Abstract
Genome-wide association studies (GWAS) have identified several susceptibility loci for bipolar disorder (BP), most notably ANK3. However, most of the inherited risk for BP remains unexplained. One reason for the limited success may be the genetic heterogeneity of BP. Clinical sub-phenotypes of BP may identify more etiologically homogeneous subsets of patients, which can be studied with increased power to detect genetic variation. Here, we report on a mega-analysis of two widely studied sub-phenotypes of BP, age at onset and psychotic symptoms, which are familial and clinically significant. We combined data from three GWAS: NIMH Bipolar Disorder Genetic Association Information Network (GAIN-BP), NIMH Bipolar Disorder Genome Study(BiGS), and a German sample. The combined sample consisted of 2836 BP cases with information on sub-phenotypes and 2744 controls. Imputation was performed, resulting in 2.3 million SNPs available for analysis. No SNP reached genome-wide significance for either sub-phenotype. In addition, no SNP reached genome-wide significance in a meta-analysis with an independent replication sample. We had 80% power to detect associations with a common SNP at an OR of 1.6 for psychotic symptoms and a mean difference of 1.8 years in age at onset. Age at onset and psychotic symptoms in BP may be influenced by many genes of smaller effect sizes or other variants not measured well by SNP arrays, such as rare alleles.
Introduction
Family, twin and adoption studies provide convincing evidence that bipolar disorder (BP) has a strong genetic component. Recently, several genome-wide association studies (GWAS) have been conducted to identify genes involved in BP susceptibility. Unfortunately, there has been little consistency in the top results reported by individual studies. The Wellcome Trust Case Control Consortium (WTCCC) (2007)reported an association on chromosome 16p12 (p=6.3×10−8). Baum et al (2008) found evidence that DGKH was associated with BP (p=1.5×10−8). Sklar et al (2008) identified MYO5B as being associated with BP. Smith et al (2009) found that the strongest BP signal was located in the region Xq27.1 in a European American sample (p=1.6×10−6) and in DPY19L3 in an African American sample (p=1.5×10−6).
The lack in consistency of top results from different studies has been a general property of GWAS, including for non-psychiatric disorders, and may be due to the modest power of individual studies. Consequently, several meta-and mega-analyses have been performed in the hopes that increasing the sample size will lead to stronger results. Scott et al (2009) reported a signal in ITIH1 in a meta-analysis of three samples from the NIMH/Pritzker, GlaxoSmithKline Research & Development, and WTCCC (p=1.8×10−7). A mega-analysis of the GWAS from the WTCCC, STEP-UCL and ED-DUB-STEP2 studies identified ANK3 and CACNA1C as genes reaching genome-wide statistical significance (Ferreira et al., 2008). Finally, in a far-reaching collaborative effort, the Psychiatric GWAS Consortium(PGC)has now performed a mega-analysis of 11 individual BP GWAS and found compelling evidence that ANK3 and SYNE1 are involved in BP (Kelsoe, 2009).
Despite these efforts, the majority of the heritability of BP remains unexplained. One reason for the lack of greater success in elucidating the inherited risk may be due to genetic heterogeneity of the disorder. We have hypothesized that clinical sub-phenotypes of BP identify more homogeneous subsets of patients that can be studied with increased power to detect genetic variation(Potash et al., 2007). This strategy was helpful with Alzheimer’s disease (Scott et al., 2000)and breast cancer (King, 1991), where age at onset was used to identify genes contributing to etiology.
Age at onset and psychotic symptoms are clinical features of BP that are heritable and thus may be especially useful markers for identifying BP related genes. Age at onset has been shown to aggregate in BP families (Lin et al., 2006)and has been incorporated into linkage analyses to identify regions which may contain genes that increase susceptibility for early or late-onset BP (Lin et al., 2005;Zandi et al., 2007). In addition, several candidate gene association studies have suggested that genes such as BDNF (Tang et al., 2008)and serotonergic genes including HTR2C (Massat et al., 2007), HTR2A (Manchia et al., 2010), and SLC6A4 (Manchia et al., 2010)might be associated with early onset BP. Psychotic symptoms have also been shown to aggregate in BP families (Potash et al., 2001). Linkage studies have identified several regions that may harbor susceptibility genes specific to psychotic BP(Kerner et al., 2007;Potash et al., 2003), and candidate gene association studies have implicated genes such as NRG1 (Green et al., 2005;Goes et al., 2009) and DAOA (Schulze et al., 2005). However, these results are inconclusive as replication is generally lacking.
Here, we report the results of a mega-analysis of combined data from three GWAS in which we incorporate information on age at onset and psychotic symptoms. The three datasets include: NIMH Bipolar Disorder Genetic Association Information Network (GAIN-BP), NIMH Bipolar Disorder Genome Study(BiGS), and a German sample.
Materials and Methods
Subjects
NIMH-BP Samples
The ascertainment and assessment procedures for the NIMH-BP sample are described elsewhere (Dick et al., 2003;Kassem et al., 2006). Briefly, the sample was collected in 5 waves. In waves 1–4 families who had a proband with bipolar I disorder and at least one other sibling with bipolar I or schizoaffective disorder, bipolar type were sought. In wave 5 unrelated bipolar I cases were recruited. All subjects were assessed with the Diagnostic Interview for Genetic Studies (DIGS) and this was combined with family informant data and medical records to assign diagnoses based on DSM-III-R or DSM-IV criteria (Nurnberger, Jr. et al., 1994). Unrelated cases were genotyped in two separate efforts described in detail elsewhere, the Genetic Association Information Network Bipolar Sample (GAIN-BP)(Smith et al., 2009)and the Bipolar Genome Study(BiGS)(manuscript in progress). Genotyping in both efforts was performed using the Affymetrix 6.0 array.
Controls for both the GAIN-BP and BiGS samples were ascertained throughout the United States and genotyped on the Affymetrix 6.0 array as part of the Molecular Genetics of Schizophrenia II (MGS2) Collaboration (Sanders et al., 2008). All control subjects completed a psychiatric questionnaire and those endorsing a history of BP, psychosis or major depression were excluded. Controls were matched by ethnicity, age and sex to the BP cases.
We included the final cleaned dataset from the primary case-control analysis of each sample. The GAIN-BP sample consisted of 1001 BP cases and 1033 controls with genotype data on 724067 SNPs. The BiGS dataset included 1190 BP cases and 401 controls with genotype data on 728187 SNPs.
German Sample
The collection and genotyping the German sample have been described elsewhere (Fangerau et al., 2004;McMahon et al., 2009). In brief, probands with bipolar I disorder were recruited through consecutive hospital admissions and assessed using a structured interview. Best estimate diagnoses were assigned according to DSM-IV criteria. A population-based control sample screened for psychiatric disorders was assembled from the PopGen (www.popgen.de), KORA (www.gsf.de/KORA) and Heinz Nixdorf Recall Study (www.recall-studie.uni-essen.de). Unrelated cases with a best-estimate diagnosis of bipolar I disorder were genotyped on the Illumina HumanHap550 array. We utilized the final cleaned dataset, which included 645 cases and 1310 controls with genotype data on 516024 SNPs.
Written informed consent was obtained from all subjects and the work described here was approved by the institutional review boards of each collection site.
Phenotype
Age at onset was defined as the year of age at which the earliest onset of an episode of depression or mania occurred. Psychotic symptoms were defined as a lifetime history of delusions and/or hallucinations with duration of at least 1 day. Where insufficient information was available to determine lifetime presence of psychotic symptoms, or where the symptom duration was judged to have lasted less than one day, cases were coded as unknown. Similarly, in cases where the interview indicated that psychotic symptoms occurred solely in the presence of drug use, psychotic status was considered unknown.
Population Stratification
We used principal components analysis to correct for population stratification in the combined sample. There were 137892 SNPs common to all three datasets (GAIN-BP, BiGS and German) that were used in this analysis. The EIGENSTRAT method in the program EIGENSOFT 3.0 was used to calculate principal components (Price et al., 2006). Based on the scree plot, we selected the top five principal components to include as covariates in our association analysis
Imputation
Imputation was performed to facilitate the mega-analysis of the three datasets, which were genotyped on two different platforms. Each dataset was imputed separately, using phased haplotype data from HapMap I & II release 24 (http://hapmap.ncbi.nlm.nih.gov/) as the reference panel. We used the program BEAGLE to ensure the orientation of our data was in the positive strand and impute allelic dosages for autosomal SNPs in the cases and controls (Browning and Yu, 2009). We applied a common quality control filter across all samples reflecting the most exclusive criteria from the individual studies. This involved excluding any SNP that had a missing data rate >5% in any of the original individual datasets, a minor allele frequency <2%, a HWE p-value <1×10−4, and an imputation R2 < 0.3.
Statistical Analysis
We combined the imputed data from all three datasets for a mega-analysis. The final combined dataset contained 2836 cases and 2744 controls with genotype data on 2373895 SNPs. Tests of association for the continuous sub-phenotype of age at onset were performed among BP cases only using the program mach2qtl (Li et al., 2009), with allelic dosages in a linear regression model. We performed tests of association for psychotic symptoms in a case-only analysis, comparing BP cases with psychotic symptoms to BP cases without psychotic symptoms. Tests of association were performed using the program mach2dat (Li et al., 2009), with allelic dosages in a logistic regression model. We included terms in each model to adjust for the top 5 principal components and a dummy-coded variable indexing the three datasets. Following convention, we considered SNPs with p-values < 5×10−8 as genome-wide significant(Hoggart et al., 2008).
In addition to the primary analysis outlined above, we performed several exploratory analyses. First, we expanded on our case-only comparison in the primary analysis, and performed a comparison of BP cases with psychotic symptoms versus unaffected controls. This analysis was performed using mach2dat with logistic regression as described above. Second, we investigated the involvement of biological pathways using the method implemented in the program Aligator (Holmans et al., 2009). This method tests for overrepresentation of pathways using the results from GWAS and corrects for linkage disequilibrium between SNPs, gene size and multiple testing. We used a liberal cutoff p-value < 0.01 in these analyses.
Replication
To validate our top findings from the GWAS of age at onset and psychotic symptoms in BP, we attempted to replicate our findings in an independent BP sample(Ferreira et al., 2008). Briefly, this replication sample consisted of 3916 BP cases with information about sub-phenotypes and 5112 controls in a combined sample from the WTCCC, STEP-BD and University College London. Age at onset and psychotic symptoms were defined as described by Ferreira et al (2008). Imputation to HapMap I & II was performed using MACH (Li and Abecasis, 2006). We prioritized results with p<0.001 in our original sample for replication. Association analysis in the replication sample was carried out under an allelic model in PLINK(Purcell et al., 2007). We also meta-analyzed the results from our sample and the replication sample using the program METAL (Willer et al., 2010). While there was some overlap in the controls included in the GAIN-BP and STEP-BD samples for the comparison of BP with psychotic symptoms versus controls, we did not account for the potential over-estimation of significance in the meta-analysis as the results were essentially null.
Results
Demographic and clinical characteristics of the three samples (GAIN-BP, BiGS, and German) are presented in Table 1. In all three studies, participants were of European Caucasian ancestry. While the GAIN-BP and German samples had similar proportions of males, the BiGS sample was predominantly female (χ2=65.6, d.f.=2, p<0.001).
Table 1.
Demographic and Clinical Characteristics of BP Cases in Three Samples
| GAIN-BP | BiGS | German | Combined GAIN-BP+BiGS+German | |
|---|---|---|---|---|
| BP Cases [N] | 1001 | 1190 | 645 | 2836 |
| Sex [% Male]1 | 50.0 | 34.1 | 48.4 | 43.0 |
| Age at Onset [mean (s.d.)]2 | 19.3 (9.3) | 18.5 (9.4) | 28.6 (11.2) | 21.0 (10.6) |
| Psychotic Symptoms [% positive]3 | 71.3 | 74.2 | 66.1 | 71.2 |
χ2=65.6, d.f.=2, p<0.001
F=124.26, d.f.=2, p<0.001
χ2=12.19, d.f.=2, p=0.002
Age at Onset
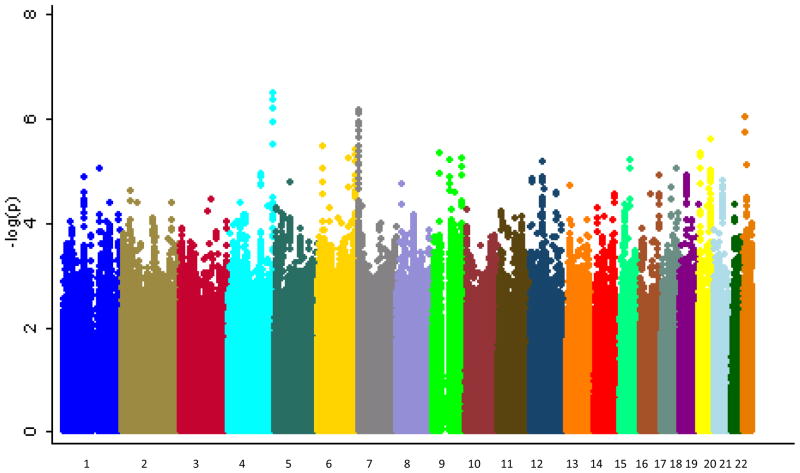
The mean age of onset differed among the three samples, with the German sample having an older age at onset than the other two (see Table 1; F=124.26, d.f.=2, p<0.001). Supplementary Figure 1 shows the Q-Q plot from the results of the primary analysis with age at onset. The corresponding inflation factor for this analysis was λ=1.04. Figure 1 shows the Manhattan plot of these results. None of the SNPs tested reached genome-wide significance. The most significant result was for the intronic SNP rs455219 in the gene FAT1 on chromosome 4 (β=0.70, p=3.04×10−7). We carried forward for replication 3464 SNPs with p<0.001, of which the direction of effect was consistent for 1599. The top ten results in our sample, the corresponding replication results and the combined meta-analytic results are shown in Table 2. Interestingly, the SNP rs2623968 in SYNE1 was among the top ten findings in this analysis (β=0.52, p=3.77×10−6). SYNE1 is one of two genes that was highlighted in the analysis by the PGC(Kelsoe, 2009)and is located in the suggestive 6q25 linkage region for early onset of mania identified by Faraone et al (2006). However, this finding did not replicate (β=0.08, p=0.7571). None of the SNPs tested in the replication sample reached genome-wide significance. In addition, none of the SNPs reached genome-wide significance in the meta-analysis of the GAIN-BP+BiGS+German and replication samples. The best meta-analytic p-value was for the SNP rs2934442 on chromosome 15(meta-analytic p=1.16×10−6; GAIN-BiGS-German: β=−0.561, p=5.18×10−6; Replication sample: β=−1.022, p=3.62×10−3). This SNP is an intronic SNP in the gene CGNL1, which codes for a cingulin-like protein that may be involved in anchoring the apical junctional complex to actin-based cytoskeletons.
Figure 1.
Manhattan plot for GWA of age at onset of BP. Genome-wide association results for observed and imputed allelic dosages using linear regression.
Table 2.
Top regions in the analysis of age at onset in the GAIN-BP+BiGS+German sample1
| GAIN-BP+BiGS+German | Replication Sample | Meta-Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Gene | BP Location | β2 | p-value | β2 | p-value | p-value3 |
| rs455219 | 4 | FAT1 | 187859058 | 0.70 | 3.04×10−7 | 0.16 | 0.5940 | 1.47×10−4 |
| rs10267593 | 7 | MAD1L1 | 1903787 | −0.70 | 6.66×10−7 | −0.16 | 0.6080 | 2.32×10−4 |
| rs4820556 | 22 | Intergenic | 22202936 | −0.69 | 8.94×10−7 | −0.62 | 0.8194 | 6.02×10−4 |
| rs393760 | 19 | Intergenic | 51627052 | −0.62 | 2.51×10−6 | 0.18 | 0.5327 | N/A |
| rs16883399 | 6 | MBOAT1 | 20239014 | −0.62 | 3.30×10−6 | −0.34 | 0.2294 | 6.75×10−5 |
| rs2623968 | 6 | SYNE1 | 152879005 | 0.52 | 3.77×10−6 | 0.08 | 0.7571 | 9.71×10−4 |
| rs1678869 | 19 | FUT5 | 5837528 | 0.76 | 4.33×10−6 | −0.26 | 0.4704 | N/A |
| rs2060409 | 9 | Intergenic | 26704614 | −0.55 | 4.34×10−6 | 0.20 | 0.4258 | N/A |
| rs17054536 | 6 | ECHDC1 | 127692059 | 1.21 | 5.62×10−6 | 0.53 | 0.3354 | 1.89×10−4 |
| rs11142517 | 9 | TRPM3 | 72434000 | 0.12 | 5.94×10−6 | −0.06 | 0.8188 | N/A |
The most significant SNP per region is listed as a representative SNP
β is calculated for the rare allele.
N/A = not applicable when the direction of effect is opposite in the two samples
We also tested whether any biological pathways were overrepresented among our top results in the analysis of age at onset. None of the pathways tested reached study-wide significance (p>0.508; see Supplementary Table 1). The top nominally overrepresented GO categories largely implicate oxygen binding and catabolic processes.
Psychotic Symptoms
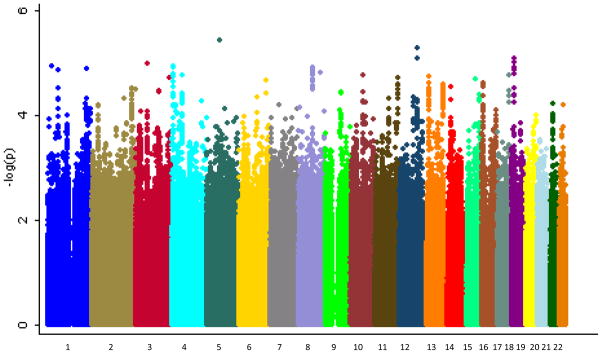
In the combined sample, 1775 BP cases met criteria for psychotic symptoms and 717 were classified as not having psychotic symptoms. The German sample had a slightly lower rate of psychotic symptoms than the other two samples (see Table 1; χ2=12.19, d.f.=2, p=0.002). Supplementary Figure 2 shows the Q-Q plot from the results of the primary analysis with psychotic symptoms. The corresponding inflation factor for this analysis was λ=1.05. Figure 2 shows the Manhattan plot of these results. None of the SNPs tested in a case-only analysis of psychotic symptoms reached genome-wide significance. The best result was for the intergenic SNP rs1010376 on chromosome 5 (see Table 3; OR=0.73, p=3.75×10−6). We carried forward for replication 2306 SNPs with p<0.001, of which 1058 showed a consistent direction of effect in the replication sample. However, none of the SNPs tested reached genome-wide significance in the replication sample or in the meta-analysis. The SNP rs7795096 on chromosome 7 had the best meta-analytic p-value at 1.94×10−6 (GAIN-BiGS-German: OR=0.79, p=2.41×10−4; Replication: OR=0.83, p=1.55×10−3). This SNP is an intronic SNP in the gene PRKAG2, a member of the AMPK gamma subunit family that modulates cellular functions by turning off enzymes involved in regulating the biosynthesis of cholesterol and fatty acid.
Figure 2.
Manhattan plot for GWA of psychotic symptoms in BP. Genome-wide association results for observed and imputed allelic dosages using logistic regression in a case-only analysis. Results are for the analysis comparing BP cases with psychotic symptoms to BP cases without psychotic symptoms.
Table 3.
Top regions from the analysis of psychotic symptoms in the GAIN-BP+BiGS+German sample1
| GAIN-BP+BiGS+German | Replication Sample | Meta-Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Gene | BP Location | OR2 | p-value | OR2 | p-value | p-value3 |
| rs1010376 | 5 | Intergenic | 71339064 | 0.727 | 3.75 × 10−6 | 0.96 | 0.4650 | 5.87 × 10−4 |
| rs7132927 | 12 | ANKS1B | 98821773 | 0.745 | 4.88 × 10−6 | 1.06 | 0.2213 | N/A |
| rs11080384 | 18 | Intergenic | 10078962 | 1.378 | 5.80 × 10−6 | 0.99 | 0.8066 | N/A |
| rs6446101 | 3 | FHIT | 59932023 | 1.323 | 8.81 × 10−6 | 1.02 | 0.7630 | 2.80 × 10−3 |
| rs607777 | 1 | Intergenic | 61171347 | 1.816 | 1.14 × 10−5 | 1.12 | 0.2676 | 4.18 × 10−4 |
| rs10903047 | 1 | IL28RA | 24381661 | 0.747 | 1.15 × 10−5 | 1.02 | 0.7900 | N/A |
| rs871938 | 4 | SORCS2 | 7617449 | 0.748 | 1.15 × 10−5 | 1.01 | 0.8645 | N/A |
| rs6472842 | 8 | GDAP1 | 75441012 | 0.744 | 1.21 × 10−5 | 1.06 | 0.2805 | N/A |
| rs714861 | 4 | PDGFRA | 54649188 | 1.329 | 1.37 × 10−5 | 0.90 | 0.0464 | N/A |
| rs4846647 | 1 | Intergenic | 218691910 | 0.717 | 1.49 × 10−5 | 1.14 | 0.0344 | N/A |
The most significant SNP per region is listed as a representative SNP
Oodds ratio is calculated for the rare allele.
N/A = not applicable when the direction of effect is opposite in the two samples
In an exploratory analysis, we tested for associations in BP cases with psychotic symptoms versus unaffected controls. None of the SNPs reached genome-wide significance in our sample, the replication sample, or in the meta-analysis of the two samples(See Supplementary Figure 1 and Supplementary Table 2). The best result in our sample was for the intergenic SNP rs6578831 on chromosome 11, with p=7.69×10−8 (OR=1.33). However, this SNP showed no association in the replication sample (OR=0.99, p=0.8245).
In our examination of biological pathways, none were significantly overrepresented among our top results at a study-wide level (study-wide p>0.362; see Supplementary Table 1). Among the top ten nominally overrepresented pathways, myeloid cell differentiation and acetylcholine processes were predominant. Acetylcholine is a neurotransmitter that is involved in aspects of synaptic plasticity, arousal and reward.
Discussion
Here we present the results from a GWAS of two clinically relevant sub-phenotypes of BP, age at onset and psychotic symptoms. No SNP reached genome-wide significance for either sub-phenotype. In addition, no SNP reached genome-wide significance in a meta-analysis with a replication sample. The full results of the primary analysis with age at onset and psychotic symptoms are available upon request.
We hypothesized a priori that the use of sub-phenotypes would facilitate the identification of loci relevant to bipolar disorder and its clinical manifestation. Although we did not identify any genome-wide significant findings, the study had several limitations that should be considered. One limitation is the loss of power inherent in the analysis of sub-phenotypes. By examining sub-phenotypes of BP, we focused on a portion of our total sample in each analysis, decreasing the power to detect associations. We hoped to overcome this limitation by combining the data from three large GWAS. As a result, our analysis of sub-phenotypes in BP is the largest to date. However, the power of the combined sample may still not have been sufficient. We estimated that the combined sample had 80% power to detect loci of genome-wide significance with a mean difference of 1.8 years for age at onset and an odds ratio of 1.6 for psychotic symptoms. By focusing on certain sub-phenotypes we sought to identify more homogeneous sub-groups of patients in which loci of such effect sizes might operate. However, our results do not support this hypothesis in relation to age at onset or psychotic symptoms in BP. It may be these sub-phenotypes are no less genetically complex than BP and that larger sample sizes of the sort being assembled by the PGC are needed to identify relevant loci. For example, we estimated that a sample of 15,775 BP cases with psychosis and 6,752 without psychosis would be needed to detect a genome-wide significant association with 80% power at an odds ratio of 1.15. It is also possible there are loci of larger effect sizes influencing these sub-phenotypes, but they are of low frequency and as a result are not adequately captured by the genotyping panels currently used in GWAS. We imputed to the HapMap phase I & II panel, which is estimated to cover 94% of the genome (2005). Thus, there could also be common loci influencing the sub-phenotypes that were not captured in our study.
Another limitation of this study is the possibility of measurement error in the sub-phenotypes. Previous studies have suggested that age at onset and psychotic symptoms can be measured reliably. One study showed that the intraclass correlation varied between 0.68 and 0.97 for different measures of age at onset(Egeland et al., 1987), and another study showed that the rater agreement for diagnosing hallucinations and delusions yielded a coefficient of reliability of 0.91(Endicott and Spitzer, 1978). However, to the extent that these sub-phenotypes are measured with error, results will likely be biased towards the null. Related to this is the fact that the samples used different diagnostic instruments to measure these sub-phenotypes. While the GAIN-BP and BiGS studies used the DIGS, the German sample used the Structured Clinical Interview for DSM-IV (SCID). Such differences may introduce error into the characterization of the sub-phenotypes across the samples, despite our efforts to harmonize their definitions. Indeed, we observed significant differences in the ages at onset and rates of psychotic symptoms in the German sample compared to GAIN-BP and BiGS. The observed differences may reflect sampling variation, differences in the measurement of the sub-phenotypes, or true differences in the patient populations across the samples. Future work aimed at matching the definition and clinical assessment of sub-phenotypes across studies may facilitate efforts to examine genotype-phenotype correlations(Potash et al., 2007). In addition, alternative approaches to defining the clinical sub-phenotypes, such as using admixture analysis to define the boundaries of age at onset classes(Bellivier et al., 2001;Bellivier et al., 2003;Lin et al., 2006), may provide further insights. Finally, we only assessed two sub-phenotypes in this study. It is possible that other clinical variables, such as response to treatment with lithium, or clusters of variables, may more robustly correlate with common genetic variation than do age at onset or psychotic symptoms. Sample sizes comparable to ours may be sufficient to detect loci relevant to these other sub-phenotypes.
Using sub-phenotypes to help resolve the complex genetic architecture of common psychiatric disorders is attractive. We sought to use this strategy to identify susceptibility genes related to age at onset and psychotic symptoms in BP, features with strong evidence of familiality and thus high likelihood of being genetically relevant. Although we were not able to definitively implicate genes in this analysis, the strategy may still have merit. It is possible that age at onset and psychotic symptoms in BP may reflect the action of genes of smaller effect sizes only detectable with larger samples. Alternatively, uncommon variants not measured well by current SNP arrays may be relevant for these sub-phenotypes and strategies such as deep sequencing may be necessary to identify them. Finally, other sub-phenotypes may be better markers of the underlying genetic heterogeneity of BP.
Supplementary Material
Acknowledgments
The authors express their profound appreciation to the families who participated in this project, and to the many clinicians who facilitated the referral of participants to the study. Data and biomaterials for the NIMH samples were collected as part of 10 projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1991 to 1998, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, U01 MH46282—John Nurnberger, MD, PhD, Marvin Miller, MD and Elizabeth Bowman, MD; Washington University, St Louis, MO, U01 MH46280—Theodore Reich, MD, Allison Goate, PhD and John Rice, PhD; Johns Hopkins University, Baltimore, MD U01 MH46274—J Raymond DePaulo Jr, MD Sylvia Simpson, MD, MPH and Colin Stine, PhD; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD—Elliot Gershon, MD, DianeKazuba, BA and Elizabeth Maxwell, MSW. From 1999 to 2003, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545—John Nurnberger, MD, PhD, Marvin J Miller, MD, Elizabeth S Bowman, MD, N Leela Rau, MD, P Ryan Moe, MD, Nalini Samavedy, MD, Rif El-Mallakh, MD (at University of Louisville), Husseini Manji, MD (at Wayne State University), Debra A Glitz, MD (at Wayne State University), Eric T Meyer, MS, Carrie Smiley, RN, Tatiana Foroud, PhD, Leah Flury, MS, Danielle M Dick, PhD and Howard Edenberg, PhD; Washington University, St Louis, MO, R01 MH059534, John Rice, PhD, Theodore Reich, MD, Allison Goate, PhD and Laura Bierut, MD; Johns Hopkins University, Baltimore, MD, R01 MH59533—Melvin McInnis, MD, J Raymond DePaulo Jr, MD, Dean F MacKinnon, MD, Francis M Mondimore, MD, James B Potash, MD, Peter P Zandi, PhD, Dimitrios Avramopoulos and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553—Wade Berrettini, MD, PhD; University of California at Irvine, CA, R01 MH60068—William Byerley, MD and Mark Vawter, MD; University of Iowa, IA, R01 MH059548—William Coryell, MD and Raymond Crowe, MD; University of Chicago, Chicago, IL, R01 MH59535—Elliot Gershon, MD, Judith Badner, PhD, Francis McMahon, MD, Chunyu Liu, PhD, Alan Sanders, MD, Maria Caserta, Steven Dinwiddie, MD, Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567—John Kelsoe, MD, Rebecca McKinney, BA; Rush University, IL, R01 MH059556—William Scheftner, MD, Howard M Kravitz, DO, MPH, Diana Marta, BA, Annette Vaughn-Brown, MSN, RN and Laurie Bederow, MA; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, Francis J McMahon, MD, Layla Kassem, PsyD, Sevilla Detera-Wadleigh, PhD, Lisa Austin, PhD, Dennis L Murphy, MD. From 2003–2007, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, M.D., Ph.D., Marvin J. Miller, M.D., Elizabeth S. Bowman, M.D., N. Leela Rau, M.D., P. Ryan Moe, M.D., Nalini Samavedy, M.D., Rif El-Mallakh, M.D. (at University of Louisville), Husseini Manji, M.D. (at Johnson and Johnson), Debra A. Glitz, M.D. (at Wayne State University), Eric T. Meyer, Ph.D., M.S. (at Oxford University, UK), Carrie Smiley, R.N., Tatiana Foroud, Ph.D., Leah Flury, M.S., Danielle M. Dick, Ph.D (at Virginia Commonwealth University), Howard Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, John Rice, Ph.D, Theodore Reich, M.D., Allison Goate, Ph.D., Laura Bierut, M.D. K02 DA21237; Johns Hopkins University, Baltimore, M.D., R01 MH59533, Melvin McInnis, M.D., J. Raymond DePaulo, Jr., M.D., Dean F. MacKinnon, M.D., Francis M. Mondimore, M.D., James B. Potash, M.D., Peter P. Zandi, Ph.D, Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini, M.D., Ph.D.; University of California at San Francisco, CA, R01 MH60068, William Byerley, M.D., and Sophia Vinogradov, M.D.; University of Iowa, IA, R01 MH059548, William Coryell, M.D., and Raymond Crowe, M.D.; University of Chicago, IL, R01 MH59535, Elliot Gershon, M.D., Judith Badner, Ph.D., Francis McMahon, M.D., Chunyu Liu, Ph.D., Alan Sanders, M.D., Maria Caserta, Steven Dinwiddie, M.D., Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, M.D., Rebecca McKinney, B.A.; Rush University, IL, R01 MH059556, William Scheftner, M.D., Howard M. Kravitz, D.O., M.P.H., Diana Marta, B.S., Annette Vaughn-Brown, M.S.N., R.N., and Laurie Bederow, M.A.; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, Francis J. McMahon, M.D., Layla Kassem, Psy.D., Sevilla Detera-Wadleigh, Ph.D, Lisa Austin, Ph.D, Dennis L. Murphy, M.D.; Howard University, William B. Lawson, M.D., Ph.D., Evarista Nwulia, M.D., and Maria Hipolito, M.D. Control subjects from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI), data and biomaterials are being collected by the “Molecular Genetics of Schizophrenia II” (MGS-2) collaboration. The investigators and coinvestigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA, MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01, MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA, MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY, MH59586, Jeremy Silverman, Ph.D. (PI). Genome-wide SNP genotyping of the NIMH samples was performed through the Genetic Association Information Network under the direction of the Bipolar Genetics Studies Collaboration. The Principal Investigators and Co-Investigators were: University of California San Diego, La Jolla, CA, John R. Kelsoe, M.D. (PI), Tiffany A. Greenwood, Ph.D., Paul D. Shilling, Ph.D., Caroline Nievergelt, Ph.D.; Scripps Research Institute, La Jolla, CA: Nicholas Schork, Ph.D. (PI), Erin N. Smith, Ph.D., Cinnamon Bloss, Ph.D.; Indiana University, Bloomington, IN, John Nurnberger, M.D. (PI), Howard J. Edenberg, Ph.D., Tatiana Foroud, Ph.D.; University of Chicago, Chicago, IL, Elliot Gershon, M.D. (PI), Chunyu Liu, Ph.D., Judith A. Badner, Ph.D.; Rush University Medical Center, Chicago, IL, William A. Scheftner, M.D.; Howard University, Washington, DC, William B. Lawson, M.D. (PI), Evaristus A. Nwulia, M.D., Maria Hipolito, M.D.; University of Iowa, Iowa City, IA, William Coryell, M.D. (PI); Washington University, St. Louis, MO, John Rice, Ph.D. (PI); University of California San Francisco, San Francisco, CA, William Byerley, M.D. (PI); National Institute of Mental Health, Bethesda, MD, Francis McMahon, M.D. (PI), Thomas G. Schulze, M.D.; University of Pennsylvania, Philadelphia, PA, Wade Berrettini, M.D., Ph.D. (PI); Johns Hopkins University, Baltimore, MD, James B. Potash, M.D. (PI), Peter P. Zandi, Ph.D., Pamela Belmonte Mahon, PhD; University of Michigan, Ann Arbor, MI, Melvin G. McInnis, M.D. (PI), Sebastian Zöllner, Ph.D.; Translation Genomic Research Institute, Phoenix, AZ, David Craig, Ph.D. (PI), Szabolics Szelinger. Data and biomaterials for the subjects in the Wellcome Trust Case-Control Consortium were collected by: University of Aberdeen, Foresterhill, Aberdeen, UK, Gerome Breen, David St Clair; Birmingham University, Birmingham, UK, Sian Caesar, Katherine Gordon-Smith, Lisa Jones; Cardiff University, Cardiff, UK, Christine Fraser, Elaine K Green, Detelina Grozeva, Marian L Hamshere, Peter A Holmans, Ian R Jones, George Kirov, Valentina Moskvina, Ivan Nikolov, Michael C O’Donovan, Michael J Owen, Nick Craddock; The Institute of Psychiatry, King’s College, London, UK, David A Collier, Amanda Elkin, Anne Farmer, Richard Williamson, Peter McGuffin; Royal Victoria Infirmary, Newcastle upon Tyne, UK, Allan H Young, I Nicol Ferrier; Supported in part by R01 MH079799 (Smoller)
Footnotes
Financial Disclosures
All authors report no competing interest.
Electronic Resources
HapMap (http://hapmap.ncbi.nlm.nih.gov/)
Heinz Nixdorf Recall Study(http://www.recall-studie.uni-essen.de)
KORA(http://www.gsf.de/KORA)
MACH(http://www.sph.umich.edu/csg/abecasis/MACH/)
PLINK(http://pngu.mgh.harvard.edu/purcell/plink/)
PopGen (http://www.popgen.de)
Reference List
- A Haplotype Map of the Human Genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome-Wide Association Study of 14,000 Cases of Seven Common Diseases and 3,000 Shared Controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum AE, Hamshere M, Green E, Cichon S, Rietschel M, Noethen MM, Craddock N, McMahon FJ. Meta-Analysis of Two Genome-Wide Association Studies of Bipolar Disorder Reveals Important Points of Agreement. Mol Psychiatry. 2008;13:466–467. doi: 10.1038/mp.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellivier F, Golmard JL, Henry C, Leboyer M, Schurhoff F. Admixture Analysis of Age at Onset in Bipolar I Affective Disorder. Arch Gen Psychiatry. 2001;58:510–512. doi: 10.1001/archpsyc.58.5.510. [DOI] [PubMed] [Google Scholar]
- Bellivier F, Golmard JL, Rietschel M, Schulze TG, Malafosse A, Preisig M, McKeon P, Mynett-Johnson L, Henry C, Leboyer M. Age at Onset in Bipolar I Affective Disorder: Further Evidence for Three Subgroups. Am J Psychiatry. 2003;160:999–1001. doi: 10.1176/appi.ajp.160.5.999. [DOI] [PubMed] [Google Scholar]
- Browning BL, Yu Z. Simultaneous Genotype Calling and Haplotype Phasing Improves Genotype Accuracy and Reduces False-Positive Associations for Genome-Wide Association Studies. Am J Hum Genet. 2009;85:847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Flury L, Bowman ES, Miller MJ, Rau NL, Moe PR, Samavedy N, El-Mallakh R, Manji H, Glitz DA, Meyer ET, Smiley C, Hahn R, Widmark C, McKinney R, Sutton L, Ballas C, Grice D, Berrettini W, Byerley W, Coryell W, DePaulo R, MacKinnon DF, Gershon ES, Kelsoe JR, McMahon FJ, McInnis M, Murphy DL, Reich T, Scheftner W, Nurnberger JI., Jr Genomewide Linkage Analyses of Bipolar Disorder: a New Sample of 250 Pedigrees From the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–114. doi: 10.1086/376562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeland JA, Blumenthal RL, Nee J, Sharpe L, Endicott J. Reliability and Relationship of Various Ages of Onset Criteria for Major Affective Disorder. J Affect Disord. 1987;12:159–165. doi: 10.1016/0165-0327(87)90009-7. [DOI] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL. A Diagnostic Interview: the Schedule for Affective Disorders and Schizophrenia. Arch Gen Psychiatry. 1978;35:837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- Fangerau H, Ohlraun S, Granath RO, Nothen MM, Rietschel M, Schulze TG. Computer-Assisted Phenotype Characterization for Genetic Research in Psychiatry. Hum Hered. 2004;58:122–130. doi: 10.1159/000083538. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Lasky-Su J, Glatt SJ, Van EP, Tsuang MT. Early Onset Bipolar Disorder: Possible Linkage to Chromosome 9q34. Bipolar Disord. 2006;8:144–151. doi: 10.1111/j.1399-5618.2006.00289.x. [DOI] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, Macintyre DJ, MacLean AW, St CD, Robinson M, van BM, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Ferrier IN, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N. Collaborative Genome-Wide Association Analysis Supports a Role for ANK3 and CACNA1C in Bipolar Disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes FS, Willour VL, Zandi PP, Belmonte PL, MacKinnon DF, Mondimore FM, Schweizer B, Gershon ES, McMahon FJ, Potash JB. Family-Based Association Study of Neuregulin 1 With Psychotic Bipolar Disorder. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:693–702. doi: 10.1002/ajmg.b.30895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Raybould R, Macgregor S, Gordon-Smith K, Heron J, Hyde S, Grozeva D, Hamshere M, Williams N, Owen MJ, O’Donovan MC, Jones L, Jones I, Kirov G, Craddock N. Operation of the Schizophrenia Susceptibility Gene, Neuregulin 1, Across Traditional Diagnostic Boundaries to Increase Risk for Bipolar Disorder. Arch Gen Psychiatry. 2005;62:642–648. doi: 10.1001/archpsyc.62.6.642. [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Clark TG, De IM, Whittaker JC, Balding DJ. Genome-Wide Significance for Dense SNP and Resequencing Data. Genet Epidemiol. 2008;32:179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O’Donovan MC, Craddock N. Gene Ontology Analysis of GWA Study Data Sets Provides Insights into the Biology of Bipolar Disorder. Am J Hum Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem L, Lopez V, Hedeker D, Steele J, Zandi P, McMahon FJ. Familiality of Polarity at Illness Onset in Bipolar Affective Disorder. Am J Psychiatry. 2006;163:1754–1759. doi: 10.1176/ajp.2006.163.10.1754. [DOI] [PubMed] [Google Scholar]
- Kelsoe JR The Psychiatric GWAS Consortium for Bipolar Disorder: Significant Association for Multiple Genes. The World Congress on Psychiatric Genetics. San Diego, CA: 2009. Ref Type: Conference Proceeding. [Google Scholar]
- Kerner B, Brugman DL, Freimer NB. Evidence of Linkage to Psychosis on Chromosome 5q33-34 in Pedigrees Ascertained for Bipolar Disorder. Am J Med Genet B NeuropsychiatrGenet. 2007;144B:74–78. doi: 10.1002/ajmg.b.30402. [DOI] [PubMed] [Google Scholar]
- King MC. Localization of the Early-Onset Breast Cancer Gene. Hosp Pract (Off Ed) 1991;26:121–126. doi: 10.1080/21548331.1991.11705309. [DOI] [PubMed] [Google Scholar]
- Li Y, Abecasis GR. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype Imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PI, McInnis MG, Potash JB, Willour V, MacKinnon DF, DePaulo JR, Zandi PP. Clinical Correlates and Familial Aggregation of Age at Onset in Bipolar Disorder. Am J Psychiatry. 2006;163:240–246. doi: 10.1176/appi.ajp.163.2.240. [DOI] [PubMed]
- Lin PI, McInnis MG, Potash JB, Willour VL, MacKinnon DF, Miao K, DePaulo JR, Zandi PP. Assessment of the Effect of Age at Onset on Linkage to Bipolar Disorder: Evidence on Chromosomes 18p and 21q. Am J Hum Genet. 2005;77:545–555. doi: 10.1086/491602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchia M, Zai CC, Squassina A, Vincent JB, De LV, Kennedy JL. Mixture Regression Analysis on Age at Onset in Bipolar Disorder Patients: Investigation of the Role of Serotonergic Genes. Eur Neuropsychopharmacol. 2010;20:663–670. doi: 10.1016/j.euroneuro.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Massat I, Lerer B, Souery D, Blackwood D, Muir W, Kaneva R, Nothen MM, Oruc L, Papadimitriou GN, Dikeos D, Serretti A, Bellivier F, Golmard JL, Milanova V, Del-Favero J, Van BC, Mendlewicz J. HTR2C (Cys23ser) Polymorphism Influences Early Onset in Bipolar Patients in a Large European Multicenter Association Study. Mol Psychiatry. 2007;12:797–798. doi: 10.1038/sj.mp.4002018. [DOI] [PubMed] [Google Scholar]
- McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, Steele CJM, Breuer R, Strohmaier J, Wendland JR, Mattheisen M, Muhleisen TW, Maier W, Nothen MM, Cichon S, Farmer A, Vincent JB, Holsboer F, Preisig M, Rietschel M Bipolar Disorder Genetics (BiGS) Consortium. Meta-Analysis of Genome-Wide Association Data Detects a Risk Locus for Major Mood Disorders on Chromosome 3p21.1. 2009 doi: 10.1038/ng.523. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic Interview for Genetic Studies. Rationale, Unique Features, and Training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- Potash JB, Toolan J, Steele J, Miller EB, Pearl J, Zandi PP, Schulze TG, Kassem L, Simpson SG, Lopez V, MacKinnon DF, McMahon FJ. The Bipolar Disorder Phenome Database: a Resource for Genetic Studies. Am J Psychiatry. 2007;164:1229–1237. doi: 10.1176/appi.ajp.2007.06122045. [DOI] [PubMed] [Google Scholar]
- Potash JB, Willour VL, Chiu YF, Simpson SG, MacKinnon DF, Pearlson GD, DePaulo JR, Jr, McInnis MG. The Familial Aggregation of Psychotic Symptoms in Bipolar Disorder Pedigrees. Am J Psychiatry. 2001;158:1258–1264. doi: 10.1176/appi.ajp.158.8.1258. [DOI] [PubMed] [Google Scholar]
- Potash JB, Zandi PP, Willour VL, Lan TH, Huo Y, Avramopoulos D, Shugart YY, MacKinnon DF, Simpson SG, McMahon FJ, DePaulo JR, Jr, McInnis MG. Suggestive Linkage to Chromosomal Regions 13q31 and 22q12 in Families With Psychotic Bipolar Disorder. Am J Psychiatry. 2003;160:680–686. doi: 10.1176/appi.ajp.160.4.680. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal Components Analysis Corrects for Stratification in Genome-Wide Association Studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, Burrell GJ, Rice JP, Nertney DA, Olincy A, Rozic P, Vinogradov S, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Crowe RR, Cloninger CR, Martinez M, Gejman PV. No Significant Association of 14 Candidate Genes With Schizophrenia in a Large European Ancestry Sample: Implications for Psychiatric Genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- Schulze TG, Ohlraun S, Czerski PM, Schumacher J, Kassem L, Deschner M, Gross M, Tullius M, Heidmann V, Kovalenko S, Jamra RA, Becker T, Leszczynska-Rodziewicz A, Hauser J, Illig T, Klopp N, Wellek S, Cichon S, Henn FA, McMahon FJ, Maier W, Propping P, Nothen MM, Rietschel M. Genotype-Phenotype Studies in Bipolar Disorder Showing Association Between the DAOA/G30 Locus and Persecutory Delusions: a First Step Toward a Molecular Genetic Classification of Psychiatric Phenotypes. Am J Psychiatry. 2005;162:2101–2108. doi: 10.1176/appi.ajp.162.11.2101. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, Tozzi F, Li JZ, Burmeister M, Absher D, Thompson RC, Francks C, Meng F, Antoniades A, Southwick AM, Schatzberg AF, Bunney WE, Barchas JD, Jones EG, Day R, Matthews K, McGuffin P, Strauss JS, Kennedy JL, Middleton L, Roses AD, Watson SJ, Vincent JB, Myers RM, Farmer AE, Akil H, Burns DK, Boehnke M. Genome-Wide Association and Meta-Analysis of Bipolar Disorder in Individuals of European Ancestry. Proc Natl Acad Sci U S A. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Grubber JM, Conneally PM, Small GW, Hulette CM, Rosenberg CK, Saunders AM, Roses AD, Haines JL, Pericak-Vance MA. Fine Mapping of the Chromosome 12 Late-Onset Alzheimer Disease Locus: Potential Genetic and Phenotypic Heterogeneity. Am J Hum Genet. 2000;66:922–932. doi: 10.1086/302828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A, de Bakker PI, Ogdie MN, Thase ME, Sachs GS, Todd-Brown K, Gabriel SB, Sougnez C, Gates C, Blumenstiel B, Defelice M, Ardlie KG, Franklin J, Muir WJ, McGhee KA, Macintyre DJ, McLean A, VanBeck M, McQuillin A, Bass NJ, Robinson M, Lawrence J, Anjorin A, Curtis D, Scolnick EM, Daly MJ, Blackwood DH, Gurling HM, Purcell SM. Whole-Genome Association Study of Bipolar Disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Bloss CS, Badner JA, Barrett T, Belmonte PL, Berrettini W, Byerley W, Coryell W, Craig D, Edenberg HJ, Eskin E, Foroud T, Gershon E, Greenwood TA, Hipolito M, Koller DL, Lawson WB, Liu C, Lohoff F, McInnis MG, McMahon FJ, Mirel DB, Murray SS, Nievergelt C, Nurnberger J, Nwulia EA, Paschall J, Potash JB, Rice J, Schulze TG, Scheftner W, Panganiban C, Zaitlen N, Zandi PP, Zollner S, Schork NJ, Kelsoe JR. Genome-Wide Association Study of Bipolar Disorder in European American and African American Individuals. Mol Psychiatry. 2009;14:755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Xiao L, Shu C, Wang G, Liu Z, Wang X, Wang H, Bai X. Association of the Brain-Derived Neurotrophic Factor Gene and Bipolar Disorder With Early Age of Onset in Mainland China. Neurosci Lett. 2008;433:98–102. doi: 10.1016/j.neulet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: Fast and Efficient Meta-Analysis of Genomewide Association Scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi PP, Badner JA, Steele J, Willour VL, Miao K, MacKinnon DF, Mondimore FM, Schweizer B, McInnis MG, DePaulo JR, Jr, Gershon E, McMahon FJ, Potash JB. Genome-Wide Linkage Scan of 98 Bipolar Pedigrees and Analysis of Clinical Covariates. Mol Psychiatry. 2007;12:630–639. doi: 10.1038/sj.mp.4002027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.