Abstract
Malaria continues to be one of the most devastating global health problems due to the high morbidity and mortality it causes in endemic regions. The search for new antimalarial targets is of high priority because of the increasing prevalence of drug resistance in malaria parasites. Malarial proteases constitute a class of promising therapeutic targets as they play important roles in the parasite life cycle and it is possible to design and screen for specific protease inhibitors. In this mini-review, we provide a phylogenomic overview of malarial proteases. An evolutionary perspective on the origin and divergence of these proteases will provide insights into the adaptive mechanisms of parasite growth, development, infection, and pathogenesis.B
Keywords: Protease, malaria, Plasmodium, phylogenomics, genomics, target, vaccine, systems biology, remote homology detection.
INTRODUCTION
Malaria is one of the most important and persistent global infectious diseases. It is re-emerging as the number one infectious killer, responsible for over one million deaths yearly. The causative agents of malaria are a group of parasites in the Plasmodium genus. Five species, P. falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi are human pathogens. P. falciparum causes the most deadly form of malaria.
The re-emergence of malaria is largely due to the growing prevalence of parasite populations that show resistance to multiple drug treatment. With the advent of high throughput genomic, transcriptomic, proteomic, metabolomic, and pharmacogenomic technologies, enormous efforts have been focused on the identification and characterization of new and effective antimalarial targets [1-7]. These targets are selected based on several common criteria: (1) They are essential for parasite biology. The disruption of these genes or gene products leads to deleterious effects on parasite growth, development, or invasion. For example, cyclin-dependent protein kinases (CDKs) play indispensible roles in cell cycle progression and signal transduction [8-17]; (2) It is feasible to design or screen for effective pharmacophores or candidate inhibitors. For example, two compounds, chalcones and tryptanthrins, were identified by rational drug design, compound screening and molecular modeling as potent and specific inhibitors for the P. falciparum CDK7 homolog, Pfmrk [18]; (3) The drugs directed at the selected targets should have no or minimal adverse effects on humans. Some of the potential targets such as 1-deoxy-D-xylulose 5-phosphate (DOXP) reducto-isomerase [19, 20] and apicoplast gyrase [21] are localized to apicoplast, an organelle uniquely present in Plasmodium parasites and other parasites in the Apicomplexa phylum. These enzymes are crucial for apicoplast metabolism, replication, transcription and translation. Because the apicoplast is of prokaryotic origin, the inhibitors of these targets may have small or no side effects on the host.
Proteases are a class of promising antimalarial targets. They are digestive enzymes that degrade peptide bonds. They have demonstrated roles in parasite nutrition, development, invasion and egress: (1) a cascade of aspartic proteases plasmepsins [22, 23], cysteine proteases falcipains [24, 25] and metalloproteases [26-28] mediate massive degradation of host hemoglobin to release amino acids for parasite nutrition; (2) serine proteases (subtilases) have been implicated in erythrocyte invasion and parasite exit from the host [29-32]; (3) proteases are active mediators for cell cycle regulation and cell signaling [33-35]. Because the mechanisms of enzymatic action for many classes of proteases are known or can be derived from structural modeling or computer-aided drug design, it is possible to design or screen for protease inhibitors. The inhibitor classes for plasmepsins and falcipains have been investigated and evaluated [36-44].
Proteases, in addition to their potential as drug targets, are a prime example of supergene families with complex evolutionary histories involving gene duplication, domain shuffling, and lateral gene transfer. In this paper, we present a phylogenomic survey of malarial proteases. A better understanding of protease evolution will bring new insights into the genetic basis of adaptive phenotypes such as pathogenesis and virulence.
PHYLOGENEOMICS FOR THE IN SILICO PREDICTION OF PROTEASES IN THE PLASMODIUM GENOMES
Phylogenomics is an emerging discipline that combines molecular evolution theory and genomics [45, 46]. One of its direct and most important applications is to make functional predictions for previously uncharacterized proteins. The major hurdle that plagues all genomics-driven efforts in antimalarial target identification is the annotation problem [47]. In Plasmodium species, sequence similarity can be low, due to mutation, insertion, deletion, shuffling and recombination events, meaning high-confidence alignments between descendant sequences are not feasible and functional assignments are obscured. Genome annotation using traditional alignment-based algorithms has failed to assign functionality to over 60% of the ORFs in P. falciparum [48]. Popular methods for building probabilistic alignment models, such as PSI-BLAST [49], hidden Markov models (HMMs) [50], COMPASS [51] and HHSearch [52] show low accuracy and coverage when sequence similarity falls below 30% [53-55].
Only a handful of proteases had been discovered and characterized prior to the completion of genome sequencing for P. falciparum [48]. Using a comparative genomic approach, we predicted that a total of 92 protease homologs were present in P. falciparum genome, and at least 88 of them were expressed at the mRNA level by microarray and RT-PCR assays [56]. Subsequent data mining on the parasite proteome revealed that 67 of these predicted proteases were expressed at the protein level at least in one stage of the life cycle [57]. Recently we extended our study to other sibling species of malaria parasites, including P. vivax [58], which is the most widely distributed human malaria parasite, and three rodent species P. berghei, P. chabaudi, and P. yoelii yoelii [59, 60], which serve as the animal models for human malaria. In addition to traditional BLAST searches, we adopted a novel support vector machine (SVM)-based, supervised machine learning approach to tackle the remote homology problem. The underlying principle for remote homology detection lies in the domain of phylogenomics: these algorithms are designed to capture subtle similarities between the unknown proteins and the annotated proteins based on the evolutionarily conserved characteristics of the genes/proteins. A SVM classifier is a function that separates the training data into two classes and also maximizes the geometric margin between them in a feature space. Unlike most alignment-based algorithms which build models only with positive sequences, SVMs also use negative sequences (proteins outside the protein family) to learn the difference between the two classes. The SVM approach discovered several putative proteases that were not detectable by PSI-BLAST. For example, one putative PPPDE protease (PFI0940c) is a member of a novel family with a papain-like fold. This family was postulated to play a role in deubiquitination and cell cycle regulation [61]. The total number of predicted proteases in P. falciparum was increased from 92 to 123.
The degradome of five malaria parasite species is comprised of 115-137 putative proteases in five distinct catalytic classes (aspartic, cysteine, metallo, serine and threonine) (See Table 2 and Table 3 in [47]), which account for 0.9-2.3% of the open reading frames (ORFs) in the genome. They form 37 protein families based on their evolutionary relationship and structure conservation, according to the MEROPS protease classification system [62], and 29 of these families are commonly shared in five species. These proteases are important players in metabolism, cell cycle regulation, invasion, stress response, transcriptional regulation, signal transduction, and trafficking. A number of these proteases are becoming targets for functional characterization and rational inhibitor design [43, 63-69].
PHYLOGENOMICS FOR FUNCTIONAL CHARACTERIZATION OF MALARIAL PROTEASES
Phylogenomic analysis provides a cost-effective means to examine the evolutionary profiles of genes and gene products for functional prediction or characterization. The procedure often involves homology identification, multiple sequence alignment, phylogenetic reconstruction, inference of function, evolutionary analysis of orthology and paralogy, and identification of lateral gene transfer events [70]. This approach is particularly useful for the studies of protein families and protein superfamilies such as kinases and transporters [71-74]. In the domain of protease researches, phylogenomics has contributed to, for example, the classification and reconstruction of evolutionary diversification of serine proteases in fungi [75], the evolutionary profiling of cystatins, which comprise a superfamily of cysteine protease inhibitors [76], and the development of a statistical framework that was able to detect site-specific functional divergence in the caspase family of cysteine proteases [77]. The annotation of the predicted malarial proteases was essentially based on phylogenomic analysis [56], which revealed an array of novel proteases that could potentially be important for parasite-specific functions. A single copy of calpain was identified in the five surveyed Plasmodium genomes. It contains active site residues (C1035-N-1371-H1391 in P. falciparum) that are conserved in known or characterized calpains. Although calpains are well-known modulators for signal transduction, differentiation, cell motility, cell cycle regulation and cell-cell communication from bacteria to humans, its physiological role in parasite biology is yet to be defined. Nevertheless, partial knockdown assays indicated that the malarial calpain is crucial for the optimal growth of the parasite and cell cycle progression [78]. Interestingly, phylogenetic analysis revealed that the malarial calpain belongs to a clade of calcium-independent calpains, a lineage restricted to alveolates; the divergence from major human calpains makes it a possible drug target.
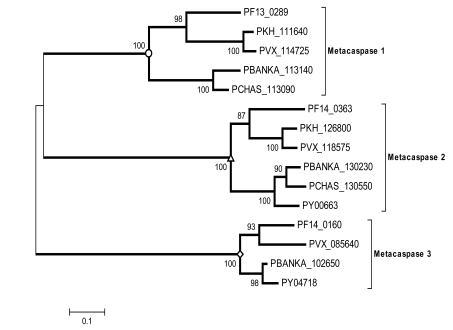
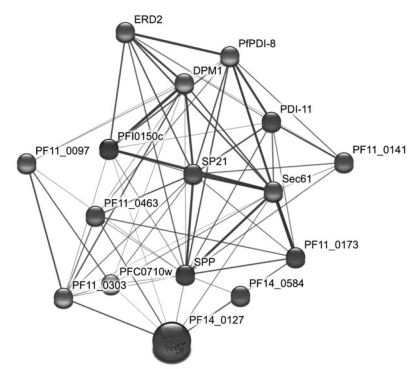
Multiple copies of metacaspases were identified in Plasmodium genomes [34, 47, 79], suggesting the existence of apoptosis or a similar signaling cascade in malaria parasites. Apoptosis-inducer and the administration of antimalarial drug chloroquine were shown to lead to DNA fragmentation and mitochondrial membrane potential disruption, indicative of the onset of programmed cell death in the parasite [80]. Phylogenetic analysis shows that this family may be generated by at least one gene duplication event: metacaspase-3 may represent an ancestral form, while metacaspase-1 and metacaspase-2 are more closely related to each other (Fig. 1). Metacaspase-1, in particular, contains the typical catalytic domain and the active site residues (histidine and cysteine dyad) that are essential for proteolytic function. In addition, it harbors a caspase recruitment domain (CARD) for apoptosis-related signaling [80]. The discovery of metacaspases has triggered the search for proteins/regulators in the parasite apoptotic network [81], which may represent a major stress-response system for parasites to survive under drug treatment and host immune challenges. Another example of potentially important proteases in malaria parasites is the signal peptide peptidase (SPP). SPP is an active player in regulated intramembrane proteolysis (RIP), which initiates signal transduction via processing the transmembrane segments of the substrates. One single copy of SPP is found in P. falciparum, P. vivax, P. yoelii yoelii, and P. chabaudi, and two copies are present in P. berghei. The putative active sites Tyr–Asp (YD) and Gly–Leu–Gly–Asp (GLGD) motifs which are universally conserved in SPPs are present in all the malarial SPPs (Fig. 2A). Phylogenetic analysis shows that the malarial SPPs form a distinct clade that is distantly related to the SPPs found in other animals from the fruit fly to the human (Fig. 2B). Data mining of the protein-protein interaction network revealed that P. falciparum SPP (PfSPP) is a highly connected protein with 54 association partners. Fig. (3) shows a schematic association map of PfSPP with representative partners with relatively high statistical support. Each association between a pair of proteins has a confidence score (S) ranging from 0.15 to 0.999 that was inferred from the evidence used to establish the association [82], including the interolog comparisons, which is rooted on phylogenomic inference of network associations among evolutionarily related organisms, yeast-2-hybrid (Y2H) assays, Gene Ontology (GO) classification for biological functions, cellular processes and sublocations, structural configuration, co-expression and co-occurrence patterns, and so on [83, 84]. Phylogenomic inference predicted that PfSPP is associated with a putative ER lumen protein retaining receptor (ERD2) [85] and a secretory protein Sec61 [86], both of which are components in the parasite translocation machinery required for the uptake of nutrients and expulsion of wastes: the SPP homolog was found to be co-expressed with the ERD2 and Sec61 homologs in three model organisms: Arabidopsis thaliana, Caenorhabditis elegans, and Drosophila melanogaster. PfAPP is also associated with signal peptidase, translation initiation and elongation factors, splicing factor, peptide chain release factor, and a variety of enzymes, suggesting that it is involved in transport, translation, posttranslational regulation and metabolism. It has recently been considered as a promising drug target since gene disruption assays indicate it is essential for parasite growth and merozoite invasion [87, 88].
Fig. (1).
The phylogenetic tree of metacaspases in Plasmodium, inferred using the neighbor-joining method based on the amino acid sequences with Poisson corrected distance [111]. Evolutionary analyses were conducted in MEGA5 [112]. The option of complete deletion of gaps was used for tree construction. 1,000 bootstrap replicates were used to infer the reliability of branching points. Bootstrap values of >50% are presented. The scale bar indicates the number of amino acid substitutions per site. The abbreviations for species names are: Pf: P. falciparum; PKH: P. knowlesi; PVX: P. vivax; PBNKA: P. berghei ANKA; PCHAS: P. chabaudi AS; PY: P. yoelli yoelii.
Fig. (2).
(A) The alignment of the catalytic region of the signal peptide peptidases (SPPs) in six Plasmodium species and other representative species. The putative active sites Tyr–Asp (YD) and Gly–Leu–Gly–Asp (GLGD) motifs are highlighted. (B) The phylogenetic tree of SPPs, inferred using the neighbor-joining method based on the amino acid sequences with Poisson corrected distance.
Fig. (3).
The protein-protein association map of Plasmodium falciparum SPP. The association partners were predicted by STRING [84]. This set of associations can be visualized in Cytoscape [113] and converted to an undirected weighted graph. Confidence scores for the interactions among the nodes (S values from STRING) were divided into three groups - low (0.150-0.399), medium (0.400-0.700) and high (0.701-0.999); the groups are represented by thin, medium and heavy lines, respectively.
PHYLOGENOMICS FOR ASSESSING THE SUITABILITY OF MALARIAL PROTEASES AS DRUG TARGETS
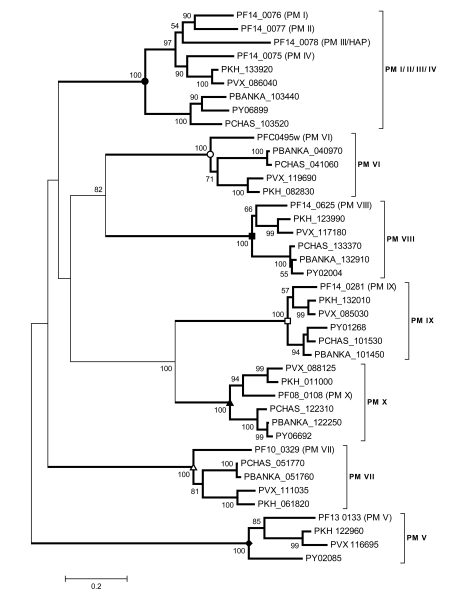
Phylogenomic analysis can reveal the complex evolutionary history of malarial proteases; their origin and relatedness to the host help researchers to assess their suitability as potential antimalaria targets. Jean et al. [89] and Coombs et al. [22] conducted elegant phylogenomic analyses on plasmepsins, a group of aspartic proteases in the pepsin (A1) family. Ten plasmepsins have been identified in P. falciparum, namely PM I-X; PM III is also known as histo-aspartic protease (HAP). They are divided into two classes (Fig. 4): (1) PM I-IV are all intronless with one single exon; They are located in adjacent positions on Chromosome 14, and are likely to be generated by tandem gene duplications. (2) PM V-X form a large clade which may represent an ancestral type of plasmepsins; Introns are present in PM VI, VII, VIII, and IX. The most extreme case is seen in PM VI, which contains 15 exons. An evolutionary model suggested that lateral gene transfer, exon shuffling and intron loss events may lead to the diverse types of plasmepsins in the parasite genome that are required for effective hemoglobin digestion [22, 89]. Inhibition of hemoglobin digestion causes starvation of the parasite and accumulation of intermediates toxic to parasites [23, 90]. Recently, two neutral aminopeptidases, M1 alanyl aminopeptidase (PfM1AAP, MAL13P1.56) and M17 leucine aminopeptidase (PfM17LAP, PF14_0439) have been characterized. Both enzymes are required for the late stage of hemoglobin digestion, which releases free amino acids for parasite nutrition and development inside the human red blood cell. Parasites were not viable in vivo and in vitro with the treatment of aminopeptidase inhibitors [91]. Because only single copy of PfM17LAP is present in P. falciparum genome, its disruption cannot be compensated for by any homolog. The X-ray structure of PfM17LAP has been resolved, opening a promising avenue for rational drug design [92].
Fig. (4).
The phylogenetic tree of plasmpepsins, inferred using the neighbor-joining method based on the amino acid sequences with Poisson corrected distance. PM I-IV contain one single exon without any introns. PM VI contains 15 exons, PM VIII contains 13 exons, PM VII and PM IX contain eight exons, and PM V and PM X contain one exon.
Gene duplication and lateral gene transfer are implicated in the evolution of other protease families such as subtilases and falcipains. Three copies of subtilases are found in P. falciparum genome and 2-5 copies are present in the other Plasmodium genomes. Subtilases are required for parasite invasion and egress from the human host [30, 93-95]. Evolutionarily, they are probably acquired via lateral gene transfer from a bacteria origin where subtilsins are commonly found. No statistically significant subtilase homologs are found in the humans, which is a desirable feature for drug targets. Similarly, falcipains are crucial for parasite biology; they may have dual roles in both hemoglobin digestion and host cell egress [30, 96-98]. They are evolutionarily closely related to the papains found in viruses and fungi. Lineage-specific expansion is evident in the evolution of rhomboid proteases (the S54 serine protease family) in Plasmodium: eight copies are present in P. falciparum, and 5-8 copies are present in the other species. They are the central players in regulated intramembrane proteolysis (RIP) and have been implicated in parasite development, invasion, cell signaling and pathogenesis [35, 99-101]. Different rhomboid proteases may have specific substrates preferences; their potential substrates include various adhesins and surface antigens [33]. Phylogenetic analysis revealed that they are closely related to the rhomboid homologs present in other apicomplexan parasites including Toxoplasma gondii, Eimeria tenella, Cryptosporidium spp. and Theileria spp [102]. There is only weak sequence similarity between the malarial rhomboids and a mitochondrial rhomboid protease, PARL, in the human genome. Targeting rhomboids and their associated signaling pathways therefore may be a novel therapeutic strategy. Other proteases with potentially important functions, such as calpain, metacaspases, and signal peptidase I, are also phylogenetically divergent from the host lineage.
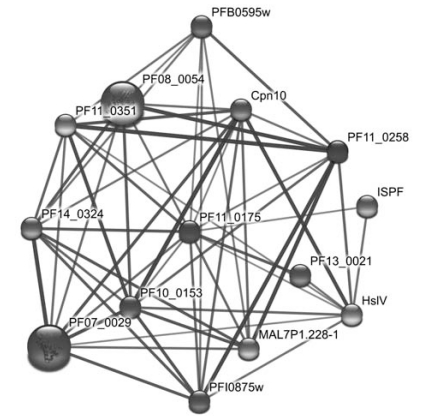
Phylogenomic analysis also revealed a group of putative malarial proteases that are destined to parasite-specific organelles of prokaryotic origin. The top target organelle for drug development is the apicoplast. It is essential for parasite life cycle, as inhibitors for apicoplast metabolism and replication resulted in the death of parasite [103, 104]. Using two independent algorithms, PATS (an artificial neural network algorithm) [105] and PlasmoAP [106], which combines signal peptide prediction [107] and rule-based classification, Ralph et al. [104] identified more than 540 genes in P. falciparum that are targeted to the apicoplast. Twenty-one of these genes encode putative proteases (Table 1). Because the apicoplast is derived from an ancient endosymbiosis in which the eukaryotic ancestor engulfed a red alga with a solitary chloroplast, its disruption does not cause significant interference with the host functions. The cyanobacterial heritage of the apicoplast enhances the potential of these apicoplast-targeted proteases as drug targets. Notably, five putative proteases from the ClpP endopeptidase family (S14) are predicted to be localized to the apicoplast (Table 1). They are the central players in the parasite heat shock response system, a key system for parasite adaptation to host environment, which involves a transmission from the Anopheles gambiae mosquito (~25°C) to the human host (~37°C), and the host's recurrent fever caused by infection. Fig. (5) shows a protein-protein association map of a putative Clp protease, PF11_0175. It is associated with various heat shock proteins (HSPs) including HslV (PFL1465c), an ATP-dependent threonine protease, Hsp60 (PF10_0153), Hsp70 homologs (PF11_0351, PF08_0054, PFI0875w, and MAL7P1.228), Hsp 90 (PF07_0029), Hsp40 (PFB0595w), a putative small Hsp (PF13_0021), a chaperonin cpn10 (PFL0740c), a putative Hsp70/Hsp90 organizing protein (PF14_0324), and a co-chaperone GrpE(PF11_0258). Inhibition assays showed that the Clp proteases and the chaperone HSPs are essential for parasite growth and development [108, 109]. It is possible to design inhibitors targeting specifically for malarial Clp proteases as the selective inhibitors for the ClpP protease complex has been developed in the bacteria system in Staphylococcus aureus [110].
Table 1.
Putative Proteases that are Predicted to Localize to the Apicoplast in P. falciparum
| Catalytic Type | Protease Family | P. falciparum protease ID | Annotation |
|---|---|---|---|
| Aspartic | A1 (pepsin family) | PF13_0133 | Plasmepsin V |
| PF14_0625 | Plasmepsin VIII | ||
| A22 (presenilin family) | PF14_0543 | signal peptide peptidase | |
| Cysteine | C1 (papain family) | PFD0230c | dipeptidyl peptidase 3 |
| Metallo | M1 (aminopeptidase N family) | MAL13P1.56 | M1-family alanyl aminopeptidase AMPN |
| M16 (pitrilysin family) | PF14_0382 | Pitrilysin | |
| M17 (leucyl aminopeptidase family) | PF14_0439 | M17 leucyl aminopeptidase | |
| M24 (methionyl aminopeptidase 1) | MAL8P1.140 | putative methionine aminopeptidase 1c | |
| PF14_0517 | aminopeptidase P | ||
| M41 (FtsH endopeptidase family) | PF14_0616 | putative ATP-dependent protease la | |
| Serine | S8 (subtilisin family) | PFE0355c | putative subtilisin-like protease 3 |
| S14 (ClpP endopeptidase family) | PFC0310c | ATP-dependent Clp protease proteolytic subunit | |
| PF14_0348 | ATP-dependent Clp protease proteolytic subunit | ||
| PF08_0063 | putative ClpB | ||
| PF14_0063 | putative ATP-dependent Clp protease | ||
| PF11_0175 | Clp protease, heat shock protein 101 | ||
| S33 (prolyl aminopeptidase) | PFC0065C | putative alpha/beta hydrolase | |
| PF14_0015 | putative aminopeptidase | ||
| S54 (Rhomboid family) | MAL8P1.16 | rhomboid protease ROM3 | |
| PF13_0312 | rhomboid protease ROM7 | ||
| Unknown | U48 (prenyl protease 2 family) | PFI0660c | putative protease |
Fig. (5).
The protein-protein association map of PF11_0175, a putative Clp protease in P. falciparum.
CONCLUSIONS
The phylogenomic approach has played an important role in the identification and in silico characterization of proteases in malaria parasites, providing a promising and largely uncharacterized set of targets for wet lab functional characterization and drug design. An evolutionary perspective on the origin and divergence of these proteases provides insights into the adaptive mechanisms of parasite growth, development, infection, and pathogenesis.
ACKNOWLEDGEMENTS
This work is supported by NIH grants GM081068, SC1AI080579, and AI067543 to YW, and the PSC-CUNY Research Award PSCREG-39-497 to JG. YW is also supported by NIH grant RR013646. We thank the Computational Biology Initiative at UTSA for providing computational support. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences, National Institute of Allergy and Infectious Diseases, or the National Institutes of Health.
ABBREVIATIONS
- CARD
= Caspase recruitment domain
- CDK
= Cyclin-dependent protein kinase
- DOXP
= 1-Deoxy-D-xylulose 5-phosphate
- ER
= Endoplasmic reticulum
- GO
= Gene Ontology
- HAP
= Histo-aspartic protease
- HSP
= Heat shock protein
- HMM
= Hidden Markov models
- ORF
= Open reading frame
- RIP
= Regulated intramembrane proteolysis
- SPP
= Signal peptide peptidase
- SVM
= Support vector machine
- Y2H
= Yeast 2-hybrid
REFERENCES
- 1.Dvorin J D, Martyn D C, Patel S D, Grimley J S, Collins C R, Hopp C S, Bright A T, Westenberger S, Winzeler E, Blackman M J, Baker D A, Wandless T J, Duraisingh M T. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010;328(5980):910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ecker A, Lakshmanan V, Sinnis P, Coppens I, Fidock D A. Evidence That Mutant PfCRT Facilitates the Transmission to Mosquitoes of Chloroquine-Treated Plasmodium Gametocytes. J. Infect. Dis. 2011;203(2):228–236. doi: 10.1093/infdis/jiq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jana S, Paliwal J. Novel molecular targets for antimalarial chemotherapy. Int. J. Antimicrob. Agents. 2007;30(1):4–10. doi: 10.1016/j.ijantimicag.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Joet T, Eckstein-Ludwig U, Morin C, Krishna S. Validation of the hexose transporter of Plasmodium falciparum as a novel drug target. Proc. Natl. Acad. Sci. USA. 2003;100(13):7476–7479. doi: 10.1073/pnas.1330865100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson D J, Fidock D A, Mungthin M, Lakshmanan V, Sidhu A B, Bray P G, Ward S A. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol. Cell. 2004;15(6):867–877. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirk K, Howitt S M, Broer S, Saliba K J, Downie M J. Purine uptake in Plasmodium: transport versus metabolism. Trends Parasitol. 2009;25(6):246–249. doi: 10.1016/j.pt.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Patel A P, Staines H M, Krishna S. New antimalarial targets: the example of glucose transport. Travel Med. Infect. Dis. 2008;6(1-2):58–66. doi: 10.1016/j.tmaid.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Rangarajan R, Bei A, Henry N, Madamet M, Parzy D, Nivez M P, Doerig C, Sultan A. Pbcrk-1, the Plasmodium berghei orthologue of P. falciparum cdc-2 related kinase-1 (Pfcrk- 1), is essential for completion of the intraerythrocytic asexual cycle. Exp. Parasitol. 2006;112(3):202–207. doi: 10.1016/j.exppara.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Dorin D, Semblat J P, Poullet P, Alano P, Goldring J P, Whittle C, Patterson S, Chakrabarti D, Doerig C. PfPK7, an atypical MEK-related protein kinase, reflects the absence of classical three-component MAPK pathways in the human malaria parasite Plasmodium falciparum. Mol. Microbiol. 2005;55(1):184–196. doi: 10.1111/j.1365-2958.2004.04393.x. [DOI] [PubMed] [Google Scholar]
- 10.Merckx A, Le Roch K, Nivez M P, Dorin D, Alano P, Gutierrez G J, Nebreda A R, Goldring D, Whittle C, Patterson S, Chakrabarti D, Doerig C. Identification and initial characterization of three novel cyclin-related proteins of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 2003;278(41):39839–39850. doi: 10.1074/jbc.M301625200. [DOI] [PubMed] [Google Scholar]
- 11.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 12.Grant K M. Targeting the cell cycle in the pursuit of novel chemotherapies against parasitic protozoa. Curr. Pharm. Des. 2008;14(9):917–924. doi: 10.2174/138161208784041042. [DOI] [PubMed] [Google Scholar]
- 13.Koyama F C, Chakrabarti D, Garcia C R. Molecular machinery of signal transduction and cell cycle regulation in Plasmodium. Mol. Biochem. Parasitol. 2009;165(1):1–7. doi: 10.1016/j.molbiopara.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlov S, Waters N C, Chavchich M. Leveraging cell cycle analysis in anticancer drug discovery to identify novel plasmodial drug targets. Infect. Disord. Drug Targets. 2010;10(3):165–190. doi: 10.2174/187152610791163354. [DOI] [PubMed] [Google Scholar]
- 15.Holland Z, Prudent R, Reiser J B, Cochet C, Doerig C. Functional analysis of protein kinase CK2 of the human malaria parasite Plasmodium falciparum. Eukaryot. Cell. 2009;8(3):388–397. doi: 10.1128/EC.00334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merckx A, Echalier A, Langford K, Sicard A, Langsley G, Joore J, Doerig C, Noble M, Endicott J. Structures of P. falciparum protein kinase 7 identify an activation motif and leads for inhibitor design. Structure. 2008;16(2):228–238. doi: 10.1016/j.str.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Waters N C, Geyer J A. Cyclin-dependent protein kinases as therapeutic drug targets for antimalarial drug development. Expert. Opin. Ther. Targets. 2003;7(1):7–17. doi: 10.1517/14728222.7.1.7. [DOI] [PubMed] [Google Scholar]
- 18.Keenan S M, Geyer J A, Welsh W J, Prigge S T, Waters N C. Rational inhibitor design and iterative screening in the identification of selective plasmodial cyclin dependent kinase inhibitors. Comb. Chem. High Throughput Screen. 2005;8(1):27–38. doi: 10.2174/1386207053328183. [DOI] [PubMed] [Google Scholar]
- 19.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler H K, Soldati D, Beck E. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285(5433):1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 20.Wiesner J, Jomaa H. Isoprenoid biosynthesis of the apicoplast as drug target. Curr. Drug Targets. 2007;8(1):3–13. doi: 10.2174/138945007779315551. [DOI] [PubMed] [Google Scholar]
- 21.Dahl E L, Rosenthal P J. Apicoplast translation, transcription and genome replication: targets for antimalarial antibiotics. Trends Parasitol. 2008;24(6):279–284. doi: 10.1016/j.pt.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Coombs G H, Goldberg D E, Klemba M, Berry C, Kay J, Mottram J C. Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends Parasitol. 2001;17(11):532–537. doi: 10.1016/s1471-4922(01)02037-2. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg D E. Hemoglobin degradation. Curr. Top. Microbiol. Immunol. 2005;295:275–291. doi: 10.1007/3-540-29088-5_11. [DOI] [PubMed] [Google Scholar]
- 24.Rosenthal P J. Hydrolysis of erythrocyte proteins by proteases of malaria parasites. Curr. Opin. Hematol. 2002;9(2):140–145. doi: 10.1097/00062752-200203000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Rosenthal P J. Cysteine proteases of malaria parasites. Int. J. Parasitol. 2004;34(13-14):1489–1499. doi: 10.1016/j.ijpara.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Eggleson K K, Duffin K L, Goldberg D E. Identification and characterization of falcilysin, a metallopeptidase involved in hemoglobin catabolism within the malaria parasite Plasmodium falciparum. J. Biol. Chem. 1999;274(45):32411–32417. doi: 10.1074/jbc.274.45.32411. [DOI] [PubMed] [Google Scholar]
- 27.Murata C E, Goldberg D E. Plasmodium falciparum falcilysin: a metalloprotease with dual specificity. J. Biol. Chem. 2003;278(39):38022–38028. doi: 10.1074/jbc.M306842200. [DOI] [PubMed] [Google Scholar]
- 28.Murata C E, Goldberg D E. Plasmodium falciparum falcilysin: an unprocessed food vacuole enzyme. Mol. Biochem. Parasitol. 2003;129(1):123–126. doi: 10.1016/s0166-6851(03)00098-7. [DOI] [PubMed] [Google Scholar]
- 29.Blackman M J. Proteases involved in erythrocyte invasion by the malaria parasite: function and potential as chemotherapeutic targets. Curr. Drug Targets. 2000;1(1):59–83. doi: 10.2174/1389450003349461. [DOI] [PubMed] [Google Scholar]
- 30.Blackman M J. Malarial proteases and host cell egress: an 'emerging' cascade. Cell Microbiol. 2008;10(10):1925–1934. doi: 10.1111/j.1462-5822.2008.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackman M J, Fujioka H, Stafford W H, Sajid M, Clough B, Fleck S L, Aikawa M, Grainger M, Hackett F. A subtilisin-like protein in secretory organelles of Plasmodium falciparum merozoites. J. Biol. Chem. 1998;273(36):23398–23409. doi: 10.1074/jbc.273.36.23398. [DOI] [PubMed] [Google Scholar]
- 32.Withers-Martinez C, Jean L, Blackman M J. Subtilisin-like proteases of the malaria parasite. Mol. Microbiol. 2004;53(1):55–63. doi: 10.1111/j.1365-2958.2004.04144.x. [DOI] [PubMed] [Google Scholar]
- 33.Baker R P, Wijetilaka R, Urban S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog. 2006;2(10):e113. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Chat L, Sinden R E, Dessens J T. The role of metacaspase 1 in Plasmodium berghei development and apoptosis. Mol. Biochem. Parasitol. 2007;153(1):41–47. doi: 10.1016/j.molbiopara.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Donnell R A, Hackett F, Howell S A, Treeck M, Struck N, Krnajski Z, Withers-Martinez C, Gilberger T W, Blackman M J. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J. Cell Biol. 2006;174(7):1023–1033. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunn B, Marzahn M, Liu P, Gutierrez-de-Teran H, Robbins A, McKenna R. Plasmepsin 9 as a New Target for Antimalarial Drug Discovery. Biopolymers. 2009;92(4):347–347. [Google Scholar]
- 37.Liu P, Marzahn M R, Robbins A H, Gutierrez-de-Teran H, Rodriguez D, McClung S H, Stevens S M, Yowell C A, Dame J B, McKenna R, Dunn B M. Recombinant Plasmepsin 1 from the Human Malaria Parasite Plasmodium falciparum: Enzymatic Characterization, Active Site Inhibitor Design, and Structural Analysis. Biochemistry. 2009;48(19):4086–4099. doi: 10.1021/bi802059r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah F, Mukherjee P, Gut J, Legac J, Rosenthal P J, Tekwani B L, Avery M A. Identification of Novel Malarial Cysteine Protease Inhibitors Using Structure-Based Virtual Screening of a Focused Cysteine Protease Inhibitor Library. J. Chem. Inf. Model. 2011;51(4):852–864. doi: 10.1021/ci200029y. [DOI] [PubMed] [Google Scholar]
- 39.Carroll C D, Patel H, Johnson T O, Guo T, Orlowski M, He Z M, Cavallaro C L, Guo J, Oksman A, Gluzman I Y, Connelly J, Chelsky D, Goldberg D E, Dolle R E. Identification of potent inhibitors of Plasmodium falciparum plasmepsin II from an encoded statine combinatorial library. Bioorg. Med. Chem. Lett. 1998;8(17):2315–2320. doi: 10.1016/s0960-894x(98)00419-3. [DOI] [PubMed] [Google Scholar]
- 40.Haque T S, Skillman A G, Lee C E, Habashita H, Gluzman I Y, Ewing T J, Goldberg D E, Kuntz I D, Ellman J A. Potent, low-molecular-weight non-peptide inhibitors of malarial aspartyl protease plasmepsin II. J. Med. Chem. 1999;42(8):1428–1440. doi: 10.1021/jm980641t. [DOI] [PubMed] [Google Scholar]
- 41.Kasam V, Zimmermann M, Maass A, Schwichtenberg H, Wolf A, Jacq N, Breton V, Hofmann-Apitius M. Design of new plasmepsin inhibitors: a virtual high throughput screening approach on the EGEE grid. J. Chem. Inf. Model. 2007;47(5):1818–1828. doi: 10.1021/ci600451t. [DOI] [PubMed] [Google Scholar]
- 42.Li R, Chen X, Gong B, Selzer P M, Li Z, Davidson E, Kurzban G, Miller R E, Nuzum E O, McKerrow J H, Fletterick R J, Gillmor S A, Craik C S, Kuntz I D, Cohen F E, Kenyon G L. Structure-based design of parasitic protease inhibitors. Bioorg. Med. Chem. 1996;4(9):1421–1427. doi: 10.1016/0968-0896(96)00136-8. [DOI] [PubMed] [Google Scholar]
- 43.Pandey K C, Barkan D T, Sali A, Rosenthal P J. Regulatory Elements within the Prodomain of Falcipain-2, a Cysteine Protease of the Malaria Parasite Plasmodium falciparum. Plos One. 2009;4(5):e5694. doi: 10.1371/journal.pone.0005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheidt K A, Roush W R, McKerrow J H, Selzer P M, Hansell E, Rosenthal P J. Structure-based design, synthesis and evaluation of conformationally constrained cysteine protease inhibitors. Bioorg. Med. Chem. 1998;6(12):2477–2494. doi: 10.1016/s0968-0896(98)80022-9. [DOI] [PubMed] [Google Scholar]
- 45.Eisen J A. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8(3):163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 46.Eisen J A, Fraser C M. Phylogenomics: intersection of evolution and genomics. Science. 2003;300(5626):1706–1707. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- 47.Kuang R, Gu J, Cai H, Wang Y. Improved prediction of malaria degradomes by supervised learning with SVM and profile kernel. Genetica. 2009;136(1):189–209. doi: 10.1007/s10709-008-9336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardner M J, Hall N, Fung E, White O, Berriman M, Hyman R W, Carlton J M, Pain A, Nelson K E, Bowman S, Paulsen I T, James K, Eisen J A, Rutherford K, Salzberg S L, Craig A, Kyes S, Chan M S, Nene V, Shallom S J, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather M W, Vaidya A B, Martin D M A, Fairlamb A H, Fraunholz M J, Roos D S, Ralph S A, McFadden G I, Cummings L M, Subramanian G M, Mungall C, Venter J C, Carucci D J, Hoffman S L, Newbold C, Davis R W, Fraser C M, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karplus K, Barrett C, Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14(10):846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 51.Sadreyev R, Grishin N. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol. 2003;326(1):317–336. doi: 10.1016/s0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- 52.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 53.Chubb D, Jefferys B R, Sternberg M J, Kelley L A. Sequencing delivers diminishing returns for homology detection: implications for mapping the protein universe. Bioinformatics. 2010;26(21):2664–2671. doi: 10.1093/bioinformatics/btq527. [DOI] [PubMed] [Google Scholar]
- 54.Qi Y, Sadreyev R I, Wang Y, Kim B H, Grishin N V. A comprehensive system for evaluation of remote sequence similarity detection. BMC Bioinformatics. 2007;8:314. doi: 10.1186/1471-2105-8-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reid A J, Yeats C, Orengo C A. Methods of remote homology detection can be combined to increase coverage by 10% in the midnight zone. Bioinformatics. 2007;23(18):2353–2360. doi: 10.1093/bioinformatics/btm355. [DOI] [PubMed] [Google Scholar]
- 56.Wu Y M, Wang X Y, Liu X, Wang Y F. Data-mining approaches reveal hidden families of proteases in the genome of malaria parasite. Genome Res. 2003;13(4):601–616. doi: 10.1101/gr.913403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Wu Y. Computer assisted searches for drug targets with emphasis on malarial proteases and their inhibitors. Curr. Drug Targets Infect. Disord. 2004;4(1):25–40. doi: 10.2174/1568005043480952. [DOI] [PubMed] [Google Scholar]
- 58.Carlton J M, Adams J H, Silva J C, Bidwell S L, Lorenzi H, Caler E, Crabtree J, Angiuoli S V, Merino E F, Amedeo P, Cheng Q, Coulson R M, Crabb B S, Del Portillo H A, Essien K, Feldblyum T V, Fernandez-Becerra C, Gilson P R, Gueye A H, Guo X, Kang'a S, Kooij T W, Korsinczky M, Meyer E V, Nene V, Paulsen I, White O, Ralph S A, Ren Q, Sargeant T J, Salzberg S L, Stoeckert C J, Sullivan S A, Yamamoto M M, Hoffman S L, Wortman J R, Gardner M J, Galinski M R, Barnwell J W, Fraser-Liggett C M. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455(7214):757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carlton J, Silva J, Hall N. The genome of model malaria parasites, and comparative genomics. Curr. Issues Mol. Biol. 2005;7(1):23–37. [PubMed] [Google Scholar]
- 60.Carlton J M, Angiuoli S V, Suh B B, Kooij T W, Pertea M, Silva J C, Ermolaeva M D, Allen J E, Selengut J D, Koo H L, Peterson J D, Pop M, Kosack D S, Shumway M F, Bidwell S L, Shallom S J, van Aken S E, Riedmuller S B, Feldblyum T V, Cho J K, Quackenbush J, Sedegah M, Shoaibi A, Cummings L M, Florens L, Yates J R, Raine J D, Sinden R E, Harris M A, Cunningham D A, Preiser P R, Bergman L W, Vaidya A B, Van Lin L H, Janse C J, Waters A P, Smith H O, White O R, Salzberg S L, Venter J C, Fraser C M, Hoffman S L, Gardner M J, Carucci D J. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419(6906):512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 61.Iyer L M, Koonin E V, Aravind L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3(11):1440–1450. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 62.Rawlings N D. A large and accurate collection of peptidase cleavages in the MEROPS database. Database (Oxford) 2009;2009 doi: 10.1093/database/bap015. bap015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boddey J A, Hodder A N, Gunther S, Gilson P R, Patsiouras H, Kapp E A, Pearce J A, de Koning-Ward T F, Simpson R J, Crabb B S, Cowman A F. An aspartyl protease directs malaria effector proteins to the host cell. Nature. 2010;463(7281):627–31. doi: 10.1038/nature08728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McGowan S, Oellig C A, Birru W A, Caradoc-Davies T T, Stack C M, Lowther J, Skinner-Adams T, Mucha A, Kafarski P, Grembecka J, Trenholme K R, Buckle A M, Gardiner D L, Dalton J P, Whisstock J C. Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases. Proc. Natl. Acad. Sci.USA. 2010;107(6):2449–2454. doi: 10.1073/pnas.0911813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moon S U, Kang J M, Kim T S, Kong Y, Sohn W M, Na B K. Plasmodium vivax: collaborative roles for plasmepsin 4 and vivapains in hemoglobin hydrolysis. Exp. Parasitol. 2011;128(2):127–132. doi: 10.1016/j.exppara.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 66.Silmon de Monerri N C, Flynn H R, Campos M G, Hackett F, Koussis K, Withers-Martinez C, Skehel J M, Blackman M J. Global identification of multiple substrates for Plasmodium falciparum SUB1, an essential malarial processing protease. Infect. Immun. 2011;79(3):1086–1097. doi: 10.1128/IAI.00902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skinner-Adams T S, Stack C M, Trenholme K R, Brown C L, Grembecka J, Lowther J, Mucha A, Drag M, Kafarski P, McGowan S, Whisstock J C, Gardiner D L, Dalton J. P. Plasmodium falciparum neutral aminopeptidases: new targets for anti-malarials. Trends Biochem. Sci. 2010;35(1):53–61. doi: 10.1016/j.tibs.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 68.Sologub L, Kuehn A, Kern S, Przyborski J, Schillig R, Pradel G. Malaria proteases mediate inside-out egress of gametocytes from red blood cells following parasite transmission to the mosquito. Cell Microbiol. 2011;13(6):897–912. doi: 10.1111/j.1462-5822.2011.01588.x. [DOI] [PubMed] [Google Scholar]
- 69.Wang F, Krai P, Deu E, Bibb B, Lauritzen C, Pedersen J, Bogyo M, Klemba M. Biochemical characterization of Plasmodium falciparum dipeptidyl aminopeptidase 1. Mol. Biochem. Parasitol. 2011;175(1):10–20. doi: 10.1016/j.molbiopara.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brown D, Sjolander K. Functional classification using phylogenomic inference. PLoS Comput. Biol. 2006;2(6):e77. doi: 10.1371/journal.pcbi.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sjolander K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20(2):170–179. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- 72.Brown J R, Auger K R. Phylogenomics of phosphoinositide lipid kinases: perspectives on the evolution of second messenger signaling and drug discovery. BMC Evol. Biol. 2011;11:4. doi: 10.1186/1471-2148-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gillespie J J, Brayton K A, Williams K P, Diaz M A, Brown W C, Azad A F, Sobral B W. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 2010;78(5):1809–1823. doi: 10.1128/IAI.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jung K H, Cao P, Seo Y S, Dardick C, Ronald P C. The Rice Kinase Phylogenomics Database: a guide for systematic analysis of the rice kinase super-family. Trends Plant Sci. 2010;15(11):595–599. doi: 10.1016/j.tplants.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 75.Hu G, Leger R J. A phylogenomic approach to reconstructing the diversification of serine proteases in fungi. J. Evol. Biol. 2004;17(6):1204–1214. doi: 10.1111/j.1420-9101.2004.00786.x. [DOI] [PubMed] [Google Scholar]
- 76.Kordis D, Turk V. Phylogenomic analysis of the cystatin superfamily in eukaryotes and prokaryotes. BMC Evol. Biol. 2009;9:266. doi: 10.1186/1471-2148-9-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y F, Gu X. Functional divergence in the caspase gene family and altered functional constraints: Statistical analysis and prediction. Genetics. 2001;158(3):1311–1320. doi: 10.1093/genetics/158.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Russo I, Oksman A, Vaupel B, Goldberg D E. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc. Natl. Acad. Sci. USA. 2009;106(5):1554–1559. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu Y, Wang X, Liu X, Wang Y. Data-mining approaches reveal hidden families of proteases in the genome of malaria parasite. Genome Res. 2003;13(4):601–616. doi: 10.1101/gr.913403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meslin B, Barnadas C, Boni V, Latour C, De Monbrison F, Kaiser K, Picot S. Features of apoptosis in Plasmodium falciparum erythrocytic stage through a putative role of PfMCA1 metacaspase-like protein. J. Infect. Dis. 2007;195(12):1852–1859. doi: 10.1086/518253. [DOI] [PubMed] [Google Scholar]
- 81.Meslin B, Zalila H, Fasel N, Picot S, Bienvenu A L. Are protozoan metacaspases potential parasite killers? Parasit. Vectors. 2011;4(1):26. doi: 10.1186/1756-3305-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.von Mering C, Jensen L J, Snel B, Hooper S D, Krupp M, Foglierini M, Jouffre N, Huynen M A, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33(Database issue):D433–7. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.LaCount D J, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth J R, Schoenfeld L W, Ota I, Sahasrabudhe S, Kurschner C, Fields S, Hughes R E. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438(7064):103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 84.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen L J, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2010;39(Database issue):D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elmendorf H G, Haldar K. Identification and localization of ERD2 in the malaria parasite Plasmodium falciparum: separation from sites of sphingomyelin synthesis and implications for organization of the Golgi. EMBO J. 1993;12(12):4763–4773. doi: 10.1002/j.1460-2075.1993.tb06165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Couffin S, Hernandez-Rivas R, Blisnick T, Mattei D. Characterisation of PfSec61, a Plasmodium falciparum homologue of a component of the translocation machinery at the endoplasmic reticulum membrane of eukaryotic cells. Mol. Biochem. Parasitol. 1998;92(1):89–98. doi: 10.1016/s0166-6851(97)00234-x. [DOI] [PubMed] [Google Scholar]
- 87.Li X, Chen H, Bahamontes-Rosa N, Kun J F, Traore B, Crompton P D, Chishti A H. Plasmodium falciparum signal peptide peptidase is a promising drug target against blood stage malaria. Biochem. Biophys. Res. Commun. 2009;380(3):454–459. doi: 10.1016/j.bbrc.2009.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li X, Chen H, Oh S S, Chishti A H. A Presenilin-like protease associated with Plasmodium falciparum micronemes is involved in erythrocyte invasion. Mol. Biochem. Parasitol. 2008;158(1):22–31. doi: 10.1016/j.molbiopara.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jean L, Long M, Young J, Pery P, Tomley F. Aspartyl proteinase genes from apicomplexan parasites: evidence for evolution of the gene structure. Trends Parasitol. 2001;17(10):491–498. doi: 10.1016/s1471-4922(01)02030-x. [DOI] [PubMed] [Google Scholar]
- 90.Banerjee R, Liu J, Beatty W, Pelosof L, Klemba M, Goldberg D E. Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc. Natl. Acad. Sci.USA. 2002;99(2):990–995. doi: 10.1073/pnas.022630099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Skinner-Adams T S, Stack C M, Trenholme K R, Brown C L, Grembecka J, Lowther J, Mucha A, Drag M, Kafarski P, McGowan S, Whisstock J C, Gardiner D L, Dalton J P. Plasmodium falciparum neutral aminopeptidases: new targets for anti-malarials. Trends Biochem. Sci. 2010;35(1):53–61. doi: 10.1016/j.tibs.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 92.McGowan S, Oellig C A, Birru W A, Caradoc-Davies T T, Stack C M, Lowther J, Skinner-Adams T, Mucha A, Kafarski P, Grembecka J, Trenholme K R, Buckle A M, Gardiner D L, Dalton J P, Whisstock J C. Structure of the Plasmodium falciparum M17 aminopeptidase and significance for the design of drugs targeting the neutral exopeptidases. Proc. Natl. Acad. Sci. USA. 2010;107(6):2449–2454. doi: 10.1073/pnas.0911813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harris P K, Yeoh S, Dluzewski A R, O'Donnell R A, Withers-Martinez C, Hackett F, Bannister L H, Mitchell G H, Blackman M J. Molecular identification of a malaria merozoite surface sheddase. Plos Pathogens. 2005;1(3):241–251. doi: 10.1371/journal.ppat.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jean L, Hackett F, Martin S R, Blackman M J. Functional characterization of the propeptide of Plasmodium falciparum subtilisin-like protease-1. J. Biol. Chem. 2003;278(31):28572–28579. doi: 10.1074/jbc.M303827200. [DOI] [PubMed] [Google Scholar]
- 95.Jean L, Withers-Martinez C, Hackett F, Blackman M J. Unique insertions within Plasmodium falciparum subtilisin-like protease-1 are crucial for enzyme maturation and activity. Mol. Biochem. Parasitol. 2005;144(2):187–197. doi: 10.1016/j.molbiopara.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 96.Shenai B R, Sijwali P S, Singh A, Rosenthal P J. Characterization of native and recombinant falcipain-2, a principal trophozoite cysteine protease and essential hemoglobinase of Plasmodium falciparum. J. Biol. Chem. 2000;275(37):29000–29010. doi: 10.1074/jbc.M004459200. [DOI] [PubMed] [Google Scholar]
- 97.Sijwali P S, Rosenthal P J. Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 2004;101(13):4384–4389. doi: 10.1073/pnas.0307720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sijwali P S, Koo J, Singh N, Rosenthal P J. Gene disruptions demonstrate independent roles for the four falcipain cysteine proteases of Plasmodium falciparum. Mol. Biochem. Parasitol. 2006;150(1):96–106. doi: 10.1016/j.molbiopara.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 99.Brossier F, Jewett T J, Sibley L D, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc. Natl. Acad. Sci. USA. 2005;102(11):4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dowse T J, Pascall J C, Brown K D, Soldati D. Apicomplexan rhomboids have a potential role in microneme protein cleavage during host cell invasion. Int. J. Parasitol. 2005;35(7):747–756. doi: 10.1016/j.ijpara.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 101.Srinivasan P, Coppens I, Jacobs-Lorena M. Distinct roles of Plasmodium rhomboid 1 in parasite development and malaria pathogenesis. PLoS Pathog. 2009;5(1):e1000262. doi: 10.1371/journal.ppat.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dowse T J, Soldati D. Rhomboid-like proteins in Apicomplexa: phylogeny and nomenclature. Trends Parasitol. 2005;21(6):254–258. doi: 10.1016/j.pt.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 103.Ralph S A, Foth B J, Hall N, McFadden G I. Evolutionary pressures on apicoplast transit peptides. Mol. Biol. Evol. 2004;21(12):2183–2194. doi: 10.1093/molbev/msh233. [DOI] [PubMed] [Google Scholar]
- 104.Ralph S A, van Dooren G G, Waller R F, Crawford M J, Fraunholz M J, Foth B J, Tonkin C J, Roos D S, McFadden G I. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2004;2(3):203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 105.Zuegge J, Ralph S, Schmuker M, McFadden G I, Schneider G. Deciphering apicoplast targeting signals--feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001;280(1-2):19–26. doi: 10.1016/s0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]
- 106.Foth B J, Ralph S A, Tonkin C J, Struck N S, Fraunholz M, Roos D S, Cowman A F, McFadden G I. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299(5607):705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- 107.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein. Eng. 1997;10(1):1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 108.El Bakkouri M, Pow A, Mulichak A, Cheung K L, Artz J D, Amani M, Fell S, de Koning-Ward T F, Goodman C D, McFadden G I, Ortega J, Hui R, Houry W A. The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum. J. Mol. Biol. 2010;404(3):456–477. doi: 10.1016/j.jmb.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 109.Rathore S, Sinha D, Asad M, Bottcher T, Afrin F, Chauhan V S, Gupta D, Sieber S A, Mohmmed A. A cyanobacterial serine protease of Plasmodium falciparum is targeted to the apicoplast and plays an important role in its growth and development. Mol. Microbiol. 2010;77(4):873–890. doi: 10.1111/j.1365-2958.2010.07251.x. [DOI] [PubMed] [Google Scholar]
- 110.Bottcher T, Sieber S A. Beta-lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J. Am. Chem. Soc. 2008;130(44):14400–1. doi: 10.1021/ja8051365. [DOI] [PubMed] [Google Scholar]
- 111.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 112.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011 doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smoot M E, Ono K, Ruscheinski J, Wang P L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]