeslundii in the formation of early microbial communities, with the V regions conferring strain specificity (Jakubovics et al., 2005). We were unable to demonstrate that L. lactis cells expressing AspA interacted with A. naeslundii. This suggests that targeting of the V region of S. pyogenes AspA in preventive strategies might not affect development of oral biofilm communities.
The differential interactions of AspA and SspB with gp-340 could be related to the putative metal ion-binding sites within the V regions. The V region of SspB has recently been modelled with the structure resolved to 2.3 Å (Forsgren et al., 2009) to contain a Ca2+ molecule. However, Mg2+ was shown to also fit this model as an alternative metal ion. Since Ca2+ and Mg2+ have a similar charge and are of a similar size, and they were not distinguishable under the ICP-OES conditions used, they could be interchangeable at this position. A main finding was that AspA, but not SspB, bound one Zn2+ ion per protein molecule. Binding of Zn2+ ions is often associated with four aa residue ligands (His, Asp, Glu or Cys) (Vallee and Auld, 1993; Auld, 2001). A putative Zn2+ binding site is found at 608–616 aa residues in AspA, and comprises three His and three Asp residues (Fig. S1). In addition, a number of potential, alternative ligand (His) aa residues are located distally, between 20 and 129 aa residues away from the metal ion-binding site, as is typical for Zn2+ binding protein domains (Andreini et al., 2006). None of these potential Zn2+ binding residues is present within SspB or within Enterococcus faecalis aggregation substance protein (Hendrickx et al., 2009), to which the V region of AspA has 35–40% identity. Thus the N-terminal region of AspA appears to have diverged such that it contains at least one new metal ion (Zn2+) binding site.
The Zn2+ could have a structural or catalytic role. Histidine residues are similarly positioned within Zn2+ binding sequences in other streptococcal proteins such as pneumococcal zinc metalloproteinase (Iga) (Bender and Weiser, 2006) and metallopeptidase (Froeliger et al., 1999). A gp-340-binding protein, designated StcE, identified in enterohaemorrhagic E. coli has a histidine-rich Zn2+ binding site necessary for interaction and enzymatic degradation of gp-340 (Lathem et al., 2002; Grys et al., 2005). We have not been able to show that AspA has proteolytic activity towards gp-340, under a wide range of conditions. Nor has it been possible to demonstrate proteolytic activity of rAspA on elastin, gelatin, collagen or fibrinogen (data not shown). We are currently investigating further the properties and functions of the V region in AspA.
If bacteria in the oral cavity are rapidly aggregated by gp-340 they may be cleared from the host before becoming established within communities on the hard or soft tissue or prosthetic surfaces available. Microorganisms that are able to better evade innate defences such as agglutination and phagocytosis will be able to colonize and develop a niche that, in the case of GAS, may lead to chronic infection. It is suggested that AspA on the S. pyogenes cell surface binds to gp-340 present on oral cavity or nasopharyngeal surfaces, as does SspB expressed by S. gordonii, promoting colonization. The SspB protein is then able to bind fluid-phase gp-340, promoting biofilm formation through intermolecular bridging of adhesins. AspA appears to promote biofilm community formation, at least in part, through cell–cell interactions mediated directly by AspA. Notably, AspA polypeptide expression was necessary for biofilm formation on gp-340 or salivary glycoproteins adsorbed to plastic (polystyrene), but not for biofilm formation on the plastic itself. This suggests that the bacteria respond to the physiological substratum (salivary glycoproteins). The mechanism by which this might occur is currently not known, but aspA could be upregulated in response to salivary glycoproteins, which would be in keeping with evidence that the S. gordonii sspA and sspB genes are upregulated in response to saliva (Dû and Kolenbrander, 2000).
The reduced ability of AspA to bind fluid-phase gp-340 could suggest that GAS cells are less likely to be subject to innate defence recognition. Since scavenger receptor-like (SRCR) sequences are also expressed on the surfaces of macrophages and of other immune cells (Sarrias et al., 2004), it could be that reduced binding to various forms of these receptors may further enable GAS to more effectively evade immune defences. This possibility is currently under investigation. In conclusion it is suggested that AspA provides S. pyogenes with a new salivary pellicle or mucosal surface adhesin that increases GAS competitiveness with indigenous or commensal microbiota to generate a niche. Although the pilus structures in GAS serotype M1 strain S370 have been shown to be necessary for biofilm formation (Manetti et al., 2007), pili do not appear to be essential for biofilm development by the two M28 strains used in this work. Direct AspA protein–protein interactions are probably responsible for enhanced biofilm formation and GAS community development. This could provide increased protection in vivo against killing by antibiotics or advance pathogenicity by protecting against phagocytic cells. Taken collectively, these studies identify AspA as a potential new colonization factor in GAS serotype M28 and other aspA-carrying strains that could be a novel target for intervention.
Experimental procedures
Bacterial strains
Strains used in this study are outlined in Table S1. E. coli strains JM109 and XL1 were used for molecular cloning experiments to generate pET vector- or pGEM-T-based constructs, and E. coli BL21/λDE3 (Novagen) for expressing recombinant proteins. E. coli was cultured in LB broth at 37°C with shaking. Streptococcus strains were grown in Todd–Hewitt broth (Oxoid) containing 0.5% yeast extract. Actinomyces strains were grown in TY-glucose medium (Jakubovics et al., 2005). S. pyogenes strains were cultured at 37°C in an atmosphere of 5% CO2 while all other Streptococcus and Actinomyces strains were grown in a candle jar at 37°C. L. lactis strains were routinely cultured in M17 medium containing 0.5% glucose in a candle jar at 30°C. Bacto agar (Difco) was added at 1.4% final concentration as appropriate. For analysis of aggregation properties, L. lactis strains were grown in C medium containing 0.2% glucose (Lyon et al., 1998). Antibiotics were added to media at the following concentrations: ampicillin, 100 µg ml−1 and kanamycin, 50 µg ml−1 for E. coli; erythromycin, 10 µg ml−1 for L. lactis; spectinomycin, 100 µg ml−1 and erythromycin, 5 µg ml−1 for S. pyogenes.
Recombinant protein production and purification
Chromosomal DNA was extracted from S. pyogenes MGAS6180 as described previously (Jenkinson, 1987). Primers (Sigma Aldrich) were designed to amplify regions of the locus spy1325 (aspA) corresponding to the full-length AspA protein excluding the leader sequence and cell wall anchor motif (AspA-F1 & AspA-R1), NAV region (AspA-F1 & AspA-R3), VP region (AspA-mF & AspA-mR) and C region (AspA-F3 & AspA-R1) of AspA (Table S2; Fig. S1). Additional sequence was incorporated at the 5′ end of both the forward (gacgacgacaagatx) and reverse primers (gaggagaagcccggtxx), to enable the PCR products to be cloned into the ligation-independent expression vector pET46-Ek-LIC (Novagen). PCR utilized a proofreading enzyme (Bio-X-Act Long; Bioline) and the PCR products were treated with T4 polymerase (Novagen) prior to mixing with pET46, according to the manufacturer's instructions. Fragments cloned into pET46 incorporated a sequence encoding a 6x His-tag at the N-terminus of the polypeptide.
Expression vectors were transformed into chemically-induced competent E. coli BL21/λDE3 (Novagen), prepared by the method of Hanahan (1983). Cells were grown with vigorous shaking at 37°C and protein expression was induced with IPTG (1 mM). Induced cultures were incubated at 30°C until early stationary phase and cells were lysed using Bugbuster (Novagen). Recombinant proteins were purified using immobilized metal ion affinity chromatography on IMAC Sepharose™ 6 Fast Flow (GE Healthcare). The imino-diacetic acid ligand was charged with 0.5 M NiSO4·6H2O and equilibrated with 50 mM HEPES (pH 7.4) containing 0.5 M NaCl, 10% glycerol, 0.1% Triton X-100 and 5 mM imidazole. Recombinant protein fragments were eluted using 150 mM imidazole in the same buffer and proteins were resolved by SDS-PAGE with 8% acrylamide. Full-length SspB was purified from inclusion bodies that were prepared using Bugbuster (Novagen) according to the manufacturer's instructions. Inclusion bodies were dissolved in 8 M urea supplemented with 0.1% Sarkosyl, then diluted to 3.4 M urea in 50 mM HEPES buffer (pH 7.4) containing 0.1% Sarkosyl. Recombinant SspB was refolded in situ on the IMAC column by washing with 50 mM HEPES (pH 7.4) containing 0.1% Sarkosyl, followed by further washes with 50 mM HEPES (pH 7.4) containing 0.1% Triton X-100 and 5 mM imidazole. SspB was eluted in 50 mM HEPES (pH 7.4), 0.1% Triton X-100 and 150 mM imidazole.
Metal ion analysis
Recombinant protein fragments of AspA and SspB (100 ng µl−1 in 50 mM HEPES, pH 7.4 containing 0.5 M NaCl, 10% glycerol and 0.1% Triton X-100) were analysed by ICP-OES (Perkin-Elmer Optima 3000). Calibration used the ICP multi-element standard solution IV (Merck) containing 23 elements (Ag, Al, B, Ba, Bi, Ca, Cd, Co, Cr, Cu, Fe, Ga, In, K, Li, Mg, Mn, Na, Ni, Pb, Sr, Tl, Zn) in dilute nitric acid. The protein preparation was assayed for the presence of Cu, Fe, Mn, Mg, Ni and Zn, the concentrations of which were calculated as molar ratios and normalized for the buffer alone.
Cell surface protein extraction and Western blotting
Proteins were extracted from the surface of S. pyogenes cells by modification of the method utilized to extract surface proteins from S. gordonii and L. lactis (Jakubovics et al., 2005). The modification involved incubating cells suspended in spheroplasting buffer with mutanolysin (500 U) and lysozyme (3 mg ml−1) for 2 h at 37°C. Proteins were solubilized in SDS-containing buffer without 2-mercaptoethanol and separated by SDS-PAGE. Polypeptides were transferred onto nitrocellulose membrane by Western electroblotting. Blots were blocked with 1% bovine serum albumin (BSA) or 1% skimmed milk powder in Tris-buffered saline supplemented with 0.1% Tween 20 (TTBS) and then incubated with primary antibody at appropriate dilution. Antibody binding was detected with HRP-linked goat anti-rabbit IgG, and blots were developed using 4-chloro-1-naphthol or by ECL (Amersham). Antibodies to AspA-VP were raised in New Zealand White rabbits (Covalab) and anti-SpaP rabbit antibodies were kindly provided by Charles Kelly (Kings College London). Both antisera were routinely used at 1:1000 dilution.
In vitro gp-340 binding assays
Gp-340 was prepared from parotid saliva samples pooled from multiple donors using a multi-step procedure including adsorption onto S. mutans as described previously (Loimaranta et al., 2005). Adherence of L. lactis cells expressing AspA or SspB to immobilized gp-340 was carried out by crystal violet assay and data converted to cell numbers as previously described (Jakubovics et al., 2005). Immulon 2HB 96-well microtitre plates were coated with 50 ng gp-340 in coating buffer (20 mM Na2CO3, 20 mM NaHCO3; pH 9.3) as this gp-340 concentration was found to be optimal for the assay. For antibody inhibition studies, L. lactis cells expressing AspA, or L. lactis MG1363 controls, were incubated with dilutions of various antisera for 30 min at 25°C before being assayed directly for binding to immobilized gp-340. Absorbance values were corrected against those obtained with corresponding L. lactis control cells taken through the entire procedure.
Transformation of L. lactis MG1363
Plasmid pKS80 expressing SspB (pKS80 sspB+) was produced by PCR (Expand Long PCR, Roche) of sspB (GenBank Accession No. U40027) using primer pairs BamH1F & BamH1Rev and BamSspF2 & BamH1Rev (Table S2) to remove the internal BamHI site. The sspB fragment was subcloned into pKS80 using BamHI and SbfI. Plasmid pKS80 aspA+ was produced by PCR amplification of the 4.5 kb region corresponding to aspA from S. pyogenes H360 (which shares 100% identity with spy1325/aspA from MGAS6180) using primers AgI/II-pKSF and AgI/II-pKSR (Table S1), and subcloning into pKS80 using BamHI and PstI.
Electrocompetent cells of L. lactis were prepared by culturing in GM17G broth (M17 broth supplemented with 10% glycine and 0.5% glucose) at 30°C to OD600 = 0.5–0.6. Cultures were incubated on ice for 10 min and cells harvested by centrifugation (3000 g, 4°C, 10 min). Cells were suspended in electroporation buffer (0.5 M glucose, 10% glycerol) and electroporated using a Bio-Rad Genepulser II at 1 kV voltage, 50 µF capacitance and 100 Ω resistance. Transformants were recovered in modified M17 medium (M17 containing 0.5% glucose, 0.5 M sucrose, 20 mM MgCl2 and 2 mM CaCl2), plated onto M17 agar supplemented with appropriate antibiotics, and incubated for up to 48 h at 30°C in a candle jar. Expression of SspB or AspA on the surface of L. lactis was verified by SDS-PAGE and Western immunoblot analyses. Expression levels of SspB and AspA were similar as visualized on Coomassie blue-stained gels.
Far-Western analysis of gp-340 binding to recombinant AspA protein fragments
Proteins were transferred from SDS-PAGE gels to PVDF membrane by electroblotting at 50 V for 2 h. Membranes were blocked for 16 h in Tris-buffered saline containing 0.1% Tween 20 and 1% BSA (TTBS-BSA). The membrane was incubated at 20°C for 1 h in TTBS-BSA containing 0.5 µg of purified gp-340 (Loimaranta et al., 2005) and washed twice with TTBS. Mouse monoclonal primary anti-gp-340 antibody (BioPorto Diagnostics, Denmark) in TTBS-BSA was incubated with the membrane for 1 h at 20°C and the membrane washed as before. Antibody binding was detected using HRP-conjugated goat polyclonal anti-mouse antibody (DakoCytomation) and blots were developed with 4-chloro-1-naphthol. The monoclonal primary anti-gp-340 antibody or secondary antibody did not react directly with the AgI/II protein bands.
Gp-340- or peptide SRCRP2-mediated aggregation assays
Lactococcus lactis strains were grown in C medium and harvested by centrifugation at 5000 g for 10 min. The OD600 was adjusted to 0.6, and aggregation assays were carried out in triplicate as previously described (Jakubovics et al., 2005). Gp-340 or SRCRP2 (16-mer peptide; QGRVEVLYRGSWGTVC) in PBS were added to cell suspensions (final concentration 0.5 µg ml−1). These were incubated at 37°C and OD600 readings taken at regular intervals over a period of 5 h. Controls containing no gp-340 or SRCRP2 were analysed simultaneously. Aggregation was expressed as a percentage decrease in OD600 relative to 100% aggregation.
Generation of aspA knockout mutants
Knockout mutants of strains H360 and MGAS6180 were generated through allelic replacement of aspA with aad9 (encoding spectinomycin resistance), and were complemented in trans with pKS80 aspA+ plasmid. The mechanism for generating the mutants is shown in Fig. S2A, based upon the MGAS6180 sequence. A 1045 bp region directly upstream of aspA and a 1051 bp region directly downstream were amplified using primers US1325-F & US1325-R, DS1325-F & DS1325-R respectively, which introduced BamHI sites into the products (Table S2). These fragments were stitched together in a second round of PCR using primers US1325-F & DS1325-R (Table S2) that generated a 2 kb fragment with a central BamHI site. This PCR product was ligated into pGEM-T (Promega), generating pGEM-T::aspA-2kb, and this plasmid was transformed into E. coli XL1. The aad9 gene, encoding spectinomycin resistance, was PCR-amplified using primers Aad9fwd & Aad9rev (Table S2) and then subcloned into pGEM-T::aspA-2kb at the BamHI site, thus interrupting the truncated aspA fragment and providing a means of positive selection of transformants (Fig. S2A). The resulting plasmid construct, designated pGEM-T::aspA-aad9, was transformed into GAS by electroporation as described above. Transformants were recovered on THY agar supplemented with spectinomycin at 37°C in 5% CO2 for up to 48 h. Correct insertions were identified by PCR of chromosomal DNA with primers US1325-F & DS1325-R, and the products were sequenced to confirm authenticity.
Saliva collection
Whole saliva was collected from five to eight healthy volunteers. Following collection, saliva was pooled and dithiothreitol added to a final concentration of 2.5 mM. Particulate matter and mucins were removed by centrifugation at 13 000 g for 10 min. Clarified saliva was diluted to 10% concentration with distilled water, passed through a 0.2 µm nitrocellulose filter and stored at −20°C. Local guidelines for collection, storage and disposal were followed in accordance with the Human Tissue Act.
Static biofilm models
Strains were initially grown in C medium for 16 h in 5% CO2, cells were harvested by centrifugation, and suspensions adjusted to OD600 = 0.1 in modified C medium (0.25% Difco Proteose Peptone #2, 0.75% yeast extract, 10 mM K2HPO4, 0.4 mM MgSO4, 17 mM NaCl, pH 7.5) containing 0.2% glucose. Biofilms were grown on sterile 13-mm-diameter glass coverslips (Menzel-Glaser, Germany) that had been incubated with 0.5 ml of 10% saliva (4°C for 16 h) in 24-well microtitre plates (Greiner). Cell suspensions (0.5 ml) were added to wells containing saliva-coated coverslips and incubated at 37°C under 5% CO2 for 24 h. To estimate biomass, biofilms on glass coverslips were washed twice with PBS and stained with crystal violet for 10 min. After washing with distilled water the biofilm-coated coverslips were transferred to a fresh 24-well plate, crystal violet was solubilized with 7% acetic acid, and absorbance at 595 nm (A595) measured as an indicator of biomass (Jakubovics et al., 2005).
In separate experiments to compare biofilm formation on different substrata, gp-340 (50 ng) or saliva (10%) was adsorbed onto the surface of Immulon 2HB 96-well microtitre plates. Non-specific binding sites were blocked with 3% BSA and the wells washed in TBS. S. pyogenes strains were grown in C medium for 16 h at 37°C in 5% CO2, harvested by centrifugation, and cell suspensions adjusted to OD600 = 1.0 in modified C medium containing 0.2% glucose. Suspensions (0.1 ml) were applied to triplicate wells and incubated for 3 h at 37°C in 5% CO2. Suspensions were then aspirated and the wells washed in TBS before fresh modified C medium/0.2% glucose (0.2 ml) was added to each well. Plates were incubated for a further 20 h, after which time wells were washed with TBS and the biofilms stained with 0.5% crystal violet for 2 min. Excess stain was removed by washing in TBS, remaining crystal violet solubilized in 10% acetic acid (0.1 ml), and A595 values were determined as before.
Confocal microscopy
Mature 24 h biofilms, grown as described above, were washed with PBS and fixed with 0.5 ml of 4% paraformaldehyde for 1 h at room temperature. Biofilm-coated coverslips were washed with PBS prior to staining with fluorescein isothiocyanate (FITC; 1.5 mM final concentration in 0.05 M Na2CO3 containing 0.1 M NaCl) for 1 h with gentle agitation in the dark. Washed biofilm-coated coverslips were mounted onto microscope slides in 5 µl of Vectashield mounting medium containing DAPI (Vectorlabs) and sealed onto the slide with varnish. Biofilms were visualized using a Leica TCS-SP2 confocal imaging system attached to a Leica DMIRBE inverted microscope. Analysis of the data was carried out using Volocity image analysis software (Improvision).
Statistical analysis
All data are reported as mean ± standard deviation (SD) unless otherwise indicated. Significance between samples was determined using the paired two-tailed Student's t-test, and a value of P < 0.05 was accepted as indicating significance. Data were analysed with GraphPad Prism v5 software.
Acknowledgments
We would like to thank Lindsay Dutton for excellent technical assistance; Simon Andrews and Mohan Rajasekaran for performing metal ion analysis; Alan Leard and Katy Jepson for training in CLSM; Shiranee Sriskandan, Andreas Podbielski and Tim Foster for kindly providing strains and vectors. This work was supported by a grant from The Wellcome Trust (#081855).
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ahn SJ, Ahn SJ, Wen ZT, Brady LJ, Burne RA. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect Immun. 2008;76:4259–4268. doi: 10.1128/IAI.00422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C, Banci L, Bertini I, Rosato A. Zinc through the three domains of life. J Proteome Res. 2006;5:3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- Bender MH, Weiser JN. The atypical amino-terminal LPTNG-containing domain of the pneumococcal human IgA1-specific protease is required for proper enzyme localization and function. Mol Microbiol. 2006;61:526–543. doi: 10.1111/j.1365-2958.2006.05256.x. [DOI] [PubMed] [Google Scholar]
- Bikker FJ, Ligtenberg AJ, Nazmi K, Veerman EC, van't Hof W, Bolscher JG, et al. Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J Biol Chem. 2002;277:32109–32115. doi: 10.1074/jbc.M203788200. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Schilling K, Giertsen E, Pearson S, Lee SF, Bleiweis A, Beeman D. Role of a cell surface-associated protein in adherence and dental caries. Infect Immun. 1991;59:4606–4609. doi: 10.1128/iai.59.12.4606-4609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Piacentini DA, Crowley PJ, Oyston PC, Bleiweis AS. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by the use of monoclonal antibodies against the major surface adhesin P1. Infect Immun. 1992;60:1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Maddocks SE, Larson MR, Forsgren N, Perssson K, Deivanayagam CC, Jenkinson HF. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol. 2010;77:276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney HS, Dale JB, Hasty DL. Mapping of the fibrinogen-binding domain of serum opacity factor of Group A Streptococci. Curr Microbiol. 2002;44:236–240. doi: 10.1007/s00284-001-0037-1. [DOI] [PubMed] [Google Scholar]
- Courtney HS, Ofek I, Penfound T, Nizet V, Pence MA, Kreikemeyer B, et al. Relationship between expression of the family of M proteins and lipoteichoic acid to hydrophobicity and biofilm formation in Streptococcus pyogenes. PLoS ONE. 2009;4:e4166. doi: 10.1371/journal.pone.0004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PJ, Brady LJ, Piacentini DA, Bleiweis AS. Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect Immun. 1993;61:1547–1552. doi: 10.1128/iai.61.4.1547-1552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D, Lam H, Cleary PP. Genetic dissection of the Streptococcus pyogenes M1 protein: regions involved in fibronectin binding and intracellular invasion. Microb Pathog. 2001;31:231–242. doi: 10.1006/mpat.2001.0467. [DOI] [PubMed] [Google Scholar]
- Daep CA, Lamont RJ, Demuth DR. Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein–protein interaction domain. Infect Immun. 2008;76:3273–3280. doi: 10.1128/IAI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doern CD, Roberts AL, Hong W, Nelson J, Lukomski S, Swords WE, Reid SD. Biofilm formation by group A Streptococcus: a role for the streptococcal regulator of virulence (Srv) and streptococcal cysteine protease (SpeB) Microbiology. 2009;155:46–52. doi: 10.1099/mic.0.021048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dû LD, Kolenbrander PE. Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect Immun. 2000;68:4834–4837. doi: 10.1128/iai.68.8.4834-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AM, Manetti AGO, Falugi F, Zingaretti C, Capo S, Buccato S, et al. Scavenger receptor gp340 aggregates group A streptococci by binding pili. Mol Microbiol. 2008;68:1378–1394. doi: 10.1111/j.1365-2958.2008.06220.x. [DOI] [PubMed] [Google Scholar]
- Fagan PK, Reinscheid D, Gottschalk B, Chhatwal GS. Identification and characterization of a novel secreted immunoglobulin binding protein from group A Streptococcus. Infect Immun. 2001;69:4851–4857. doi: 10.1128/IAI.69.8.4851-4857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren N, Lamont RJ, Persson K. Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB. Prot Sci. 2009;18:1896–1905. doi: 10.1002/pro.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren N, Lamont RJ, Persson K. Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain. J Mol Biol. 2010;397:740–751. doi: 10.1016/j.jmb.2010.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froeliger EH, Oetjen J, Bond JP, Fives-Taylor P. Streptococcus parasanguis pepO encodes an endopeptidase with structure and activity similar to those of enzymes that modulate peptide receptor signaling in eukaryotic cells. Infect Immun. 1999;67:5206–5214. doi: 10.1128/iai.67.10.5206-5214.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, et al. Genome sequence of a serotype M28 strain of Group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- Grys TE, Siegel MB, Lathem WW, Welch RA. The StcE protease contributes to intimate adherence of enterohaemorrhagic E. coli O157:H7 to host cells. Infect Immun. 2005;73:1295–1303. doi: 10.1128/IAI.73.3.1295-1303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Koga T, Russell MW. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994;73:1493–1502. doi: 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hartford O, O'Brien L, Schofiled K, Wells J, Foster TJ. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology. 2001;147:2545–2552. doi: 10.1099/00221287-147-9-2545. [DOI] [PubMed] [Google Scholar]
- Hendrickx APA, Willems RJL, Bonten MJM, van Schaik W. LPxTG surface proteins of enterococci. Trends Microbiol. 2009;17:423–430. doi: 10.1016/j.tim.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Hryniewicz W, Lipinski B, Jeljaszewicz J. Nature of the interaction between M protein of Streptococcus pyogenes and fibrinogen. J Infect Dis. 1972;125:626–630. doi: 10.1093/infdis/125.6.626. [DOI] [PubMed] [Google Scholar]
- Jakubovics NS, Strömberg N, van Dolleweerd CJ, Kelly CG, Jenkinson HF. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol Microbiol. 2005;55:1591–1605. doi: 10.1111/j.1365-2958.2005.04495.x. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF. Novobiocin resistant mutants of Streptococcus sanguis with reduced cell hydrophobicity and defective in coaggregation. J Gen Microbiol. 1987;133:1909–1918. doi: 10.1099/00221287-133-7-1909. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Demuth DR. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Terry SD, McNab R, Tannock GW. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect Immun. 1993;61:3199–3208. doi: 10.1128/iai.61.8.3199-3208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RH. Characterization of group A streptococcal R-28 antigen purified by hydroxylapatite column chromatography. Infect Immun. 1975;12:901–909. doi: 10.1128/iai.12.4.901-909.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CG, Todryk S, Kendal HL, Munro GH, Lehner T. T-cell, adhesion, and B-cell epitopes of the cell surface Streptococcus mutans protein antigen I/II. Infect Immun. 1995;63:3649–3658. doi: 10.1128/iai.63.9.3649-3658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Okahashi N, Takahashi I, Kanamoto T, Asakawa H, Iwaki M. Surface hydrophobicity, adherence, and aggregation of cell surface protein antigen mutants of Streptococcus mutans serotype c. Infect Immun. 1990;58:289–296. doi: 10.1128/iai.58.2.289-296.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller NM, Moritz K, Ribardo D, Jonas L, McIver KS, Sumitomo T, et al. Mode of expression and functional characterization of FCT-3 pilus region-encoded proteins in Streptococcus pyogenes serotype M49. Infect Immun. 2009;77:32–44. doi: 10.1128/IAI.00772-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MR, Rajashankar KR, Patel MH, Robinette RA, Crowley PJ, Michalek S, et al. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices. Proc Natl Acad Sci USA. 2010;107:5983–5988. doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathem WW, Grys TE, Witowski SE, Torres AG, Kaper JB, Tarr PI, Welch RA. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol Microbiol. 2002;45:277–288. doi: 10.1046/j.1365-2958.2002.02997.x. [DOI] [PubMed] [Google Scholar]
- Leito JTD, Ligtenberg AJM, Nazmi K, de Blieck-Hogervorst JMA, Veerman ECI, Nieuw Amerongen AV. A common binding motif for various bacteria of the bacteria binding peptide SRCRP2 of DMBT1/gp-340/salivary agglutinin. Biol Chem. 2008;389:1193–1200. doi: 10.1515/BC.2008.135. [DOI] [PubMed] [Google Scholar]
- Lembke C, Podbielski A, Hidalgo-Grass C, Jonas L, Hanski E, Kreikemeyer B. Characterisation of biofilm formation by clinically relevant serotypes of group A streptococci. Appl Environ Microbiol. 2006;72:2864–2875. doi: 10.1128/AEM.72.4.2864-2875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligtenberg AJ, Bikker FJ, De Blieck-Hogervorst JM, Veerman EC, Nieuw Amerongen AV. Binding of salivary agglutinin to IgA. Biochem J. 2004;383:159–164. doi: 10.1042/BJ20040265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligtenberg AJ, Veerman EC, Nieuw Amerongen AV, Mollenhauer J. Salivary agglutinin/glycoprotein-340/DMBT1: a single molecule with variable composition and with different functions in infection, inflammation and cancer. Biol Chem. 2007;388:1275–1289. doi: 10.1515/BC.2007.158. [DOI] [PubMed] [Google Scholar]
- Loimaranta V, Jakubovics NS, Hytönen J, Finne J, Jenkinson HF, Strömberg N. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect Immun. 2005;73:2245–2252. doi: 10.1128/IAI.73.4.2245-2252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loimaranta V, Hytonen J, Pulliainen AT, Sharma A, Tenuvuo AJ, Stromberg N, Finne J. Leucine rich repeats of bacterial surface proteins serve as common pattern recognition motifs of human scavenger receptor gp340. J Biol Chem. 2009;284:18614–18623. doi: 10.1074/jbc.M900581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen J, Mollenhauer J, Holmskov U. Review: gp-340/DMBT1 in mucosal innate immunity. Innate Immun. 2010;16:160–167. doi: 10.1177/1753425910368447. [DOI] [PubMed] [Google Scholar]
- Manetti AG, Zingaretti C, Falugi F, Capo S, Bombaci M, Bagnoli F, et al. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol. 2007;64:968–983. doi: 10.1111/j.1365-2958.2007.05704.x. [DOI] [PubMed] [Google Scholar]
- Moisset A, Schatz N, Lepoivre Y, Amadio S, Wachsmann D, Scholler M, et al. Conservation of salivary glycoprotein-interacting and human immunoglobulin G-cross-reactive domains of antigen I/II in oral streptococci. Infect Immun. 1994;62:184–193. doi: 10.1128/iai.62.1.184-193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci USA. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro GH, Evans P, Todryk SD, Buckett P, Kelly CG, Lehner T. A protein fragment of streptococcal cell surface antigen I/II which prevents adherence of Streptococcus mutans. Infect Immun. 1993;61:4590–4508. doi: 10.1128/iai.61.11.4590-4598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCIRD (National Center for Immunization and Respiratory Diseases: Division of Bacterial Diseases) 2008. Group A Streptococcus (GAS) Disease [www document]. URL http://www.cdc.gov/ncidod/dbmd/diseaseinfo/groupastreptococcal_g.htm.
- Nobbs AH, Shearer BH, Drobni M, Jepson MA, Jenkinson HF. Adherence and internalization of Streptococcus gordonii by epithelial cells involves β1 integrin recognition by SspA and SspB (antigen I/II family) polypeptides. Cell Microbiol. 2007;9:65–83. doi: 10.1111/j.1462-5822.2006.00768.x. [DOI] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahashi N, Takahashi I, Nakai M, Senpuku H, Nisizawa T, Koga T. Identification of antigenic epitopes in an alanine-rich repeating region of a surface protein antigen of Streptococcus mutans. Infect Immun. 1993;61:1301–1306. doi: 10.1128/iai.61.4.1301-1306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Terao Y, Hasuike K, Hamada S, Kawabata S. A novel streptococcal leucine zipper protein (Lzp) binds to human immunoglobulins. Biochem Biophys Res Commun. 2008;377:1128–1134. doi: 10.1016/j.bbrc.2008.10.126. [DOI] [PubMed] [Google Scholar]
- Palmer RJ, Jr, Gordon SM, Cisar JO, Kolenbrander PE. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J Bacteriol. 2003;185:3400–3409. doi: 10.1128/JB.185.11.3400-3409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecharki D, Petersen FC, Assev S, Scheie AA. Involvement of antigen I/II surface proteins in Streptococcus mutans and Streptococcus intermedius biofilm formation. Oral Microbiol Immunol. 2005;20:366–371. doi: 10.1111/j.1399-302X.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- Ryan PA, Pancholi V, Fischetti VA. Group A streptococci bind to mucin and human pharyngeal cells through sialic acid-containing receptors. Infect Immun. 2001;69:7402–7412. doi: 10.1128/IAI.69.12.7402-7412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrias MR, Grønlund J, Padilla O, Madsen J, Holmskov U, Lozano F. The Scavenger Receptor Cysteine-Rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol. 2004;24:1–37. doi: 10.1615/critrevimmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Mann K, Cooney J, Köhler W. Multiple binding of type 3 streptococcal M protein to human fibrinogen, albumin and fibronectin. FEMS Immunol Med Microbiol. 1993;7:135–143. doi: 10.1111/j.1574-695X.1993.tb00392.x. [DOI] [PubMed] [Google Scholar]
- Silverman RJ, Nobbs AH, Vickerman MM, Jenkinson HF. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 2010;78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soell M, Hemmerlé J, Hannig M, Haïkel Y, Sano M, Selimovic D. Molecular force probe measurement of antigen I/II-matrix protein interactions. Eur J Oral Sci. 2010;118:590–595. doi: 10.1111/j.1600-0722.2010.00785.x. [DOI] [PubMed] [Google Scholar]
- Stålhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol Microbiol. 1999;33:208–219. doi: 10.1046/j.1365-2958.1999.01470.x. [DOI] [PubMed] [Google Scholar]
- Takahashi I, Okahashi N, Matsushita K, Tokuda M, Kanamoto T, Munekata E, et al. Immunogenicity and protective effect against oral colonization by Streptococcus mutans of synthetic peptides of a streptococcal surface protein antigen. J Immunol. 1991;146:332–336. [PubMed] [Google Scholar]
- Troffer-Charlier N, Ogier J, Moras D, Cavarelli J. Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4 Å resolution suggests a sugar preformed binding site. J Mol Biol. 2002;318:179–188. doi: 10.1016/S0022-2836(02)00025-6. [DOI] [PubMed] [Google Scholar]
- Vallee BL, Auld DS. Cocatalytic zinc motifs in enzyme catalysis. Proc Natl Acad Sci USA. 1993;90:2715–2718. doi: 10.1073/pnas.90.7.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lei Y, Nobbs AH, Khammanivong A, Herzberg MC. Inactivation of Streptococcus gordonii SspAB alters expression of multiple adhesin genes. Infect Immun. 2005;73:3351–3357. doi: 10.1128/IAI.73.6.3351-3357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Green NM, SitkiewiczI I, Lefebvre RB, Musser JM. Identification and characterization of an antigen I/II family protein produced by group A Streptococcus. Infect Immun. 2006;74:4200–4213. doi: 10.1128/IAI.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


