Abstract
Deoxyhypusine synthase catalyzes an unusual protein modification reaction. A portion of spermidine is covalently added to one specific lysine residue of one eukaryotic protein, eIF5A (eukaryotic initiation factor 5A) to form a deoxyhypusine residue. The assay measures the incorporation of radioactivity from [1,8-3H]spermidine into the eIF5A protein. The enzyme is specific for the eIF5A precursor protein and does not work on short peptides (<50 amino acids). Optimum conditions for the reaction and four detection methods for the product, deoxyhypusine-containing eIF5A, are described in this chapter. The first, and most specific, method is the measurement of the amount of [3H]deoxyhypusine in the protein hydrolysate after its separation by ion exchange chromatography. However, this method requires some specialized equipment. The second method is counting the radioactivity in TCA-precipitated protein after thorough washing. The third method involves determining the radioactivity in the band of [3H] deoxyhypusine-containing eIF5A after separation by SDS-PAGE. The fourth method is a filter-binding assay. It is important to minimize nonspecific binding of [3H]spermidine to proteins in the assay mixture, especially for methods 2 and 4, as illustrated in a comparison figure in the chapter.
Keywords: Polyamine, Spermidine, Deoxyhypusine synthase, Deoxyhypusine, Hypusine, eIF5A, Posttranslational modification
1. Introduction
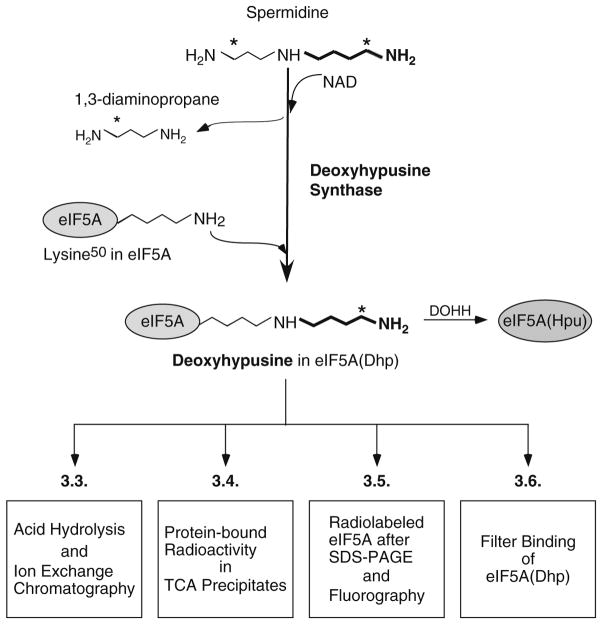
Deoxyhypusine synthase (DHS) (EC 2.5.1.46) is an essential protein found in all eukaryotes. It catalyzes the synthesis of deoxyhypusine [Nε-(4-aminobutyl)-lysine], the first step in the post-translational synthesis of an unusual amino acid hypusine [Nε-(4-amino-2-hydroxybutyl)-lysine] (see a recent review (1)). This enzyme uses two substrates, spermidine (butylamine donor) and eIF5A precursor (butylamine acceptor) and requires a cofactor, NAD (2–5). It catalyzes a NAD-dependent cleavage between N4 and C5 of spermidine and the transfer of the butylamine moiety to the ε-amino group of a specific lysine residue of the eIF5A precursor, eIF5A(Lys), to form the deoxyhypusine residue (Fig. 1). 1,3-Diaminopropane is released as a byproduct from spermidine cleavage. This protein modification is strictly specific for one single lysine residue of one cellular protein, eukaryotic initiation factor 5A (eIF5A) (6). DHS also exhibits a remarkable specificity toward its amine substrate, spermidine (7).
Fig. 1.
Biosynthesis of deoxyhypusine from the polyamine spermidine and detection methods for deoxyhypusine-containing eIF5A, eIF5A(Dhp). Deoxyhypusine synthase catalyzes the cleavage of spermidine and conjugation of its 4-aminobutyl moiety (in bold) to the ε-amino group of a specific lysine residue of eIF5A precursor to form a deoxyhypusine residue. [1,8-3H]spermidine is used in the assay and the positions of radioactivity in spermidine, 1,3-diaminopropane and deoxyhypusine are indicated by asterisk. The aminobutyl moiety derived from spermidine is indicated in bold. Four different methods for detection of radiolabeled eIF5A(Dhp) are presented in boxes.
Cultured cells or tissues normally contain the mature hypusine-containing form of eIF5A, but little or no eIF5A precursor, eIF5A(Lys) (8, 9). Cells or tissues can serve as a source of deoxyhypusine synthase, but eIF5A(Lys) must be added as a substrate (4). The DHS assay protocol can be designed either for detection of the enzyme or of eIF5A(Lys) (4, 5, 10). The cDNAs for both eIF5A (11) and DHS (12) have been cloned and recombinant proteins can be purified by simple methods (12–14). Since the in vitro assay uses a low concentration of [1,8-3H]spermidine, it is critical to use a high pH buffer (pH 9.2–9.5) (2) (since the percentage of the properly charged spermidine substrate (15) with the unprotonated secondary amino group is highest at this pH range (16)). Because only a very small portion of [1,8-3H]spermidine in the assay is converted to deoxyhypusine and because spermidine tightly adheres to proteins in the assay mixture, it is most critical to remove all the unreacted radiolabeled spermidine from the labeled protein product.
Typical reaction conditions for deoxyhypusine synthesis are described under Subheading 3.2. The [3H]deoxyhypusine formed in eIF5A can be measured by four different methods (Fig. 1, Subheading 3.3–3.6): (1) determination of radioactive deoxyhypusine after its ion exchange chromatographic separation (4, 5, 17, 18), (2) determination of protein-bound radioactivity by TCA precipitation after repeated washing, (3) determination of radioactivity in eIF5A after SDS-PAGE (3, 10, 15, 19), (4) filter binding assay of radiolabeled eIF5A (20). The first method (Subheading 3.3) gives the most accurate estimate of the amount of [3H]deoxyhypusine formed, since it is measured after its complete separation from [1,8-3H]spermidine. However, it requires an amino acid analyzer (or HPLC system) with an ion exchange column with specific buffers. For those who do not have access to such a separation system, one of the other methods can be employed for an estimation of enzyme activity. The sensitivity (signal) and the background of the DHS assay are compared for the four detection methods (Fig. 2).
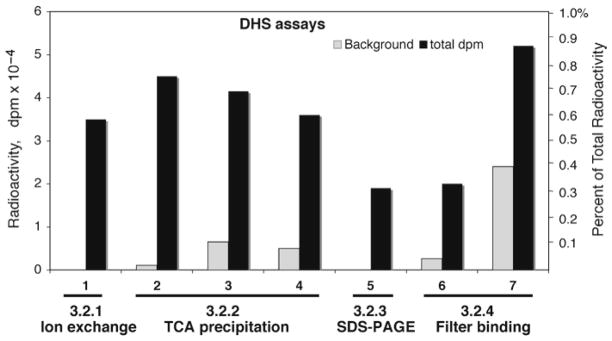
Fig. 2.
Comparison of different methods of measurement of deoxyhypusine formed. Approximately 2.5 units of purified DHS (lanes 1, 2, 5–7 ) or 50 μg of total HeLa cell protein (lanes 3 and 4) was used as enzyme source. Purified eIF5A(Lys) (2.5 μg) was used in each case. 1, Measurement of [3H]deoxyhypusine after its separation by ion exchange chromatography, accurate estimate with no background. 2–4, Estimation of TCA precipitable radioactivity after thorough TCA wash. The background is low when pure DHS and eIF5A(Lys) were used with no carrier BSA in the reaction mix (lane 2 ). However, when crude HeLa extract was used as an enzyme source, the amount of [1,8-3H]spermidine noncovalently bound to proteins increases (lanes 3 and 4 ). This background binding was estimated when DHS is completely inhibited in the presence of 0.1 mM of a DHS inhibitor, N1-guanyl-1,7-diaminoheptane (GC7). 5, Radioactivity in the 18 kDa eIF5A after its separation by SDS-PAGE. 6, A filter-binding assay with purified DHS and eIF5A(Lys) without BSA. 7, A filter-binding assay that contained BSA in the reaction mix shows high background level of [1,8-3H]spermidine binding to the filter. Total dpm per assay is indicated on the left axis, and the percent of the total radioactivity in the reaction mix is shown on the right axis.
2. Materials
Those required for a specific protocol are indicated by the method number.
2.1. Equipment
Plastic Eppendorf tubes (1.5 ml size) with screw caps, resistant to heating at 110°C (Subheading 3.3).
Dry heating block (108°C) for acid hydrolysis (Subheading 3.3).
SpeedVac concentrator (e.g., Savant) with a NaOH trap (Subheading 3.3).
An amino acid analyzer with an ion exchange column (e.g., 0.4 × 7.5 cm, DC-6A cation exchange resin) (Subheading 3.3) (see Note 1).
Liquid scintillation fluid that tolerates high salt buffers (Optiphase High Safe 3 from PerkinElmer works well) (Subheading 3.3).
Whatman 3 MM filter discs (2.4 cm diameter) (Subheading 3.6).
2.2. Reagent Stock Solutions
Use deionized or ultrapure water for preparation of all solutions.
Buffer A: 50 mM Tris–HCl buffer (pH 7.5) and 1 mM DTT.
Glycine/NaOH buffer: 1 M, pH 9.2 buffer. Dissolve 7.5 g of glycine in 50 ml of water, and adjust pH to 9.2 by adding NaOH solution (2 N), adjust the final volume to 100 ml with water and store at 4°C.
eIF5A(Lys) solution (5 mg/ml) (see Note 2 for pure recombinant protein or alternate source of eIF5A precursor). Store aliquots of solutions in 50 mM Tris–HCl pH 7.5, 0.3 M KCl, 1 mM DTT buffer in a −70°C freezer.
20 mM NAD: Dissolve 13.7 mg/ml of NAD sodium salt in water and store at −20°C in aliquots.
DTT (100 mM): the solution of 100 mM DTT (15.4 mg/ml) in water is stored frozen at −20°C in aliquots.
BSA (50 mg/ml in water), Store frozen at −20°C in aliquots.
[1,8-3H]spermidine (16–20 Ci/mmol) from PerkinElmer Life Sciences is stored at 4°C.
TCA polyamine solution (10% trichloroacetic acid (TCA) containing 1 mM each of putrescine, spermidine, and spermine), Store at 4°C.
0.1 N NaOH containing 1 mM each of putrescine, spermidine, and spermine.
6 N HCl solution (Subheading 3.2).
PA buffer B: 13.2 mM sodium citrate buffer, 1.5 N NaCl, pH 5.55, 0.1% Phenol.
PA buffer C: 53 mM sodium citrate buffer, 3.0 N NaCl, pH 5.55, 0.1% Phenol. Buffers filtered and stored at room temperature; Brij is added (to 0.1%) just before use (Subheading 3.3).
SDS-PAGE buffers, sample buffer and running buffers from Invitrogen (Subheading 3.5).
Coomassie-Blue staining solution: 10% acetic acid, 50% methanol, 1.25 g/l Brilliant Blue R dye.
Destaining solution: 10% acetic acid, 10% methanol (Subheading 3.5).
Amplify (GE Healthcare) for soaking gels prior to fluorography (Subheading 3.5).
PCA polyamine solution: 4% perchloric acid solution containing 1 mM each of putrescine, spermidine, and spermine (Subheading 3.6).
3. Methods
3.1. Preparation of Enzyme Extract from Cultured Cells or Tissues
For preparation of a crude enzyme extract, sonicate freshly harvested cell pellets or homogenize fresh tissues in Buffer A containing protease inhibitor cocktail (0.2 ml buffer for 107 cells, or 1 ml per 1 g wet tissue). Centrifuge the lysates in a refrigerated microfuge (at 15,000 × g) for 20 min and take the supernatants for protein determination and enzyme assays (see Note 3). Use 20–100 μg of total protein per assay as an enzyme source.
3.2. DHS Assay for Measurement of DHS Activity (see Note 4)
-
Just before the assay, make a master mixture by multiplying each volume by the sample number (n) and keep on ice in an Eppendorf tube.
Stock solution volume per assay final concentration1. 1 M Glycine NaOH buffer pH 9.2 4 μL 0.2 M 2. 100 mM DTT 0.2 μL 1 mM 3. 20 mM NAD 1 μL 1 mM 4. 50 mg/ml BSA (optional, see Note 5) 1 μL 2.5 mg/ml 5. [1,8-3H]spermidine (see Note 6) 3 μL (~150 pmol) 7.5 μM 6. 6 mg/ml eIF5A(Lys) 0.5 μL 9.0 μM 7. H2O 5.3 μL 8. Volume of master mix per tube 15 μL 9. Enzyme solution/lysate 5 μL 10. Total volume 20 μL Set up screw cap plastic Eppendorf tubes and number them. Add an enzyme sample in 5 μl to each. Use 5 μl of Buffer A as a negative control and 2–10 units of purified DHS (if available) as a positive control (see Note 7).
Start the reaction by adding 15 μl of the master mix to each tube. Mix gently and cap each tube.
Incubate the reaction mixtures in a 37°C water bath for 60 min.
3.3. Determination of Deoxyhypusine Formed After Ion Exchange Chromatographic Separation
Stop the reaction by adding 5 μl of spermidine (0.1 M), 5 μl (250 μg) of carrier BSA to each tube. Immediately add 1 ml of ice-cold 10% TCA polyamine solution. Gently mix and keep on ice for 30 min.
Centrifuge the TCA precipitated samples in a refrigerated microfuge (4°C) at 15,000 × g for 5 min and remove all the supernatant by careful suction using a fine 10 μl tip.
Add 1 ml of the TCA polyamine solution to each sample. Resuspend the pellet by pipetting up and down using 200 μl tips and centrifuge again to remove all the supernatant.
Dissolve the TCA pellet in 0.1 ml 0.1 N NaOH containing 1 mM each of polyamines. Immediately add 1 ml of the TCA polyamine solution and keep on ice for 30 min to precipitate the proteins. Repeat this step if needed, until the radioactivity in the 0.1 ml of the final wash is sufficiently low (<200 dpm) (see Note 8).
After removing all the TCA supernatant, resuspend the pellet in 0.4 ml of 6 N HCl, tighten the cap and place in a heating block at 108°C overnight.
Dry the hydrolyzed samples using a SpeedVac concentrator set up with a NaOH trap.
Add 0.1 ml of water to each tube to dissolve dried residues. Count a 5 μl aliquot and apply an aliquot containing up to 50,000 dpm to the amino acid analyzer (see Note 9).
Separate deoxyhypusine by running 13 min of PA Buffer B at the speed of 0.75 ml per min. Collect 13 tubes of 1 min fractions and count the total radioactivity in a liquid scintillation counter. In our machine, the elution peak time for deoxyhypusine is approximately 10 min (fractions 9–11 contain deoxyhypusine). After running five samples, the column should be washed with PA buffer C for 30 min and reequilibrated with PA buffer B (20 min) for the next cycle of five samples (see Note 10).
Sum the radioactivity of the deoxyhypusine peak (usually in four fractions, 9–12), after subtraction of background radioactivity in the negative control, and multiply the number by a factor (100/volume injected) to estimate the total amount of deoxyhypusine formed in each reaction (see Note 11).
3.4. Determination of Protein-Bound Radioactivity by TCA Precipitation
If an amino acid analyzer or HPLC system is not available, one can follow the Subheading 3.3, steps 1–4 described above and count radioactivity in the TCA precipitates (see Note 12).
Dissolve the TCA precipitates (from Subheading 3.3, step 4) in 0.1 ml of 0.1 N NaOH and count the radioactivity of the dissolved proteins.
3.5. Determination of Radiolabeled eIF5A(Dhp) After SDS-PAGE (see Note 13)
Mix 10 μl of each reaction sample with 10 μl of SDS sample buffer (2×) and separate proteins by PAGE on 4–12 % gradient NuPAGE Bis–Tris gel and MES electrophoresis buffer.
Stain for 1 h, destain the gel for 20 h with two changes of destaining solution, and identify the eIF5A band (18 kDa) (see Note 14). Scan the stained gel for a record.
Cut out the stained eIF5A band and add to a screw cap Eppendorf tube. Add 0.5 ml of 30% hydrogen peroxide solution and incubate at 70°C overnight. After the gel pieces dissolve, count the radioactivity by liquid scintillation counting.
The radioactivity incorporated into eIF5A can also be determined by fluorography. Wash the destained gel with deionized water for 15 min and soak it in Amplify (GE Healthcare) for 30 min. Expose the gel to X-Ray film (BioMax XAR film) at –70°C. Develop the film after 2–20 days (see Note 15) of exposure and scan the intensity of the labeled eIF5A band using a densitometer.
3.6. Filter Binding Assay of Radiolabeled eIF5A(Dhp) Formed
Set up the reaction as in Subheading 3.2, but with omission of BSA (see Note 16). Stop the reaction by adding 5 μl of 100 mM spermidine.
Spot 10 μl of the reaction mixtures on Whatman 3 MM filter paper discs in duplicate. Mark numbers on the discs with a pencil.
Let the filters dry for 5 min, then wash the filters in the PCA polyamine solution (50 ml per ten filters in a beaker, heated at 90°C) for 5 min.
Wash the filters in fresh hot PCA polyamine solution and heat for 5 min. Repeat washing, if necessary, until the radioactivity in the wash is sufficiently low (see Note 17).
Rinse the filters briefly in 0.2 N HCl and in ethanol. Count radioactivity of the filters after letting the filters sit in the liquid scintillation fluid for 20 h at room temperature (see Note 18).
Acknowledgments
This research was supported by the Intramural Research Program of the NIDCR, National Institutes of Health.
Footnotes
We use a Dionex D-400 analyzer using a column (0.4 × 7.5 cm) of cation exchange resin, DC-6A (17, 18). Elution conditions on our analyzer are temperature 66°C, flow rate 0.75 ml/min, and we collect 0.5 or 1 min fractions directly into 7 ml glass scintillation vials. An amino acid analyzer or HPLC system can be modified to separate basic compounds (e.g., hypusine, deoxyhypusine, spermidine, and 1,3-diaminopropane (17, 18)) using the high salt PA buffers given in step 12 in Subheading 2.2.
The human recombinant protein substrate is purified from BL21(DE3) cells transformed with pET11a/heIF5A as described in ref. (14). Crude extracts prepared from mammalian cells, that are depleted of spermidine and thereby have accumulated eIF5A(Lys), can be used as a source of substrate protein (2–4).
Crude extracts from cells or tissues contain spermidine (~10 nmol per mg protein) and will dilute the specific activity of [1,8-3H]spermidine added to the assay. This dilution has to be calibrated for accurate estimation of DHS activity. Crude extract from fresh HeLa cells contain ~1 unit of activity per 10 μg of total proteins. If not used immediately, the crude extracts should be frozen on dry ice and stored at −80°C.
The assay can be set up for detection of DHS activity (as in Subheading 3.2) or for detection of substrate, eIF5A(Lys) (4, 10, 14, 15, 19, 21). In the latter case, add 10 units of DHS per assay (if purified enzyme is available) instead of eIF5A(Lys) in the master mix and samples containing eIF5A(Lys) in 5 μl is assayed by addition of master mix containing DHS. If a crude cell lysates sample containing both eIF5A(Lys) and DHS are assayed (2–4, 19), the master mix lacking both protein substrate and enzyme can be used. If accurate quantitative measurement is required, set up the samples in duplicate and/or in larger volume (40–100 μl per assay).
Addition of carrier BSA (25 μg per assay) prevents loss of eIF5A(Lys) and DHS by their adsorption to the plastic tubes, and increases the yield of products. If the product is separated from radioactive spermidine by ion exchange chromatographic separation (Subheading 3.3) or SDS-PAGE (Subheading 3.3, step 3), it is better to include carrier BSA in the reaction mix. Since BSA binds [1,8-3H]spermidine tightly and markedly increases the background (Fig. 2), its addition is not recommended when counting TCA precipitable radioactivity (Subheading 3.4) or the filter-binding assay (Subheading 3.6).
One could use [14C]spermidine (100–124 mCi/mmol) instead of [1,8-3H]spermidine. However, its specific activity is ~200 times lower than that of [1,8-3H]spermidine and is much more costly. The concentration of [1,8-3H]spermidine is based on a batch with specific activity of 20 Ci/mmol. The amount added can be adjusted if another batch with a different specific activity is used.
1 unit of activity is defined as that generating 1 pmol of deoxyhypusine in 1 h at 37°C (22). Human recombinant DHS freshly prepared from BL21(DE3) cells has ~1 unit per 1 ng of enzyme (12), but its activity gradually decays upon storage at 70°C.
If one removes the wash supernatant completely, one repeat is sufficient, but it is useful to count the wash supernatant (0.1 ml) at each step of washing.
By estimating total radioactivity in 100 μl of hydrolyzed sample, one can calculate the volume of sample to apply to the column. It is recommended not to overload the column with too high radioactivity (>50,000 dpm) to avoid machine contamination and carry-over to the next run.
In our system, spermidine elutes at ~67 min in PA buffer B. Therefore, after a cycle of five runs of 13 min each with B buffer, the column should be washed with a high salt buffer (PA buffer C) (30 min) and reequilibrated with buffer B (20–25 min) to elute all the radioactive spermidine accumulated in the previous runs.
The radioactivity in deoxyhypusine fraction in dpm can be converted to pmol as follows. For a batch of [1,8-3H]spermidine with 20 Ci/mmol specific activity, 1 pmol of spermidine is 4 × 104 dpm. Since only the aminobutyl side of [1,8-3H] spermidine is incorporated into deoxyhypusine, the specific activity of [3H]deoxyhypusine is 2 × 104 dpm per pmol.
[1,8-3H]spermidine binds very tightly to BSA or other cell proteins, and it is impossible to remove completely the [1,8-3H]spermidine noncovalently bound to TCA precipitates. One can minimize this background by following the wash protocol in Subheading 3.2, step 1, point 8.
It is convenient to use precast NuPAGE gels (4–12%) from Invitrogen and readymade electrophoresis buffers. However, any other SDS-PAGE system that resolves eIF5A (18 kDa) will work. When the radioactive reaction mix is applied to the SDS gel, the buffer, staining and destaining solutions all contain radioactive spermidine and need to be disposed of properly.
eIF5A(18 kDa) can be easily identified on a stained gel, if purified DHS and eIF5A are used. If crude eIF5A or DHS preparations are used, the eIF5A band can be identified after fluorography.
The period required to detect the labeled eIF5A band depends on the amount of radioactivity. The radiolabeled band with >2,000 dpm of 3H-labeled protein of a SDS gel treated with Amplify is visible after 3-day exposure to X-ray film at −80°C.
As shown in Fig. 1, this assay works well when purified enzyme and eIF5A(Lys) are used, for example, for screening of DHS inhibitors. However, in assays containing a large amount of crude proteins or BSA carrier which traps [1,8-3H]spermidine, the background filter binding is quite high, regardless of wash methods. So it is appropriate only when a large enzyme activity is used (high signal/noise ratio) or when purified enzyme and substrate proteins are used.
Count 0.1 ml of the second and third wash until the radioactivity in the wash is low (<100 dpm) and constant.
Since 3H is low energy beta emitter, its counting efficiency on the filter bound form is low. However, after sitting in the liquid scintillation fluid for 18 h, a stable level of radioactivity is obtained, probably due to the release of radiolabeled protein into the liquid phase.
References
- 1.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem. 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphey RJ, Gerner EW. Hypusine formation in protein by a two-step process in cell lysates. J Biol Chem. 1987;262:15033–15036. [PubMed] [Google Scholar]
- 3.Chen KY, Dou QP. NAD+ stimulated the spermidine-dependent hypusine formation on the 18 kDa protein in cytosolic lysates derived from NB-15 mouse neuroblastoma cells. FEBS Lett. 1988;229:325–328. doi: 10.1016/0014-5793(88)81149-9. [DOI] [PubMed] [Google Scholar]
- 4.Park MH, Wolff EC. Cell-free synthesis of deoxyhypusine. Separation of protein substrate and enzyme and identification of 1, 3-diaminopropane as a product of spermidine cleavage. J Biol Chem. 1988;263:15264–15269. [PubMed] [Google Scholar]
- 5.Wolff EC, Park MH, Folk JE. Cleavage of spermidine as the first step in deoxyhypusine synthesis. The role of NAD. J Biol Chem. 1990;265:4793–4799. [PubMed] [Google Scholar]
- 6.Wolff EC, Kang KR, Kim YS, Park MH. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33:341–350. doi: 10.1007/s00726-007-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakus J, Wolff EC, Park MH, Folk JE. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies Effective inhibition by bis- and monoguanylated diamines and polyamines. J Biol Chem. 1993;268:13151–13159. [PubMed] [Google Scholar]
- 8.Duncan RF, Hershey JW. Changes in eIF-4D hypusine modification or abundance are not correlated with translational repression in HeLa cells. J Biol Chem. 1986;261:12903–12906. [PubMed] [Google Scholar]
- 9.Park MH. Regulation of biosynthesis of hypusine in Chinese hamster ovary cells. Evidence for eIF-4D precursor polypeptides. J Biol Chem. 1987;262:12730–12734. [PubMed] [Google Scholar]
- 10.Byers TL, Wiest L, Wechter RS, Pegg AE. Effects of chronic 5′-([(Z)-4-amino-2-butenyl]methylamino)-5′-deoxy-adenosine (AbeAdo) treatment on polyamine and eIF-5A metabolism in AbeAdo-sensitive and -resistant L1210 murine leukaemia cells. Biochem J. 1993;290:115–121. doi: 10.1042/bj2900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smit-McBride Z, Dever TE, Hershey JW, Merrick WC. Sequence determination and cDNA cloning of eukaryotic initiation factor 4D, the hypusine-containing protein. J Biol Chem. 1989;264:1578–1583. [PubMed] [Google Scholar]
- 12.Joe YA, Wolff EC, Park MH. Cloning and expression of human deoxyhypusine synthase cDNA Structure-function studies with the recombinant enzyme and mutant proteins. J Biol Chem. 1995;270:22386–22392. doi: 10.1074/jbc.270.38.22386. [DOI] [PubMed] [Google Scholar]
- 13.Smit-McBride Z, Schnier J, Kaufman RJ, Hershey JW. Protein synthesis initiation factor eIF-4D Functional comparison of native and unhypusinated forms of the protein. J Biol Chem. 1989;264:18527–18530. [PubMed] [Google Scholar]
- 14.Joe YA, Park MH. Structural features of the eIF-5A precursor required for post-translational synthesis of deoxyhypusine. J Biol Chem. 1994;269:25916–25921. [PubMed] [Google Scholar]
- 15.Dou QP, Chen KY. Characterization and reconstitution of a cell free system for NAD(+)-dependent deoxyhypusine formation on the 18 kDa eIF-4D precursor. Biochim Biophys Acta. 1990;1036:128–137. doi: 10.1016/0304-4165(90)90024-q. [DOI] [PubMed] [Google Scholar]
- 16.Lee YB, Joe YA, Wolff EC, Dimitriadis EK, Park MH. Complex formation between deoxyhypusine synthase and its protein substrate, the eukaryotic translation initiation factor 5A (eIF5A) precursor. Biochem J. 1999;340:273–281. [PMC free article] [PubMed] [Google Scholar]
- 17.Folk JE, Park MH, Chung SI, Schrode J, Lester EP, Cooper HL. Polyamines as physiological substrates for transglutaminases. J Biol Chem. 1980;255:3695–3700. [PubMed] [Google Scholar]
- 18.Park MH, Cooper HL, Folk JE. Chromatographic identification of hypusine [Nε-(4-amino-2-hydroxyl)lysine] and deoxyhypusine [Nε-(4-aminobutyl)lysine) Methods Enzymol. 1983;94:458–462. [Google Scholar]
- 19.Byers TL, Ganem B, Pegg AE. Cytostasis induced in L1210 murine leukaemia cells by the S-adenosyl-L-methionine decarboxylase inhibitor 5′-([(Z)-4-amino-2-butenyl]methylamino)-5′-deoxyadenosine may be due to hypusine depletion. Biochem J. 1992;287(Pt 3):717–724. doi: 10.1042/bj2870717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki K, Abid MR, Miyazaki M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;384:151–154. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- 21.Byers TL, Lakanen JR, Coward JK, Pegg AE. The role of hypusine depletion in cytostasis induced by S-adenosyl-L-methionine decarboxylase inhibition: new evidence provided by 1-methylspermidine and 1, 12-dimethylspermine. Biochem J. 1994;303(Pt 2):363–368. doi: 10.1042/bj3030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolff EC, Lee YB, Chung SI, Folk JE, Park MH. Deoxyhypusine synthase from rat testis: purification and characterization. J Biol Chem. 1995;270:8660–8666. doi: 10.1074/jbc.270.15.8660. [DOI] [PubMed] [Google Scholar]