Abstract
Objective
Gene expression alterations, especially in target tissues of insulin, have been associated with type 2 diabetes (T2D). In this study, we examined if genes involved in oxidative phosphorylation (OXPHOS) show differential gene expression and DNA methylation in pancreatic islets from patients with T2D compared with non-diabetic donors.
Design and methods
Gene expression was analyzed in human pancreatic islets from 55 non-diabetic donors and nine T2D donors using microarray.
Results
While the expected number of OXPHOS genes with reduced gene expression is 7.21, we identified 21 downregulated OXPHOS genes in pancreatic islets from patients with T2D using microarray analysis. This gives a ratio of observed over expected OXPHOS genes of 26.37 by a χ2-test with P=2.81×10−7. The microarray data was validated by qRT-PCR for four selected OXPHOS genes: NDUFA5, NDUFA10, COX11, and ATP6V1H. All four OXPHOS genes were significantly downregulated in islets from patients with T2D compared with non-diabetic donors using qRT-PCR (P≤0.01). Furthermore, HbAlc levels correlated negatively with gene expression of NDUFA5, COX11, and ATP6V1H (P<0.05). Gene expression of NDUFA5, NDUFA10, COX11, and ATP6V1H correlated positively with glucose-stimulated insulin secretion (P<0.03). Finally, DNA methylation was analyzed upstream of the transcription start for NDUFA5, COX11, and ATP6V1H. However, none of the analyzed CpG sites in the three genes showed differences in DNA methylation in islets from donors with T2D compared with non-diabetic donors.
Conclusion
Pancreatic islets from patients with T2D show decreased expression of a set of OXPHOS genes, which may lead to impaired insulin secretion.
Introduction
Type 2 diabetes (T2D) is characterized by hyperglycemia due to insulin resistance in peripheral tissues and pancreatic β-cell dysfunction. Altered gene expression in target tissues for T2D might influence cellular functions and biological pathways and hence promote the disease. Indeed, previous studies have shown that a set of genes involved in oxidative phosphorylation (OXPHOS) genes is downregulated in skeletal muscle and adipose tissue from patients with T2D compared with non-diabetic subjects using microarray technology (1–3). Similar results have been found in pancreatic islets from a diabetic mouse model, where several OXPHOS genes show decreased expression using genome-wide transcriptome and proteome analysis (4). However, whether OXPHOS genes show differential gene expression in human pancreatic islets from patients with T2D compared with non-diabetic control donors has not been examined to a similar extent. Moreover, although previous studies from our group have shown that epigenetic factors such as DNA methylation can regulate OXPHOS genes in human skeletal muscle (5–8), this has not been studied in human pancreatic islets. On the other hand, a previous study from our group have shown that increased DNA methylation of PPARGC1A (encoding peroxisome proliferator-activated receptor γ, coactivator 1 α (PGC1α)) is associated with decreased PPARGC1A mRNA expression in pancreatic islets from patients with T2D, which subsequently contributed to impaired insulin secretion (9). PPARGC1A is a transcriptional coactivator that coordinates expression of genes involved in mitochondrial oxidative metabolism. The aim of this study was to examine if the level of expression and/or DNA methylation of OXPHOS genes is changed in pancreatic islets from patients with T2D compared with non-diabetic donors.
Subjects and methods
Study populations
Human pancreatic islets from 55 non-diabetic and nine T2D deceased donors were obtained from the Human Tissue Laboratory at Lund University Diabetes Centre (Table 1). Islets were prepared by collagenase digestion and density gradient purification. After isolation, islets were cultured free floating in CMRL 1066 culture medium (ICN Biomedicals, Costa Mesa, CA, USA) supplemented with 10 mmol/l HEPES, 2 mmol/l l-glutamine, 50 μg/ml gentamicin, 0.25 μg/ml Fungizone (Gibco-BRL), 20 μg/ml Ciprofloxacin (Bayer Healthcare), and 10 mmol/l Nicotinamide at 37 °C (5% CO2) before RNA and DNA preparation. The islet purity was assessed by dithizone staining and the purity was similar for non-diabetic and T2D donors (P=0.15). To further examine the quality of islet isolation, gene expression of endocrine (somatostatin and glucagon) and exocrine markers (pancreatic lipase, amylase α 2A, and chymotrypsin 2) was also used to determine islet purity. Based on this method there were neither any differences in islet purity between non-diabetic and T2D donors (72 vs 68%, P=0.29). Glucose-stimulated insulin secretion (GSIS) from the human islets was measured in vitro in static incubations. Islets were preincubated for 30 min at 37 °C in Krebs Ringer bicarbonate (KRB) buffer (pH 7.4) containing (in mm) 120 NaCl, 25 NaHCO3, 4.7 KCl, 1.2 MgSO4, 2.5 CaCl2, 1.2 KH2PO, 10 HEPES supplemented with 1 mg/ml albumin, N-2 hydroxyethylpiperazine-N′-2-ethanesulfonic acid (10 mM), and 1 mM glucose. Each incubation vial contained 12 islets in 1.0 ml KRB buffer solution and was treated with 95% O2–5% CO2 to obtain constant pH and oxygenation. After preincubation, the buffer was changed to a KRB buffer containing either 1 mM (basal secretion) or 16.7 mM glucose (stimulated secretion; GSIS). The islets were then incubated for 1 h at 37 °C in a metabolic shaker (30 cycles per minute). Immediately after incubation an aliquot of the medium was removed for analysis of insulin using an RIA kit (Euro-Diagnostica, Malmo, Sweden) as described previously (10, 11).
Table 1.
Characteristics of human pancreatic donors. Data is presented as median (IQR).
| Non-diabetic donors | Type 2 diabetic donors | P value | |
|---|---|---|---|
| n (male/female) | 55 (29/26) | 9 (5/4) | |
| Age (years) | 57.0 (54.0–63.0) | 57.0 (43.5–70.0) | 0.99 |
| BMI (kg/m2) | 26.0 (23.9–27.5) | 28.7 (24.0–33.7) | 0.13 |
| HbAlc | 5.6 (5.3–6.0) | 6.9 (6.8–7.6) | 0.00010 |
| BIS (ng/islet per h) | 0.21 (0.14–0.50) | 0.22 (0.085–0.26) | 0.58 |
| GSIS (ng/islet per h) | 1.05 (0.56–1.45) | 0.48 (0.29–0.63) | 0.029 |
P values were between P values and calculated with Mann–Whitney U test. BIS, basal insulin secretion; GSIS, glucose-stimulated insulin secretion.
The donor before death or her/his relatives upon admission to intensive care unit had given their consent to donate organs for medical research. All procedures were approved by the ethics committees at Uppsala and Lund Universities.
Gene expression in human pancreatic islets
Total RNA was isolated with the AllPrep DNA/RNA Mini Kit (Qiagen GmbH). RNA quality and concentration was measured using an Agilent 2100 bioanalyzer and Nanodrop ND-1000 equipment respectively.
The microarrays were performed following the Affymetrix standard protocol. Briefly, total RNA (200 ng) was processed following the GeneChip Expression 3′-Amplification Reagents One-cycle cDNA synthesis kit instructions (Affymetrix, Inc., Santa Clara, CA, USA) to produce double-stranded cDNA. This was used as a template to generate biotin-targeted cRNA following manufacturer's specifications; 15 μg of the biotin-labeled cRNA was fragmented to strands between 35 and 200 bases in length, 10 μg of which was hybridized onto the GeneChip Human Gene 1.0 ST whole transcript based assay overnight in the GeneChip Hybridization oven 6400 using standard procedures. The arrays were washed and stained in a GeneChip Fluidics Station 450. Scanning was carried out with the GeneChip Scanner 3000 and image analysis was performed by GeneChip Operating Software. The array data was summarized and normalized with Robust Multi-array analysis method by the software ‘Expression Console’ (Affymetrix). The complete array data will be reported elsewhere (Jalal Taneera, Stefan Lang, Amitabh Sharma, Emma Ahlqvist, Yuedan Zhou, Anna Jonsson, Valeriya Lyssenko, Petter Vikman, Ola Hansson, Albert Salehi, Erik Renström, and Leif Groop, unpublished results).
Quantitative RT-PCR
Quantitative RT-PCR was performed for selected genes to technically validate the results obtained in the microarray study. cDNA was synthesized using QuantiTect RT kit (Qiagen). Quantitative analysis of gene expression was performed by the TaqMan Real-Time PCR with an ABI Prism 7900 HT System (Applied Biosystems, Foster City, CA, USA) using gene-specific probes and primer pairs (Assays-on-Demand, Applied Biosystems) for NDUFA5 (Hs01634019_g1), NDUFA10 (Hs00190004_m1), COX11 (Hs01680112_mH), and ATP6V1H (Hs00210960_m1). The transcript quantity was normalized to the mRNA level of cyclophilin A (4326316E, Applied Biosystems) and quantified with the ΔΔCt method.
DNA methylation analysis
Genomic DNA was isolated with the AllPrep DNA/RNA Mini Kit (Qiagen GmbH); 500 ng of genomic DNA was bisulfite treated using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA). DNA methylation analysis was performed with EpiTYPER using Sequenom MassARRAY (Sequenom, San Diego, CA, USA) system as described previously (11). Three EpiTYPER assays were designed for NDUFA5, COX11, and ATP6V1H using the online EpiDesigner tool (http://www.epidesigner.com/). Primer information and information about target sequence position is given in Supplementary Table 1, see section on Supplementary data given at the end of this article. Owing to high or low mass of the cleavage product or that the sites did not generate data successfully; a number of CpG sites were excluded from this study. In total six CpG sites in NDUFA5, 12 CpG sites in COX11, and 16 CpG sites in ATP6V1H generated DNA methylation data that were included in this study. For COX11 and ATP6V1H, a number of CpG sites were analyzed as CpG units including CpG sites −201/197, −168/−160, and −96/−89 for COX11 and CpG sites −259/−255, −211/−208, −198/−192, −187/−183/−180, and −80/−77 for ATP6V1H.
Statistical analysis
Data are shown as median with range. Differences in OXPHOS gene expression and DNA methylation in pancreatic islets of non-diabetic and T2D donors were analyzed by Mann–Whitney U test.
A χ2-test was used to analyze if a set of OXPHOS genes show significantly reduced gene expression in pancreatic islets from patients with T2D. The test examined if the observed number of OXPHOS genes with differential gene expression in human pancreatic islets due to T2D (n=21) is significantly more than the expected number of OXPHOS genes (n=7.20). The expected number of OXPHOS genes is calculated based on the total number of probe IDs annotated to OXPHOS genes analyzed using the microarray (n=147), the total number of probe IDs analyzed using the microarray (n=28869), and the total number of probe IDs that show reduced gene expression in the T2D human islets with P<0.05 (n=1416). The KEGG database (http://www.genome.jp/kegg) was used as reference set for identification of genes in the OXPHOS pathway.
A gene set enrichment analysis performed in DAVID (12, 13) was used to identify key biological pathways of all downregulated probe IDs in human T2D islets. Downregulated probe IDs in the microarray analysis of T2D and non-diabetic islets with P<0.05 were assigned to KEGG pathway analysis to rank the enrichment of the pathways in the gene set. Affymetrix HuGene-1_0-st-v1 was used as background reference to the uploaded gene set. The P value indicating the significance of enrichment was corrected with the Benjamini–Hochberg method.
Correlations were calculated using Spearman correlation coefficient. All P values were two-tailed and P values <0.05 were considered significant. The presented P values have not been corrected for multiple testing. Statistical calculations were performed by PASW Statistics 18 for Windows (SPSS, Chicago, IL, USA).
Results
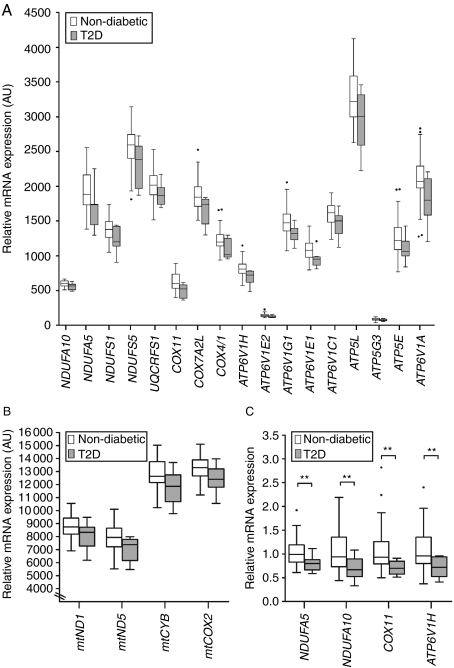
To examine if OXPHOS gene expression is downregulated in human islets from patients with T2D, we analyzed microarray data of human pancreatic islets from 55 non-diabetic and nine T2D donors (Table 1). We identified 21 OXPHOS genes (17 nuclear-encoded OXPHOS genes and four mitochondrial-encoded OXPHOS genes) with nominally reduced expression and an individual range in fold change of 6–22% and range in P values of 0.0041–0.045 (Fig. 1A and B and Supplementary Table 2, see section on Supplementary data given at the end of this article). Since the expected number of OXPHOS genes with reduced gene expression is 7.20, the χ2 of observed over expected OXPHOS genes is 26.37 with P=2.81×10−7.
Figure 1.
Box and Whiskers plots showing downregulation of OXPHOS gene expression in human pancreatic islets from patients with type 2 diabetes (T2D). (A) 17 nuclear-encoded OXPHOS genes and (B) four mitochondrial-encoded OXPHOS genes showed reduced mRNA expression with P≤0.05 in human pancreatic islets from patients with T2D (n=9; gray boxes) compared with non-diabetic donors (n=55; white boxes) using microarray. Since the expected number of OXPHOS genes with reduced gene expression is 7.21, the χ2 of observed over expected OXPHOS genes is 26.37 with P=2.81×10−7. (C) Technical validation of the microarray data for NDUFA5, NDUFA10, COX11, and ATP6V1H in human pancreatic islets of non-diabetic donors (n=47; white boxes) and T2D patients (n=9; gray boxes) using quantitative RT-PCR. **P≤0.01.
When performing a gene set enrichment analysis by DAVID (12, 13) and with KEGG biochemical pathway as annotation, we observed that the key pathway of all downregulated genes in T2D islets was OXPHOS (number of genes=21; fold enrichment=3.0; and P value=3.0×10−3). No other pathway falls out as significantly enriched. We next performed a technical validation of the microarray data using qRT-PCR. Based on ranking of P values from the microarray analysis and the fold change of the 17 nuclear-encoded OXPHOS genes (Supplementary Table 2, see section on Supplementary data given at the end of this article), we selected two genes from complex I (NDUFA5 and NDUFA10), one gene from complex IV (COX11), and one gene involved in ATPase activity (ATP6V1H) for the validation analysis. In concordance with our microarray analysis results, NDUFA5, NDUFA10, COX11, and ATP6V1H were downregulated in T2D compared with non-diabetic donors using qRT-PCR (Fig. 1B and Supplementary Table 3, see section on Supplementary data given at the end of this article).
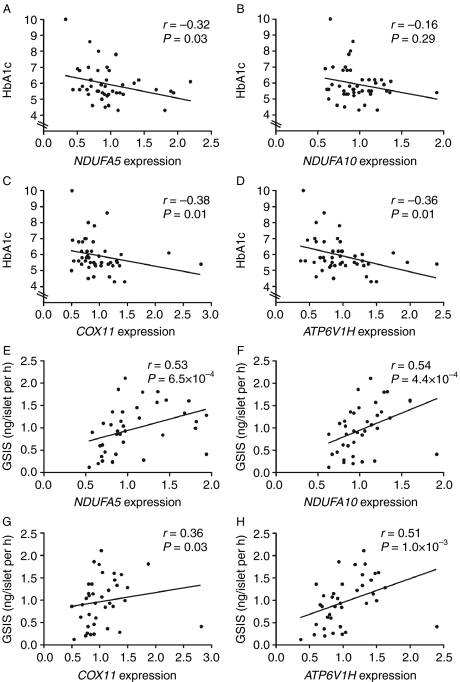
We next examined whether expression levels of NDUFA5, NDUFA10, COX11, and ATP6V1H correlated with HbAlc levels. When including both non-diabetic and T2D islets, HbAlc levels correlated negatively with the mRNA expression of NDUFA5, COX11, and ATP6V1H, but not with NDUFA10 (Fig. 2A–D). On the other hand, we could not detect any significant correlation between OXPHOS expression and HbAlc levels when including only non-diabetic donors (data not shown).
Figure 2.
Expression levels of OXPHOS genes correlated with HbAlc and GSIS. (A–D) Correlations between HbAlc levels and OXPHOS gene expression analyzed with qRT-PCR in human pancreatic islets, including both type 2 diabetic (T2D) patients and non-diabetic donors. (E–H) Correlations between OXPHOS gene expression analyzed with qRT-PCR and GSIS in human pancreatic islets of non-diabetic donors.
We further tested if the expression levels of NDUFA5, NDUFA10, COX11, and ATP6V1H correlated with glucose-stimulated insulin secretion in the human islets. The expression levels of all four OXPHOS genes correlated positively with GSIS (Fig. 2E–H). The same patterns were observed when both non-diabetic and T2D donors were included (data not shown).
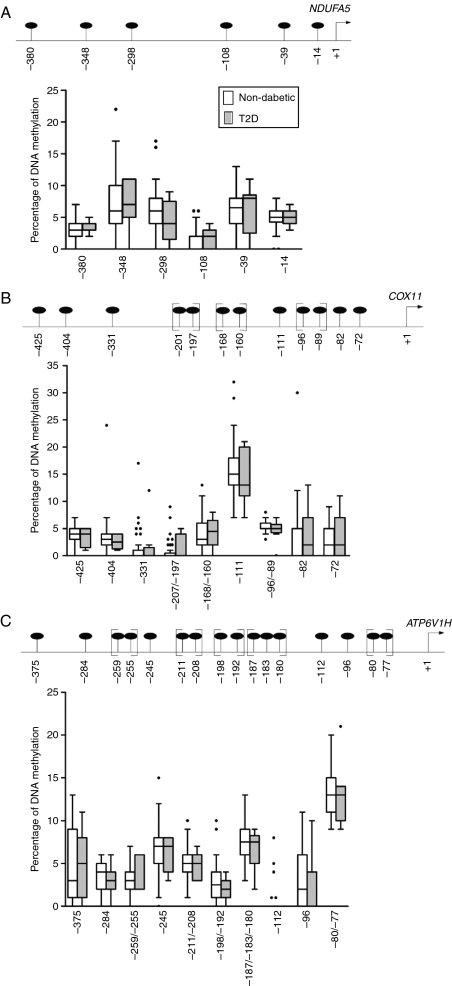
We obtained DNA methylation levels for six CpG sites for NDUFA5, 12 CpG sites for COX11, and 16 CpG sites for ATP6V1H, all located upstream of respective transcription start site, in the human pancreatic islets (Fig. 3A–C). The three analyzed regions were hypomethylated and DNA methylation was below 10% for the majority of the analyzed CpG sites. Moreover, none of the analyzed CpG sites in any of the three promoter regions showed differences in DNA methylation in islets from donors with T2D compared with non-diabetic donors (Fig. 3A–C). We next examined whether the degree of methylation of the CpG sites in the analyzed promoter regions correlated with mRNA expression of its respective gene, age, body mass index (BMI), and/or HbAlc. DNA methylation of CpG unit −96/−89, upstream of the transcription start site for COX11, correlated negatively with COX11 mRNA expression (r=−0.33, P=0.030) and positively with age (r=0.35, P=0.012) in non-diabetic donors. However, none of the other analyzed CpG sites in any of the three gene regions showed any significant correlation to gene expression, age, BMI, or HbAlc. To study if the general methylation pattern correlates with gene expression, the average methylation of each promoter was calculated using all of the analyzed CpG sites. Neither here were any differences in DNA methylation found between T2D and non-diabetic donors nor any significant correlation with gene expression (data not shown).
Figure 3.
No differences in DNA methylation in islets from patients with type 2 diabetes (T2D) compared with non-diabetic donors. (Upper panel A–C) is a schematic representation of the gene regions analyzed for DNA methylation for (A) NDUFA5, (B) COX11, and (C) ATP6V1H. Positions of analyzed CpG sites in relation to the transcription start sites are indicated in black circles. CpG sites within a bracket were analyzed as a CpG unit. (Lower panel A–C) are Box and Whiskers plots showing DNA methylation levels in percentage for the analyzed CpG sites of (A) NDUFA5, (B) COX11, and (C) ATP6V1H in human pancreatic islets from non-diabetic donors (n=55; white boxes) and T2D patients (n=9; gray boxes) using EpiTYPER.
Discussion
Mitochondrial dysfunction has been suggested to contribute to both impaired insulin secretion and action (5–7, 9, 14–20). This study demonstrates that a set of genes involved in OXPHOS is downregulated in pancreatic islets from patients with T2D. Although alterations in gene expression of individual OXPHOS genes were of relatively small magnitude, the number of genes enriched was significant based on a χ2-test and included 21 OXPHOS genes. Also when performing a gene enrichment analysis, OXPHOS was the most significant enriched pathway within all downregulated genes in T2D islets. Our result is in agreement with previous studies reporting reduced expression of a set of OXPHOS genes in human skeletal muscle from patients with T2D (1, 2). It is possible that a coordinated downregulation of multiple OXPHOS genes affects the overall ATP production. Since mitochondrial ATP production by OXPHOS in the respiratory chain is a key requirement for glucose-stimulated insulin release by pancreatic β-cells, a reduced OXPHOS gene expression in T2D islets may result in reduced insulin secretion (21). Indeed, in this study, islet expression of multiple OXPHOS genes correlated positively with GSIS, supporting the hypothesis that appropriate OXPHOS gene expression is required for sufficient insulin secretion. Based on the data from this study, we cannot rule out that the changes we find in insulin secretion and OXPHOS gene expression in T2D islets are due to altered cell composition in islets from T2D donors. However, although some researchers have found reductions in β-cell number in human type 2 diabetic islets (22), others have not observed any changes (23). Moreover, our previous animal and cell data support the finding that reduced OXPHOS levels in both pancreatic islets and clonal β-cells leads to reduced OXPHOS enzyme activity and hence impaired glucose-stimulated ATP production and insulin secretion (19). It has been shown that mitochondrially encoded subunits play a crucial function during modular assembly of complex I (24, 25). In our previous study, we also suggested that activity of complex I is most profoundly impaired by reduced mitochondrially encoded OXPHOS expression (19). This suggests that the downregulation of mitochondrially encoded ND1 and ND5 found in islets from patients with T2D may play an important role in the contribution to impaired insulin secretion. On the other hand, OXPHOS was the most predominant upregulated gene set in islets of a prediabetic mouse model in response to diabetogenic high fat (26). It is known that insulin resistance in peripheral tissues in the prediabetic state, force the pancreatic β-cells to compensate by secreting more insulin. In the paper by Dreja et al. (26), the β-cells may be in a state where they compensate for insulin resistance by enhancing β-cell function through upregulating OXPHOS expression. In addition, we have previously shown that mice fed a high-fat diet, which are in a prediabetic state and do not develop frank diabetes, have increased mitochondrial volume and enhanced insulin secretion when stimulated with fuels that require mitochondrial metabolism (27). In contrast, the present study suggests that once diabetes has developed, islet OXPHOS function and insulin secretion are impaired.
Environmental risk factors for T2D, such as BMI and age, may contribute to the decreased expression observed in OXPHOS genes. By multivariate regression analysis correcting for age, sex, and BMI, we confirmed that the differences in expression of OXPHOS genes are mainly due to diabetic status (data not shown). Moreover, HbAlc levels correlated negatively with gene expression of NDUFA5, COX11, and ATP6V1H in the human islets. These data suggest that hyperglycemia may be involved in the downregulation of OXPHOS genes.
Recent studies propose that epigenetic factors including DNA methylation can regulate mRNA expression of genes involved in OXPHOS and contribute to impaired insulin secretion and action observed in patients with T2D (5, 6, 9). In this study, we were unable to detect any differences in the level of DNA methylation between T2D and non-diabetic donors for the analyzed CpG sites upstream of NDUFA5, COX11, and ATP6V1H. However, DNA methylation levels of a CpG unit upstream of COX11 correlated negatively with COX11 mRNA expression and positively with age. This result is in line with previous findings that ageing is associated with increased DNA methylation and reduced expression of OXPHOS genes in human skeletal muscle (5, 6). One limitation with our study is the small number of CpG sites analyzed since the assays only cover ∼400–500 bp of the respective gene. We can therefore not exclude the fact that some of the OXPHOS genes and CpG sites not analyzed will show differential DNA methylation in pancreatic islets from T2D donors. Future genome-wide methylation studies may further dissect the role of DNA methylation in pancreatic islets of patients with T2D.
In conclusion, islets from patients with T2D showed decreased expression of OXPHOS genes compared with non-diabetic individuals. Although decreased OXPHOS expression in pancreatic islets may contribute to T2D by impaired GSIS additional studies are needed dissecting this relationship further.
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/EJE-11-0282.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This investigation was supported by grants from the Swedish Research Council (319-2008-135 and 522-2006-6480), SFO EXODIAB (Dnr 2009-1039), Region Skåne, Malmö University Hospital, Knut and Alice Wallenberg Foundation, Lundberg foundation, Novo Nordisk, Söderberg, Påhlsson, and Linné grant (B31 5631/2006).
Acknowledgements
Human pancreatic islets were obtained from the Nordic Islet Transplantation Program by the courtesy of Prof. Olle Korsgren, Uppsala University, Uppsala, Sweden. We thank Mona Svärdh for skilled technical assistance and SCIBLU at Lund University for analyzing gene expression.
References
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. PNAS. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Dahlman I, Forsgren M, Sjogren A, Nordstrom EA, Kaaman M, Naslund E, Attersand A, Arner P. Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-alpha. Diabetes. 2006;55:1792–1799. doi: 10.2337/db05-1421. [DOI] [PubMed] [Google Scholar]
- Lu H, Yang Y, Allister EM, Wijesekara N, Wheeler MB. The identification of potential factors associated with the development of type 2 diabetes: a quantitative proteomics approach. Molecular and Cellular Proteomics. 2008;7:1434–1451. doi: 10.1074/mcp.M700478-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Poulsen P, Simonsson S, Ronn T, Holmkvist J, Almgren P, Hagert P, Nilsson E, Mabey AG, Nilsson P, Vaag A, Groop L. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. Journal of Clinical Investigation. 2007;117:3427–3435. doi: 10.1172/JCI30938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronn T, Poulsen P, Hansson O, Holmkvist J, Almgren P, Nilsson P, Tuomi T, Isomaa B, Groop L, Vaag A, Ling C. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia. 2008;51:1159–1168. doi: 10.1007/s00125-008-1018-8. [DOI] [PubMed] [Google Scholar]
- Ronn T, Poulsen P, Tuomi T, Isomaa B, Groop L, Vaag A, Ling C. Genetic variation in ATP5O is associated with skeletal muscle ATP50 mRNA expression and glucose uptake in young twins. PLoS ONE. 2009;4:e4793. doi: 10.1371/journal.pone.0004793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons C, Jacobsen S, Nilsson E, Ronn T, Jensen CB, Storgaard H, Poulsen P, Groop L, Ling C, Astrup A, Vaag A. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. Journal of Clinical Endocrinology and Metabolism. 2010;95:3048–3056. doi: 10.1210/jc.2009-2413. [DOI] [PubMed] [Google Scholar]
- Ling C, Del Guerra S, Lupi R, Ronn T, Granhall C, Luthman H, Masiello P, Marchetti P, Groop L, Del Prato S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren AH, Jokubka R, Tojjar D, Granhall C, Hansson O, Li DQ, Nagaraj V, Reinbothe TM, Tuncel J, Eliasson L, Groop L, Rorsman P, Salehi A, Lyssenko V, Luthman H, Renstrom E. Overexpression of alpha 2A-adrenergic receptors contributes to type 2 diabetes. Science. 2010;327:217–220. doi: 10.1126/science.1176827. [DOI] [PubMed] [Google Scholar]
- Yang BT, Dayeh TA, Kirkpatrick CL, Taneera J, Kumar R, Groop L, Wollheim CB, Nitert MD, Ling C. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA (1c) levels in human pancreatic islets. Diabetologia. 2011;54:360–367. doi: 10.1007/s00125-010-1967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Mulder H, Ling C. Mitochondrial dysfunction in pancreatic beta-cells in type 2 diabetes. Molecular and Cellular Endocrinology. 2009;297:34–40. doi: 10.1016/j.mce.2008.05.015. [DOI] [PubMed] [Google Scholar]
- Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- Anello M, Lupi R, Spampinato D, Piro S, Masini M, Boggi U, Del Prato S, Rabuazzo AM, Purrello F, Marchetti P. Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia. 2005;48:282–289. doi: 10.1007/s00125-004-1627-9. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. New England Journal of Medicine. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Poulsen P, Carlsson E, Ridderstrale M, Almgren P, Wojtaszewski J, Beck-Nielsen H, Groop L, Vaag A. Multiple environmental and genetic factors influence skeletal muscle PGC-1 alpha and PGC-1 beta gene expression in twins. Journal of Clinical Investigation. 2004;114:1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck T, Olsson AH, Nitert MD, Sharoyko VV, Ladenvall C, Kotova O, Reiling E, Ronn T, Parikh H, Taneera J, Eriksson JG, Metodiev MD, Larsson NG, Balhuizen A, Luthman H, Stancakova A, Kuusisto J, Laakso M, Poulsen P, Vaag A, Groop L, Lyssenko V, Mulder H, Ling C. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metabolism. 2011;13:80–91. doi: 10.1016/j.cmet.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Olsson AH, Ronn T, Ladenvall C, Parikh H, Isomaa B, Groop L, Ling C. Two common genetic variants near nuclear-encoded OXPHOS genes are associated with insulin secretion in vivo. European Journal of Endocrinology. 2011;164:765–771. doi: 10.1530/EJE-10-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A, Wollheim CB. Minireview: implication of mitochondria in insulin secretion and action. Endocrinology. 2006;147:2643–2649. doi: 10.1210/en.2006-0057. [DOI] [PubMed] [Google Scholar]
- Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- Clark A, Jones LC, de Koning E, Hansen BC, Matthews DR. Decreased insulin secretion in type 2 diabetes: a problem of cellular mass or function? Diabetes. 2001;50(Supplement 1):S169–S171. doi: 10.2337/diabetes.50.2007.S169. [DOI] [PubMed] [Google Scholar]
- Perales-Clemente E, Fernandez-Vizarra E, Acin-Perez R, Movilla N, Bayona-Bafaluy MP, Moreno-Loshuertos R, Perez-Martos A, Fernandez-Silva P, Enriquez JA. Five entry points of the mitochondrially encoded subunits in mammalian complex I assembly. Molecular and Cellular Biology. 2010;30:3038–3047. doi: 10.1128/MCB.00025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A. Mitochondrial genetic control of assembly and function of complex I in mammalian cells. Journal of Bioenergetics and Biomembranes. 2001;33:251–257. doi: 10.1023/A:1010791204961. [DOI] [PubMed] [Google Scholar]
- Dreja T, Jovanovic Z, Rasche A, Kluge R, Herwig R, Tung YC, Joost HG, Yeo GS, Al-Hasani H. Diet-induced gene expression of isolated pancreatic islets from a polygenic mouse model of the metabolic syndrome. Diabetologia. 2010;53:309–320. doi: 10.1007/s00125-009-1576-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fex M, Nitert MD, Wierup N, Sundler F, Ling C, Mulder H. Enhanced mitochondrial metabolism may account for the adaptation to insulin resistance in islets from C57BL/6J mice fed a high-fat diet. Diabetologia. 2007;50:74–83. doi: 10.1007/s00125-006-0464-4. [DOI] [PubMed] [Google Scholar]