Abstract
Background
Seropositivity to HPV16 and 18 antibodies is used as a measure of cumulative HPV exposure and as a stratifier of HPV exposure for vaccine efficacy analyses. Overall performance of these assays, as a measure of HPV exposure, has not been evaluated.
Methods
Using data from the enrollment phase of the HPV16/18 vaccine trial in Costa Rica, we evaluated the performance of the polyclonal ELISA HPV16 and 18 serological assays as a measure of HPV exposure. Biological (for eg. HPV infection at the cervix) and behavioral characteristics (for eg. lifetime number of sexual partners) with known associations with current and past HPV infection were used to define cases and controls (HPV exposed vs. not exposed). Pre-vaccination serum was measured for antibodies against HPV16 and HPV18 by ELISA; cervical samples were tested for HPV DNA using PCR SPF10/LiPA25. ELISA results were analyzed using receiver-operator-characteristic curves (ROC); performance was evaluated at the manufacturer set cutpoint (HPV16 =8, HPV18 =7) and at cutpoints chosen to optimize sensitivity and specificity (HPV16 =34, HPV18 =60).
Results
Defining cases as type-specific HPV DNA positive with high-grade abnormal cytolzogy (i.e. combined molecular and microscopic markers of infection), HPV16-ELISA gave sensitivity that was lower at the optimal cutpoint than the manufacturer cutpoint (62.2 compared with 75.7, respectively; p=0.44). However, specificity was higher (85.3 compared with 70.4, respectively; p<0.0001). Similarly, HPV18-ELISA gave sensitivity that was lower at the optimal cutpoint than the manufacturer cutpoint (34.5 compared with 51.7, respectively; p=0.40), with higher specificities (94.9 compared with 72.6, respectively; p<0.0001).
Conclusions
Modifying cutpoints did not improve the low sensitivity. The low sensitivity of this assay does not support its use for risk stratification or clinical settings.
Keywords: Human Papillomavirus, serology, polyclonal ELISA
INTRODUCTION
Human papillomavirus (HPV) is a common sexually transmitted infection (STI), often acquired shortly after sexual initiation (1, 2). In epidemiologic research, current HPV DNA infections are detected using sensitive polymerase chain reaction (PCR) assays on cervical exfoliated samples (3). HPV DNA based assays are highly sensitive and specific for measuring current infections. However, they are limited as they provide information on current infection only and HPV clearance (i.e. loss of DNA detectability) occurs within months to a few years in the majority of infections (4). Thus, DNA based assays do not provide information on lifetime, cumulative HPV exposure.
Several serological assays with different properties are currently available for research purposes; they measure a wide range of anti-HPV16 and 18 antibodies. Seropositivity to HPV16 and 18 antibodies is being used as an epidemiologic measure of cumulative HPV exposure, a marker of immunity or protection from subsequent infections (5), and in conjunction with DNA-based measures, to define or stratify subgroups of potentially HPV-naïve women for vaccine efficacy analyses. Despite the use of HPV serological assays in many studies, the overall performance of the assays as a measure of HPV exposure have not been evaluated.
The aim of this analysis was to evaluate the performance of the polyclonal ELISA HPV assay as a measure of HPV exposure and to determine whether alternative cutpoints would improve the sensitivity/specificity of the ELISA HPV serology assay as a biomarker of HPV exposure. We also compared the correlates of HPV16 and HPV18 seropositivity (separately) at the manufacturer cutoff and the cutpoint determined to maximize sensitivity and specificity.
MATERIALS AND METHODS
Study population
Data are from the enrollment/pre-vaccination phase of the on-going publicly funded, community-based randomized phase III HPV16/18 vaccine trial in Costa Rica (CVT). The study has been described elsewhere (6). Briefly, the main objectives of the trial are to evaluate the efficacy of a prophylactic HPV16/18 vaccine manufactured by GlaxoSmithKline for prevention of HPV16/18 infection and related precancerous lesions compared to women receiving a control hepatitis A vaccine. A total of 7,466 women provided written, informed consent at enrollment.
At enrollment participants were asked detailed questions regarding demographics, sexual practices, contraceptive use, reproductive and menstrual history, and smoking. For sexually experienced women, a pelvic exam was performed and exfoliated cervical cells were collected for cytology, HPV DNA, Chlamydia trachomatis (Ct) DNA, and Neisseria gonorrhoeae (GC) DNA testing. ThinPrep slides were prepared to obtain a Pap stain for cervical cytology interpretation.
All testing was done masked to the results of randomization arm and other test results. Protocols were approved by the US National Cancer Institute and a Costa Rican institutional review board.
HPV serological measurements
Serum collected at enrollment was used to determine HPV16 and -18 IgG serostatus at GSK Biologicals (Rixensart, Belgium) using a VLP-based direct enzyme linked immunoabsorbent assay (ELISA) developed by GSK that measures polyclonal antibodies as described previously (7, 8). All research and development of the assay and testing of the samples was conducted at GSK. Briefly, ELISA microtiter plates were separately coated with 2.7 µg/mL of either HPV16 or HPV18 VLPs that were produced in a baculovirus expression system. The plates were blocked with PBS containing 4% skim milk with 0.2% Tween-20. Serum samples from participants were serially diluted in the blocking solution starting at 1:100 in twofold increments. Serial dilutions of samples, standard, and quality control specimens were added to the microtiter plates. After incubation and washing steps, a peroxidase-conjugated anti-human polyclonal antibody was added. Following incubation and washing, enzyme substrate and chromogen were added to allow color development. Reactions were stopped, and optical density (OD) read at 450 and 620 nm, with background measured at 620 nm and subtracted from the OD reading at 450 nm. Antibody levels, expressed as ELISA units (EU)/mL, were calculated by interpolation of OD values from the standard curve by averaging the calculated concentrations from all dilutions that fell within the working range of the reference curve. The seropositivity cutpoints were determined by GSK and calculated from antibody titer values three standard deviations above the geometric mean titers taken from two groups of known HPV-negative individuals. These groups included: 1) human serum samples previously incubated with corresponding VLP to remove specific antibodies, and 2) human serum taken at day 0 before vaccination from women who did not show an increased immune response after 7 days following the first vaccine (8). Cutpoints were set at OD≥8 EU/ml for anti-HPV16 and OD≥7 EU/ml for anti-HPV18 (8).
HPV DNA- SPF10/DEIA/LiPA25
HPV DNA detection and genotyping was performed at DDL Diagnostic Laboratory (Voorburg, Netherlands), as described previously (9, 10). Extracted DNA was used for PCR amplification with the SPF10 primer sets (9, 10). The samples were run through an HPV DNA enzyme immunoassay (DEIA) to obtain an OD reading, and categorized as HPV DNA negative, positive, or borderline. The same SPF10 amplimers were used on SPF10-DEIA-positive samples to identify HPV genotype by reverse hybridization on a line probe assay (LiPA) (SPF10-DEIA/HPVLiPA25,version 1; Labo Bio-Medical Products, Rijswijk, Netherlands), which detects 25 HPV genotypes.
Since CVT uses the bivalent HPV16/18 vaccine, to ensure detection for these types, HPV16 and 18 type-specific PCR (TS-PCR) primer sets were used to selectively amplify HPV16 and HPV18 from specimens tested SPF10 DEIA-positive, but LiPA25 HPV16 and/or HPV18 negative (9). Amplimers from the TS-PCRs were detected by DEIA similar to the method used for SPF10 amplimer detection (9–11).
Statistical analysis
All analyses were conducted separately for HPV16 and HPV18. We note that the results from the HPV16 and HPV18 models cannot be directly compared to one another because the HPV16 and 18 ELISAs are not identical. We excluded virginal women (n=1,592) from this analysis because they did not have a pelvic examination and thus we could not assess their HPV DNA or cytology status. However, for completeness, we also performed the analyses including virginal women and assumed they were HPV DNA and cytology negative. As expected specificity increased; however, the main conclusions of the analysis were not altered by restricting to sexually active women (data not shown). We also excluded three women who were outside the trial age range.
We used nonparametric empirical receiver operating characteristic (ROC) analysis to calculate the area under the curve (AUC) (12, 13). We sought to evaluate HPV serology as a biomarker of exposure to HPV. Because HPV DNA is not sensitive for detecting prior infections, we used both biological and behavioral characteristics known to be associated with current and past HPV infection to define cases and controls. Specifically, in separate analyses, cases were defined as either: (1) current type-specific HPV DNA positive at cervix (HPV16 DNA positive for the HPV16 analysis; HPV18 DNA positive for HPV18 analysis); (2) abnormal cytology/histology, defined as ≥HSIL/CIN2+ or ASC-H; (3) Lifetime number of sexual partners >4 (that is the 90th percentile among non-virginal women); (4) Years since sexual debut >4 (median among non-virginal women); and, (5) combined current DNA infection with abnormal cytology/histology (≥HSIL/CIN2+/ASC-H). Given that we cannot attribute specific HPV type to a cytological lesion, for definition #5, we conditioned first on HPV16 DNA positivity (or 18 DNA positivity for the HPV18 analysis) to increase the likelihood of type-specificity. We chose the 90th percentile of number of lifetime sexual partners and median years since sexual debut to increase the probability of accurately classifying women as HPV exposed. The main conclusions were not altered by dichotomizing at different points (data not shown). A priori, we considered the last definition to provide the strongest evidence of past exposure because it incorporated both molecular and microscopic evidence of infection. For each of the above five case-definitions, controls were accordingly defined as women not having the specific case-definition.
We estimated how well continuous levels of HPV16 serology (and HPV18 serology for the HPV18 analysis) collected at enrollment discriminates between cases and controls. The y-axis of the ROC graph represents sensitivity, or the true positive rate, i.e. the proportion correctly discriminated or predicted as such by serology among cases. The x-axis represents the complement of specificity, or the false positive rate, which is the proportion incorrectly discriminated by serology amongst non-cases. An area under the ROC curve (AUC) is a commonly used summary measure. An AUC of 1.0 represents a perfect test, while an AUC of 0.5 represents random classification (12). Using the continuous measures of HPV16 and HPV18 serology, we determined the cutpoint that maximized the sum of sensitivity and specificity. This method implicitly weights sensitivity and specificity equally.
In addition, we compared the sensitivity and specificity at the current standard cutpoints (8 EU/mL and 7 EU/mL, for HPV16 and 18 respectively) to three a priori specified cutpoints (the 25th, 50th and 75th percentiles among non-virginal seropositive women) and the cutpoint that maximizes the sum of sensitivity and specificity to show the trade-offs in loss and gain of sensitivity and specificity at different cutpoints.
Finally, to investigate whether different serology cutpoints would alter our previous estimates of enrollment correlates of HPV16/18 seropositivity (14), we compared multivariate logistic regression models using the current cutpoints for HPV16 and 18 (8 EU/mL and 7 EU/mL, respectively), to the cutpoint that maximized the sum of sensitivity and specificity.
RESULTS
Of the 7,466 women enrolled in the trial, 5,871 (78.7%) sexually experienced subjects had a pelvic examination, constituting the population for this analysis. The median age of the women included in the analysis was 21 years (interquartile range [IQR], 19 to 23 years); the median time since sexual debut was 4 years (IQR, 3 to 7 years); and the median number of lifetime sexual partners was 2 (IQR, 1 to 3). HPV16 DNA prevalence was 8.3% (n=488/5,868) and HPV18 was 3.2% (n=188/5,868).
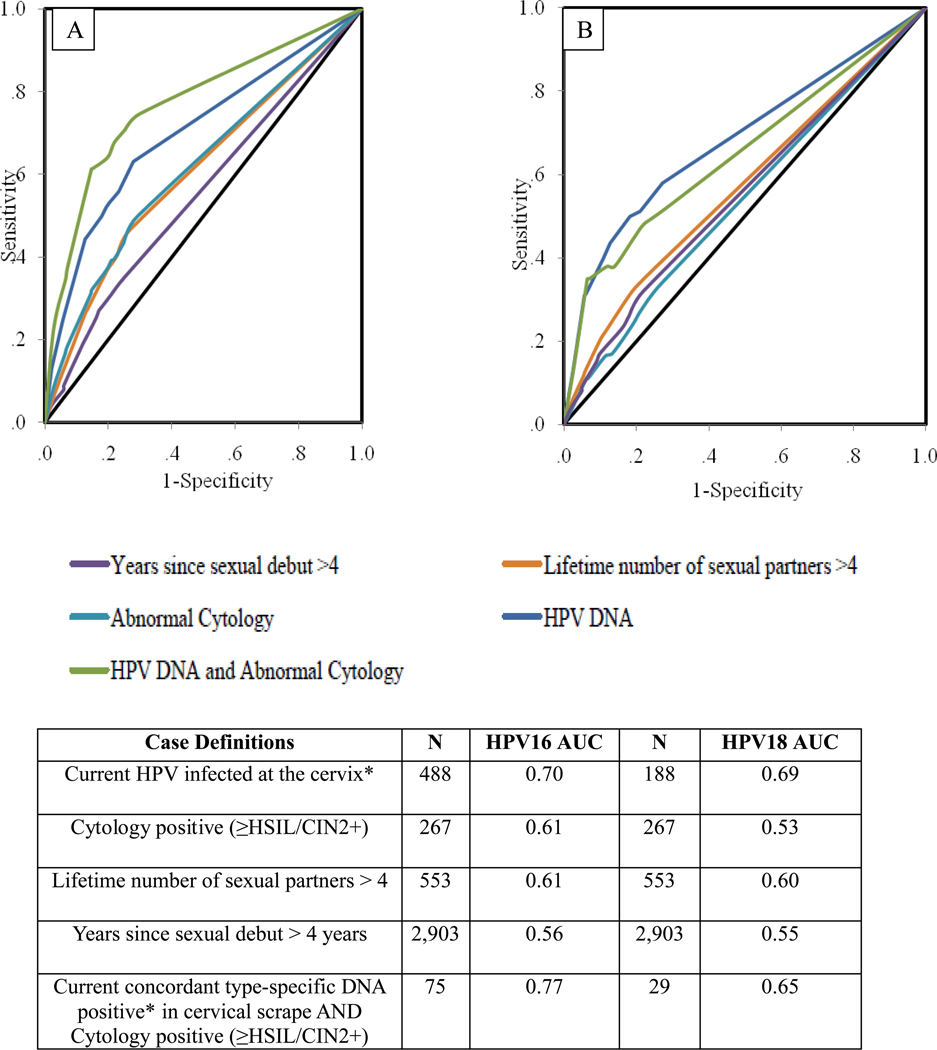
Figure 1 shows the ROC graphs and corresponding AUCs comparing how this polyclonal ELISA discriminated cases and controls based on the five different case definitions. For each analysis, the biomarker discriminated significantly better than a completely uninformative model (P <0.001 for each test of the null hypothesis of AUC=0.5). The AUC for the HPV16 DNA only model was 0.70; adding cytology increased AUC to 0.77. For HPV18, the AUC for the HPV18 DNA only model was 0.69, while for combined DNA and cytology was 0.65.
Figure 1.
Receiver Operating Characteristics (ROCs) for the Detection of HPV16 (A) and HPV18 (B) According to the Five Different Case Definitions by Serology; HPV16 and HPV18 Area Under the Curve (AUC) for Each Case Definition
NOTE: AUC, Area under the curve; N, number of women positive for outcome
*HPV16 DNA-positive at the cervix for the HPV16 analysis (HPV16 AUC); HPV18-positive at the cervix for the HPV18 analysis (HPV18 AUC).
Focusing on the most stringent case definition ‘current DNA infection and abnormal cytology’, using the continuous serology measure, and acknowledging that sensitivity and specificity are weighted equally, for HPV16 a cutpoint of 34 EU/mL yielded the best combination of sensitivity and specificity (sensitivity: 62.2%; specificity: 85.3%); for HPV18, a cutpoint of 60 EU/ mL yielded the best sum of sensitivity and specificity (sensitivity: 34.5%; specificity: 94.9%).
Using the same case definition of ‘current DNA infection and abnormal cytology’, from the lowest to highest cutpoint, sensitivity ranged from 75.7%–37.8%; specificity ranged from 70.4%–92.8% (Table 1A). Comparing 34 EU/mL to 8 EU/mL, there was not a statistically significant difference in sensitivity (P 0.44), but a statistically significant difference in specificity (P <0.0001). Similarly for HPV18 (Table 1B), sensitivity ranged from 51.7%–34.5%; specificity ranged from 72.6%–94.9%. Comparing 60 EU/mL to 7 EU/mL, there was a statistically significant difference in specificity (P <0.0001), but a non-statistically significant difference in sensitivity (P 0.40).
Table 1.
Ability of HPV16 Serology to Detect Women With a Current HPV16 DNA infection and Women With a Current HPV16 DNA Infection and an Abnormal Cytology (≥HSIL/CIN2+) (A); Ability of HPV18 Serology to Detect Women With a Current HPV18 DNA infection and Women With a Current HPV18 DNA Infection and an Abnormal Cytology (≥HSIL/CIN2+) (B)
| (A) HPV16 | ||||
|---|---|---|---|---|
| HPV16 DNA Positive (n=488) |
HPV16 DNA Positive AND Abnormal Cytology1 (n=75) |
|||
| Cut-Point | Sensitivity2 | Specificity3 | Sensitivity2 | Specificity3 |
| 84 | 62.5 | 72.6 | 75.7 | 70.4 |
| 15 | 53.1 | 79.8 | 68.9 | 77.8 |
| 345 | 44.3 | 87.3 | 62.2 | 85.3 |
| 93 | 25.4 | 94.0 | 37.8 | 92.8 |
| (B) HPV18 | ||||
|
HPV18 DNA Positive (n=181) |
HPV18 DNA Positive AND Abnormal Cytology1(n=29) |
|||
| Cut-Point | Sensitivity2 | Specificity3 | Sensitivity2 | Specificity3 |
| 74 | 58.0 | 72.8 | 51.7 | 72.6 |
| 10 | 51.1 | 78.9 | 48.2 | 77.9 |
| 18 | 43.6 | 87.2 | 37.9 | 86.3 |
| 43 | 31.4 | 93.9 | 34.5 | 93.2 |
| 605 | 26.7 | 95.4 | 34.5 | 94.9 |
Abnormal cytology defined as: ASC-H, HSIL-CIN2, HSIL-CIN3
Sensitivity defined as: True Positives/ (True Positives + False Negatives)
Specificity defined as: True Negatives/ (True Negatives + False Positives)
Current ELISA-based HPV serological cutpoint as determined by antibody titer values three standard deviations above the geometric mean titers taken from groups of HPV-negative individuals (n=278) and established by GSK.
Cutpoint chosen from the continuous serology measure that optimized combined sensitivity and specificity; for HPV16, also one of the three a priori specified cutpoints
Finally, to investigate whether these new cutpoints would alter our previous estimates of correlates of HPV16/18 seropositivity (14), we compared models using the current cutpoints of 8 EU/mL and 7 EU/mL to the cutpoints that maximized combined sensitivity and specificity, for HPV16 and 18, respectively (Table 2). Although the adjusted odds ratios of the correlates of HPV16 seropositivity we reported previously (14) did not change substantially using the higher cutpoint, some correlates lost statistical significance, possibly due to loss of power. Similar results were seen for HPV18 seropositivity (Table 2). As expected, HPV16 and HPV18 seroprevalence among virgins decreased using the higher cutpoints; however, virginal seroprevalence did not reach 0% (data not shown).
Table 2.
Adjusted Odds Ratios (aOR) and 95% Confidence Intervals (95% CI) for the Multivariate Model of the Correlates of HPV16 and HPV18 Seropositivity at Two HPV16 and Two HPV18 Serology Cutpoints:
| HPV16 Seropositivity | HPV18 Seropositivity | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | 8 EU/mL1 aOR |
8 EU/mL1 95% CI |
342 EU/mL aOR |
342 EU/mL 95% CI |
7 EU/mL1 aOR |
7 EU/mL1 95% CI |
602 EU/mL aOR |
602 EU/mL 95% CI |
| Years Since Sexual Debut | 1.06 | 1.03, 1.09 | 1.07 | 1.04, 1.11 | 1.07 | 1.04, 1.10 | 1.07 | 1.00, 1.15 |
|
Lifetime Sexual Partners 1 2–3 4+ |
1.00 1.59 2.30 |
---- 1.38, 1.83 1.91, 2.76 |
1.00 1.67 2.45 |
---- 1.39, 2.01 1.95, 3.07 |
1.00 1.38 1.60 |
----- 1.20, 1.59 1.33, 1.93 |
1.00 1.76 2.60 |
----- 1.19, 2.58 1.65, 4.10 |
|
Number of Pregnancies 0 1 2+ |
1.00 1.16 1.26 |
---- 1.00, 1.37 1.02, 1.55 |
1.00 1.21 1.27 |
---- 0.98, 1.48 0.98, 1.65 |
1.00 1.10 1.19 |
----- 0.93, 1.30 0.96, 1.30 |
1.00 1.41 1.41 |
----- 0.95, 2.10 0.85, 2.32 |
|
Hormonal Contraceptives Neither Ever Only OC3 Ever Only Inj3 Ever Both OC and Inj3 |
1.00 1.11 1.45 1.20 |
---- 0.91, 1.34 1.08, 1.94 0.97, 1.49 |
1.00 1.03 1.02 1.06 |
---- 0.81, 1.32 0.70, 1.48 0.81, 1.38 |
1.00 1.04 1.10 1.12 |
----- 0.86, 1.26 0.82, 1.48 0.90, 1.38 |
1.00 1.39 1.18 1.04 |
----- 0.84, 2.28 0.56, 2.50 0.60, 1.80 |
|
Condom use Never Ever |
1.00 0.86 |
---- 0.76, 0.98 |
1.00 0.87 |
---- 0.74, 1.02 |
1.00 0.98 |
----- 0.86, 1.12 |
1.00 0.89 |
----- 0.66, 1.21 |
|
Smoking Never Former Current |
1.00 1.20 1.26 |
---- 0.94, 1.50 1.04, 1.53 |
1.00 1.02 1.19 |
---- 0.77, 1.36 0.95, 1.51 |
1.00 1.10 1.15 |
----- 0.87, 1.39 0.94, 1.41 |
1.00 0.61 1.28 |
----- 0.32, 1.19 0.84, 1.94 |
|
Current and/or Past STI4 No Yes |
1.00 1.48 |
---- 1.27, 1.72 |
1.00 1.35 |
---- 1.13, 1.61 |
1.00 1.40 |
----- 1.21, 1.63 |
1.00 1.29 |
----- 0.92, 1.81 |
|
HC2/Cytology HC2 Neg/Normal HC2 Neg/ASCUS or LSIL HC2 Neg/ASC-H or HSIL-CIN2 or HSIL-CIN3 or AGC NOS HC2 Pos/Normal HC2 Pos/ASCUS or LSIL HC2 Pos/ASC-H or HSIL-CIN2 or HSIL-CIN3 or AGC NOS |
1.00 1.00 1.55 1.84 2.02 3.07 |
---- 0.66, 1.50 0.82, 2.92 1.59, 2.13 1.66, 2.47 2.29, 4.11 |
1.00 1.14 1.33 1.93 2.36 3.59 |
---- 0.67, 1.94 0.58, 3.06 1.61, 2.31 1.87, 2.98 2.63, 4.89 |
1.00 1.33 0.94 1.48 1.83 1.62 |
----- 0.90, 1.97 0.47, 1.89 1.27, 1.72 1.50, 2.23 1.20, 2.18 |
1.00 2.60 0.85 1.80 2.46 2.57 |
----- 1.21, 5.55 0.11, 6.28 1.26, 2.58 1.60, 3.77 1.44, 4.59 |
NOTE: HPV16 and HPV18 multivariate models were simultaneously adjusted for: years since sexual debut, lifetime number of sexual partners, number of pregnancies, hormonal contraceptive use, condom use, smoking history, current and/or past STIs, and HC2/cytology result.
Current cutpoint as defined by GSK
Cutpoint maximizing the sum of sensitivity and specificity as determined using the continuous enrollment HPV16 and HPV18 serology and the most stringent outcome definition of ‘HPV DNA positive and abnormal cytology’
Ever only OC = Ever only used oral contraceptives; Ever only Inj = Ever only used injectable contraceptives; Ever OC and Inj = Ever used oral and injectable contraceptives
Current and/or Past STI = Reported ever being diagnosed with warts, herpes, syphilis, Gonorrhea, or other; and/or currently diagnosed with Chlamydia and/or Gonorrhea infection
DISCUSSION
HPV serology (mainly against HPV16 and 18, the two most carcinogenic HPV types and the main targets of the prophylactic vaccines) is being used in research settings to identify current and past HPV-exposed individuals. However, the utility and performance of the different serological assays to discriminate HPV exposure is currently unknown. Using data from a large study of unvaccinated, sexually active, young adult women at peak exposure to HPV, we evaluated the sensitivity and specificity of the polyclonal HPV16 and HPV18 ELISA assay developed by GSK over a wide range of antibody levels and at various pre-specified case definitions. Our results showed that the ELISA assays provided moderate discriminative ability to detect cases with a current HPV DNA infection (AUC= 0.7 for both HPV16 and 18), and for HPV16, it improved slightly with the addition of abnormal cytology (AUC=0.77). Interestingly, addition of cytology to HPV18 DNA decreased the AUC relative to HPV18 DNA alone. While reasons for these discrepancies are unclear, it could be as a result of the differences in the HPV18 serology assay and also detection of HPV18 DNA or cytology. It is not possible to directly compare the two assays, however it is important to note that the HPV18 VLPs in general are more difficult to make and reproduce. It is also noteworthy that HPV18 related lesions are harder to detect with cytology (15–18) compared with HPV16 related lesions. Finally, using a case definition of HPV DNA-positive and abnormal cytology, serology at cutpoints identified by the ROC analysis (34 EU/mL and 60 EU/mL, for HPV16 and HPV18 respectively) had lower sensitivity and statistically significantly higher specificity than manufacturer set cutpoints (8 and 7 EU/mL, for HPV16 and HPV18 respectively).
Accurate classification of both HPV DNA and serostatus are important considerations for epidemiologic studies of HPV infection and vaccine efficacy. For HPV16, using a case definition of HPV16 DNA positive and abnormal cytology, a cutpoint of 8 EU/mL set by the manufacturer had the highest sensitivity (75.7%), resulting in a 24.3% false negative rate. Similarly for HPV18, the set cutpoint of 7 EU/mL (51.7% sensitivity), results in a 48.3% false negative rate. Therefore, even at the highest sensitivity, there is misclassification if HPV serology is used as a biomarker of HPV exposure. This could have implications for studies investigating co-factors influencing progression of HPV-infected cells to precancer and cancer, whereby associations found could be due to residual confounding by HPV positivity. As an example, some studies that have found an association between Chlamydia trachomatis (Ct) and cervical cancer, in the absence of HPV DNA exposure, have used HPV serology to account or adjust for HPV status (19–21). Based on our results, given that they used an assay with similar misclassification of HPV exposure as the one we evaluated in this report, 25% of HPV16 and 48% of HPV18 exposed women would be classified as HPV-negative. Thus, an observed positive association could be due to HPV-positive women who were misclassified as negative by the serology assay. In addition, it is premature to use this assay clinically to distinguish HPV exposed from un-exposed.
Our correlates of HPV16 and 18 seropositivity found in a previous analysis did not change substantially at the higher cutpoints, which indicates that the cutpoint set by the manufacturer is specific for epidemiologic studies of correlates of seropositivity. While some correlates lost statistical significance at the higher cutpoints possibly due to decreased power, risk estimates remained similar. Additionally, we found virginal seroprevalences never reached 0%, which is not unexpected as virginal seroprevalence can reflect the inherent biases in self-report of virginity, a technical artifact of non-specific binding, possible transmission through other non-penetrative sexual contact, or perinatal transmission, which we are unable to assess in this analysis.
There are some limitations to be considered. Currently there is no gold standard to classify HPV exposure, and we know from natural history studies that the majority of HPV infections clear (4). Additionally, natural history studies of HPV prevalence and incidence consistently show that the strongest correlates/predictors of HPV infection are those associated with increased exposure and persistence of the virus, such as higher lifetime sexual partners, increased years since sexual debut, abnormal cytology, and a current infection. Taken together, because DNA-positivity does not reflect cumulative infection, we used the aforementioned predictors of past exposure when defining our cases. In addition, not all HPV infected women develop antibody response to natural infection; some seroconvert at very low antibody levels, often below the assays’ limit of detection; and, it may take time and repeated exposures to develop such a detectable response (22). Therefore, it is unknown whether the moderate AUCs we obtained are due to poor assay, poor “gold standard”, a combination of the two, or due to biology of HPV infection. Thus, restricting HPV exposed women to biological and behavioral characteristics could have resulted in misclassification of our outcome.
Our analysis is also limited by the cross-sectional nature of our data. Only longitudinal studies can evaluate the serological response to HPV and its relation to HPV exposure, infection, persistence, and immunity. It is also important to note that the results from this study are not applicable to studies using other HPV serology assays. Finally, this study was performed in a young, healthy population; whether the assay performance may be different in older women is unknown.
In summary, this polyclonal ELISA assay provided moderate discriminative ability to detect HPV exposure. In addition, for HPV16 and HPV18 respectively, serology at cutpoints of 34 EU/mL and 60 EU/mL yielded the best combination of sensitivity and specificity, and had lower sensitivity and statistically higher specificity than the current cutpoints (8 and 7 EU/mL) to discriminate HPV exposed women. It is also important to note that even at the highest sensitivity (current cutpoints), a substantial proportion of women are still misclassified. The application of the polyclonal ELISA HPV assays should be considered in the context of the outcome of interest and balance between sensitivity and specificity. If the intent is measuring exposure, the goal may be to maximize sensitivity; thus, the current cutpoints are appropriate as the higher cutpoints yielded higher specificity, but at a detriment to sensitivity. However, if the objective is to measure immunity, it may be more appropriate to maximize specificity and positive predictive value; thus, a higher cutpoint may be more appropriate.
Present evidence does not support use of the ELISA serological assays in risk stratification or clinical setting. Future research on this topic should consider application of the ELISA assays and the important balance between sensitivity and specificity in the specific research setting.
Summary.
The low sensitivity of the polyclonal ELISA HPV16 and 18 serological assays to detect HPV exposure highlights the limitations of using this assay for risk stratification or clinical settings.
Acknowledgements
We would like to extend a special thanks to the women of Guanacaste and Puntarenas, Costa Rica, who gave of themselves in participating in this effort. We also acknowledge the tremendous effort and dedication of the staff in Costa Rica involved in this project, including Bernardo Blanco and his team (census), Ricardo Cerdas and Ana Hernández (blood processing), José Miguel González, Osman López, Johnny Matamoros, Manuel Sánchez, Rafael Thompson and Jorge Umaña (field activity coordinators), Su Yen Araya, Hazel Alvarez, Hayleen Campos, Muriel Grijalba, Ana Cristina Monge, Ana Peraza, Diana Robles, María Fernanda Sáenz, Dorita Vargas, and Jessica Vindas (clinic coordinators), Paola Alvarez, Dinia Angulo, Ana Live Arias, Betzaida Barrantes, Andrea Bolaños, Diego Bonilla, Marianela Bonilla, Mary José Calvo, Loretto Carvajal, Jessenia Chinchilla, Blanca Cruz, Marianela Herrera, Andrea Interiano, Fabiola Jiménez, Erick Lagos, Viviana Loría, Andrea Messeguer, Rebeca Ocampo, Ana Cristina Ocampo, Silvia Padilla, Angie Ramírez, Daniela Romero, Byron Romero, Yessenia Ruiz, Daniela Ruiz, Genie Saborío, Sofía Soto, Malena Salas, Adriana Torrez, Natalia Ugalde, Adriana Vallejos, Yesenia Vásquez, Maricela Villegas (clinicians), Marta Alvarado, Ana Cristina Arroyo, Gloriana Barrientos, Diana Díaz, Marlen Jara, Maureen Matarrita, María Ester Molina, Elida Ordóñez, Adriana Ramírez, Gina Sánchez, and Sihara Villegas (nurses), Arianne Castrillo and Vivian López (education and outreach effort coordinators), Karla Coronado (appointment coordinator), Ricardo Alfaro (quality control coordinator), Yenory Estrada (pharmacist) Charles Sánchez and Livia Romero (document center coordinators), Cristian Montero (quality assurance, regulatory) and Carlos Avila and Eric Alpízar (IT coordinators). Special recognition is also extended to Sofía Elizondo, Executive Director of Fundación INCIENSA and her staff for their administrative support. In the United States we would like to extend our appreciation to the team from Information Management Services (IMS) responsible for the development and maintenance of the data system used in the trial and who serve as the data management center for this effort. We would like to specifically acknowledge the invaluable contributions made by Julie Buckland and Laurie Rich. We acknowledge the contributions made by individuals at Westat, Inc., who provided project development and/or monitoring support, including Maribel Gomez, Kirk Midkiff, Isabel Trejos and Susan Truitt. We acknowledge the assistance provided by Carla Chorley, Troy Moore, Kathi Shea, Mindy Collins and Heather Siefers in the establishment of a specimen and vaccine repository for our trial and in their continued assistance with the handling and shipment of specimens. From GSK Biologicals, we would like to acknowledge the contributions of Gary Dubin, Anne Schuind, Kelechi Lawrence, Darrick Fu, and Bruce Innis for their contribution to discussions regarding trial conduct and Francis Dessy and Catherine Bougelet for HPV-16/18 antibody testing. We would like to thank members of the Data and Safety Monitoring Board charged with protecting the safety and interest of participants in our trial (Steve Self, Chair, Adriana Benavides, Luis Diego Calzada, Ruth Karron, Ritu Nayar, and Nancy Roach) and members of the external Scientific HPV Working Group who have contributed to the success of our efforts over the years (Joanna Cain, Chair, Diane Davey, Francisco Fuster, Ann Gershon, Elizabeth Holly, Silvia Lara, Wasima Rida, Henriette Raventós, Luis Rosero-Bixby, and Sarah Thomas).
Funding/Support
The Costa Rican Vaccine Trial is a longstanding collaboration between investigators in Costa Rica and NCI. The trial is sponsored and funded by NCI (N01-CP-11005) with support from the NIH Office of Research on Women’s Health and conducted in agreement with the Ministry of Health of Costa Rica. The NCI and Costa Rica investigators are responsible for the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation of the manuscript. Vaccine was provided for our trial by GSK Biologicals, under a Clinical Trials Agreement with NCI. GSK also provided support for aspects of the trial associated with regulatory submission needs of the company under FDA BB-IND 7920. Douglas Lowy and John Schiller are named inventors on U.S. government owned HPV vaccine patents that are licensed to GSK and Merck, and so are entitled to limited royalties as specified by federal law.
Cervarix is a registered trade mark of the Glaxo Smith Kline Biologicals group of companies.
Gardasil is a registered trade mark of Merck and Co. Inc.
Names and Affiliations of investigators in the Costa Rica Vaccine Trial (CVT) group are as follows
Proyecto Epidemiológico Guanacaste, Fundación INCIENSA, San José, Costa Rica
Mario Alfaro (Cytopathologist)
Manuel Barrantes (Field Supervisor)
M. Concepción Bratti (co-Investigator)
Fernando Cárdenas (General Field Supervisor)
Bernal Cortés (Specimen and Repository Manager)
Albert Espinoza (Head, Coding and Data Entry)
Paula González (co-Investigator)
Diego Guillén (Pathologist)
Rolando Herrero (co-Principal Investigator)
Silvia E. Jiménez (Trial Coordinator)
Jorge Morales (Colposcopist)
Lidia Ana Morera (Head Study Nurse)
Elmer Pérez (Field Supervisor)
Carolina Porras (co-Investigator)
Ana Cecilia Rodríguez (co-Investigator)
Libia Rivas (Clinician´s coordinator)
Luis Villegas (Colposcopist)
University of Costa Rica, San José, Costa Rica
Ivannia Atmella (Microbiologist, Immunology Laboratory)
José Bonilla (Head, HPV Immunology Laboratory)
Enrique Freer (Director, HPV Diagnostics Laboratory)
Alfonso García-Piñeres (Immunologist)
Margarita Ramírez (Microbiologist, Immunology Laboratory)
Sandra Silva (Head Microbiologist, HPV Diagnostics Laboratory)
United States National Cancer Institute, Bethesda, MD, USA
Allan Hildesheim (co-Principal Investigator & NCI co-Project Officer)
Aimee Kreimer (Investigator)
Douglas R. Lowy (HPV Virologist)
Nora Macklin (Trial Coordinator)
Mark Schiffman (Medical Monitor & NCI co-Project Officer)
John T. Schiller (HPV Virologist)
Mark Sherman (QC Pathologist)
Diane Solomon (Medical Monitor & QC Pathologist)
Sholom Wacholder (Statistician)
SAIC, NCI-Frederick, Frederick, MD, USA
Ligia Pinto (Head, HPV Immunology Laboratory)
Troy Kemp (Scientist, HPV Immunology Laboratory)
Womens and Infants’ Hospital, Providence, RI, USA
Claire Eklund (QC Cytology)
Martha Hutchinson (QC Cytology)
Georgetown University, Washington DC
Mary Sidawy (Histopathologist)
DDL Diagnostic Laboratory, The Netherlands
Wim Quint (Virologist, HPV DNA Testing)
Leen-Jan van Doorn (HPV DNA Testing)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential Conflicts of Interest:
Wim Quint and Leen-Jan van Doorn are employees of DDL Diagnostic Laboratory; Catherine Bougelet is an employee of GSK Biologicals. None of the authors have any potential conflicts of interest to report.
References
- 1.Cogliano V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005;6:204. doi: 10.1016/s1470-2045(05)70086-3. [DOI] [PubMed] [Google Scholar]
- 2.Dunne EF, Unger ER, Sternberg M, et al. Prevalence of HPV infection among females in the United States. JAMA. 2007;297:813–819. doi: 10.1001/jama.297.8.813. [DOI] [PubMed] [Google Scholar]
- 3.Schiffman M, Herrero R, Hildesheim A, et al. HPV DNA Testing in Cervical Cancer Screening: Results from Women in a High-Risk Province in Costa Rica. JAMA. 2000;283:87–93. doi: 10.1001/jama.283.1.87. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez AC, Schiffman M, Herrero R, et al. Longitudinal study of human papillomavirus persistence and cervical intraepithelial neoplasia grade 2/3: critical role of duration of infection. JNCI. 2010;102(5):315–324. doi: 10.1093/jnci/djq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiffman M, Safaeian M, Wentzensen N. The Use of Human Papillomavirus Seroepidemiology to Inform Vaccine Policy. Sex Trans Dis. 2009;36(11):675–679. doi: 10.1097/OLQ.0b013e3181bce102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrero R, Hildesheim A, Rodriguez AC, et al. Rationale and design of a community-based double-blind randomized clinical trial of an HPV 16 and 18 vaccine in Guanacaste, Costa Rica. Vaccine. 2008;26:4795–4808. doi: 10.1016/j.vaccine.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dessy FJ, Giannini SL, Bougelet CA, et al. Correlation between direct ELISA, single epitope-based inhibition ELISA and pseudovirion-based neutralization assay for measuring anti-HPV-16 and anti-HPV-18 antibody response after vaccination with the AS04-adjuvanted HPV-16/18 cervical cancer vaccine. Hum Vaccin. 2008;4:425–434. doi: 10.4161/hv.4.6.6912. [DOI] [PubMed] [Google Scholar]
- 8.Harper DM, Franco EL, Wheeler C, et al. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human Papillomavirus types 16 and 18 in young women: a randomised controlled trial. Lancet. 2004;364:1757–1765. doi: 10.1016/S0140-6736(04)17398-4. [DOI] [PubMed] [Google Scholar]
- 9.van Doorn LJ, Molijn A, Kleter B, Quint W, Colau B. Highly effective detection of human papillomavirus 16 and 18 DNA by a testing algorithm combining broad-spectrum and type-specific PCR. J Clin Microbiol. 2006;44:3292–3298. doi: 10.1128/JCM.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safaeian M, Herrero R, Hildesheim A, et al. Comparison of the SPF10-LiPA system to the Hybrid Capture 2 Assay for detection of carcinogenic human papillomavirus genotypes among 5,683 young women in Guanacaste, Costa Rica. J Clin Microbiol. 2007;45:1447–1454. doi: 10.1128/JCM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baay MF, Quint WG, Koudstall J, et al. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J Clin Microbiol. 1996;34:745–747. doi: 10.1128/jcm.34.3.745-747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanley JA, McNeil BJ. The Meaning and Use of the Area under a Receiver Operating Characteristic (ROC) Curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 13.Gonen M. Receiver Operating Characteristic (ROC) Curves; SUGI 31: Statistics and Data Analysis Paper 210-31; 2006. [Google Scholar]
- 14.Coseo S, Porras C, Hildesheim A, et al. Seroprevalence and Correlates of Human Papillomavirus 16/18 Seropositivity among Young Women in Costa Rica. Sex Trans Dis. 2010;37(11):706–714. doi: 10.1097/OLQ.0b013e3181e1a2c5. [DOI] [PubMed] [Google Scholar]
- 15.Clifford GM, Smith JS, Aquado T, Franceschi S. Comparison of HPV type distribution in high-grade cervical lesions and cervical cancer: a meta-analysis. BJC. 2003;89:101–105. doi: 10.1038/sj.bjc.6601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JS, Lindsay L, Hoots B, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121:621–632. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- 17.Kovacic MB, Castle PE, Herrero R, et al. Relationships of human papillomavirus type, qualitative viral load, and age with cytologic abnormality. Cancer Res. 2006;66:10112–10119. doi: 10.1158/0008-5472.CAN-06-1812. [DOI] [PubMed] [Google Scholar]
- 18.Safaeian M, Schiffman M, Gage J, Solomon D, Wheeler CM, Castle PE. Detection of precancerous cervical lesions is differential by human papillomavirus type. Cancer Res. 2009;69(8):3262–3266. doi: 10.1158/0008-5472.CAN-08-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anttila T, Saikku P, Koskela P, et al. Serotypes of Chlamydia trachomatis and risk for development of cervical squamous cell carcinoma. JAMA. 2001;285(1):47–51. doi: 10.1001/jama.285.1.47. [DOI] [PubMed] [Google Scholar]
- 20.Koskela P, Anttila T, Bjorge T, et al. Chlamydia trachomatis infection as a risk factor for invasive cervical cancer. Int J Cancer. 2000;85(1):35–39. doi: 10.1002/(sici)1097-0215(20000101)85:1<35::aid-ijc6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto K, Yasugi T, Oki A, et al. Area smoking and chlamydial infection risk factors for CIN? Different results after adjustment for HPV DNA and antibodies. Br J Cancer. 2003;89(5):831–833. doi: 10.1038/sj.bjc.6601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley M. Immunobiology of HPV and HPV vaccines. Gynecol Oncol. 2008;109:S15–S21. doi: 10.1016/j.ygyno.2008.02.003. [DOI] [PubMed] [Google Scholar]