Abstract
It is well known that drug disposition and response are greatly determined by the activities of drug metabolizing enzymes, which are polymorphic. Some of these polymorphisms are clinically relevant and presented an ethnic-dependent pattern of distribution. The characterization of the genetic distribution of different populations allows the selection of therapeutic options in accordance with the genetic background, with the objective to avoid adverse reactions and inefficacy of the treatment. In this work, we studied selected genetic polymorphisms in drug metabolizing enzymes in three different ethnic groups – Portugal, Mozambique and Colombia. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) genotyping methods were developed for drug metabolizing enzymes, namely, cholesterol 7α-hydroxylase (CYP7A1) (–203A>C, –346C>T, –496C>T, N233S, G347S), sterol 27-hydroxylase (CYP27A1) (R164W, A169V, D273N, V400A) and oxysterol 7α-hydroxylase (CYP7B1) (–116C>G, R324H, 1774C>T) to characterize the allelic distribution of these polymorphisms among three different ethnic/geographic origins. A total of 12 CYP7A1, CYP27A1 and CYP7B1 genetic variants were genotyped in a sample of 92 Portuguese, 151 Mozambican and 91 Colombian subjects. The variants N233S in CYP7A1 and 1774C>T in CYP7B1 were not detected in any population studied. The promoter polymorphisms in CYP7A1 (–203A>C, –346C>T, –496C>T) had high frequency in the three ethnic groups. G347S (CYP7A1), R164W, A169V and V400A (CYP27A1) were present in a low frequency but with a similar distribution in the three ethnic groups. Significant differences were observed for D273N (CYP27A1), –346C>T (CYP7A1), –116C>G and R324H (CYP7B1)Our results demonstrate a high variability of drug metabolizing enzymes between the different populations analyzed, indicating that at least some of these polymorphisms are ethnic specific.
Keywords: Cholesterol 7α-hydroxylase, drug metabolizing enzymes, oxysterol 7α-hydroxylase, sterol 27-hydroxylase
The main promise of pharmacogenetics is the individualized therapy, giving the patient a dug selection and dosage in accordance with his genetic profile. This kind of treatment should prevent adverse reactions or treatment failure and provide more effective and safe therapeutics for each person.
Although many polymorphic transporters and receptors have been associated with clinical consequences, drug metabolizing enzymes are the most likely reason for inter-individual variation in drug response.[1]
Cytochromes P450 (CYP) play a key role in drug response and also in the metabolism of endogenous compounds. Several polymorphisms have been described for these enzymes and their functional consequences may result in a moderate reduction, a lack or an increase of enzymatic activity.
Cardiovascular diseases are the most prevalent cause of death and morbidity in developed countries and excess of cholesterol is a known factor for the progression of cardiovascular events. Cytochromes P450 are involved in cholesterol homeostasis, playing key roles in cholesterol metabolism, either in its synthesis or elimination through biosynthesis of bile acids, the main products of cholesterol catabolism.[2] Besides the role of CYPs in the homeostasis of endogenous substrates, these enzymes may be of crucial importance in the metabolism of drugs used to treat cardiovascular diseases.[3] The cholesterol lowering drugs in use are mainly 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitors (statins), fibrates, ezetimibe and bile acid binding resins that act in reducing the levels of cholesterol by different pathways.[4]
Clinically relevant drugs are transported and metabolized by polymorphic proteins. Some of these single nucleotide polymorphisms (SNPs) are associated with altered function of the protein.[5,6]
Cholesterol 7α-hydroxylase (CYP7A1) is the rate-limiting enzyme in the classical pathway for the synthesis of bile acids and its activity is highly variable among healthy individuals.[7] CYP7A1 is highly regulated by bile acids via Farnesoid X Receptor (FXR).[8] It has narrow substrate specificity for cholesterol, which makes this gene an important candidate in disorders of cholesterol and bile acid metabolism. In fact, some polymorphisms of CYP7A1 have been associated with gallstone disease and familiar hypertriglyceridemia[9–11] and recently were associated with increased plasma low density lipoprotein (LDL)-cholesterol concentrations.[12]
Sterol 27-hydroxylase (CYP27A1) and oxysterol 7α-hydroxylase (CYP7B1) are involved in bile acid synthesis in the alternative pathway.[13]
CYP7B1 has high homology with CYP7A1 and it was hypothesized that the main function of this enzyme is the inactivation of oxysterols, protecting the liver from cholestasis.[7] Mutations in CYP7B1 are related with spastic paraplegia autosomal recessive type 5A (SPG5A), which is a neurodegenerative disorder characterized by a slow, gradual, progressive weakness and spasticity of the lower limbs.[14,15] Absence of CYP7B1 is the cause of congenital bile acid synthesis defect type 3 (CBAS3), which is characterized by severe cholestasis, cirrhosis and liver synthetic failure.[16]
CYP27A1 is a ubiquitous enzyme involved in the 27-hydroxylation of cholesterol and in the formation of potentially important regulatory oxysterols,[17] indicating a potential regulatory role for CYP27A1. Mutations in CYP27A1 were identified in patients with cerebrotendinous xanthomatosis, a rare sterol storage disorder characterized clinically by progressive neurologic dysfunction, premature atherosclerosis and cataracts.[18]
CYP7A1, CYP27A1 and CYP7B1 are repressed by bile acids via nuclear receptors.[19]
An evaluation of SNP frequencies among different populations with variable ethnic background will certainly be useful to characterize the changes in drug pharmacokinetics and predict differential susceptibility to possible adverse effects.
In the present study, we examined the allelic frequencies of 12 CYP7A1, CYP27A1 and CYP7B1 SNPs that may have an important role in drug metabolism, in populations from three different ethnic/geographic origins.
Materials and Methods
Subjects
The populations under study consisted of 92 Portuguese, 151 Mozambican and 91 Colombian subjects. The populations from Mozambique and Portugal have been described elsewhere.[20,21] Colombian subjects were from the Northwest region, mainly from Antioquia and Chocó Departments (62 men, 27 women). This study followed the recommendations of the Declaration of Helsinki, promulgated in 1964 (http://ohsr.od.nih.gov/helsinki.php3). Genomic DNA was extracted from whole peripheral blood using the Qiamp DNA Extraction Kit (Qiagen, Hilden, Germany) or Chelex (BioRad, Berkeley, USA) according to the manufacturer's protocols.
Pharmacogenetic analysis of polymorphic variants
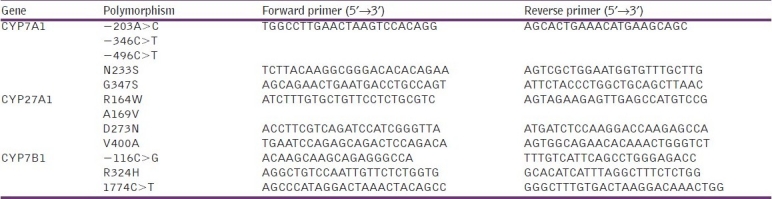
Each of the 12 SNPs analyzed in CYP7A1, CYP7B1 and CYP27A1 (NC_000008.10, NC_000008.10 and NC_000002.11, respectively) were identified by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assays. Table 1 lists the primers employed in performing PCR for the analyzed SNPs.
Table 1.
Primers used in polymerase chain reaction
Genomic DNA was added to PCR mixtures of 25 μl consisting of 1 × PCR buffer, 1–4 mM MgCl2 (Promega, Wisconsin, USA), 0.2 mM dNTPs (Promega, Wisconsin, USA), and 1.5 U of Go Taq polymerase (Promega, Wisconsin, USA). PCR amplifications were conducted (Tpersonal Themocycler, Biometra, Göttingen, Germany) using one denaturation step at 94°C for 2 min, followed by 30–35 cycles of 94°C for 30 s, 40–55°C for 30 s and 72°C for 1 min and a final PCR extension of 3 mins at 72°C.
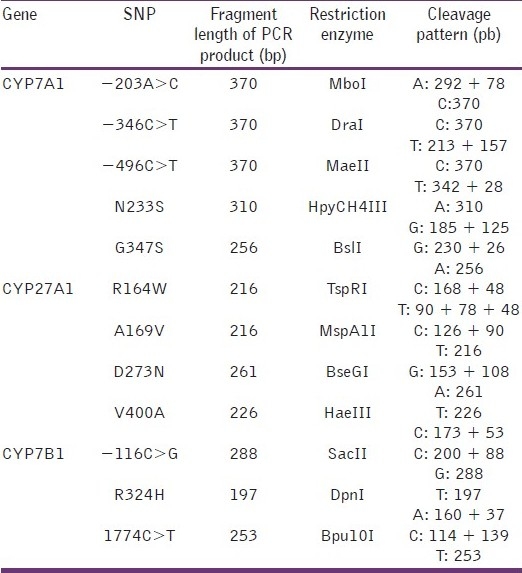
After amplification by PCR, genotype assignment was performed by digestion of amplification products with specific restriction endonucleases [Table 2], separation by electrophoresis on 8% or 10% (W/V) polyacrylamide gels and visualized with ethidium bromide staining under UV light (AlphaImager, AlphaInnotech, San Leandro, USA).
Table 2.
Restriction endonucleases used in PCR-RFLP
Statistical analysis
Fisher's exact test was used to assess Hardy–Weinberg equilibrium. Statistical analyses were performed using the Student's t-test and the analysis of variance (ANOVA) one-way test.
All analyses were performed using the Primer software.
Results
A total of 12 CYP7A1, CYP7B1 and CYP27A1 genetic variants were genotyped in a sample of 92 Portuguese, 151 Mozambican and 91 Colombian subjects [Figure 1]. The allelic frequencies of CYP7A1, CYP7B1 and CYP27 in different ethnic groups are shown in Table 3 and in Figures 2–4.
Figure 1.
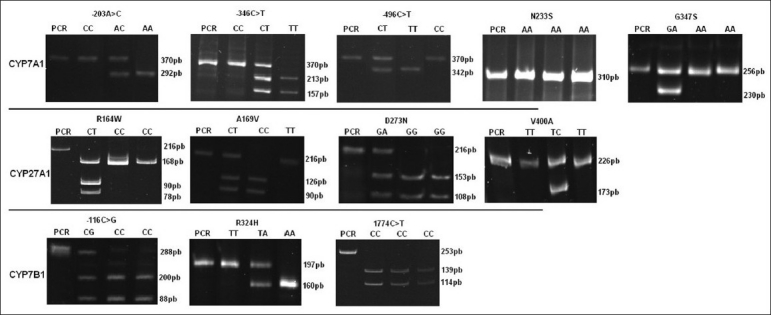
CYP7A1, CYP27A1 and CYP7B1 polymorphisms. Patterns of DNA fragments obtained for each genotype identified. PCR-RFLP analysis was performed using specific primers and enzymes described in Tables 1 and 2
Table 3.
CYP7A1, CYP7B1 and CYP27A1 allelic frequencies (%) in subjects of different ethnic groups
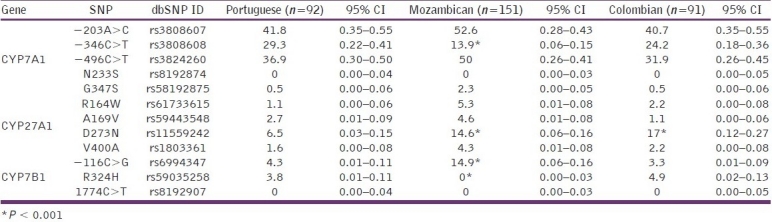
Figure 2.
CYP7A1 polymorphisms. Patterns of DNA fragments obtained for each genotype identified. PCR-RFLP analysis was performed using specific primers and enzymes described in Tables 1 and 2
Figure 4.
CYP7B1 polymorphisms. Patterns of DNA fragments obtained for each genotype identified. PCR-RFLP analysis was performed using specific primers and enzymes described in Tables 1 and 2
Figure 3.
CYP27A1 polymorphisms. Patterns of DNA fragments obtained for each genotype identified. PCR-RFLP analysis was performed using specific primers and enzymes described in Tables 1 and 2
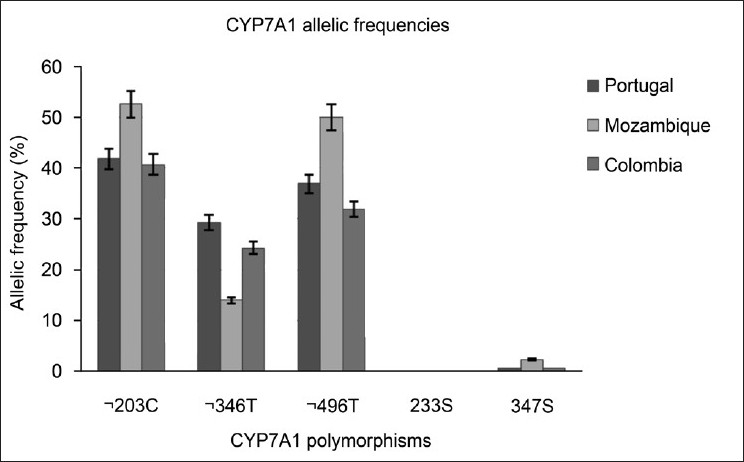
The variants N233S (CYP7A1) and 1774C>T (CYP7B1) were not detected between the individuals analyzed from Portugal, Mozambique and Colombia. In contrast, the CYP7A1 promoter variants had a high frequency between the three populations studied. While the frequencies for the –203C>T and –496C>T variants were similar between the three populations, the –346C>T variant was significantly lower for the population of Mozambique (P < 0.001) compared with Portuguese and Colombian individuals. The frequency of G347S was similar between individuals from different ethnic backgrounds, but with a low allele frequency (0.5% for Portuguese and Colombian and 2.3% for Mozambican individuals).
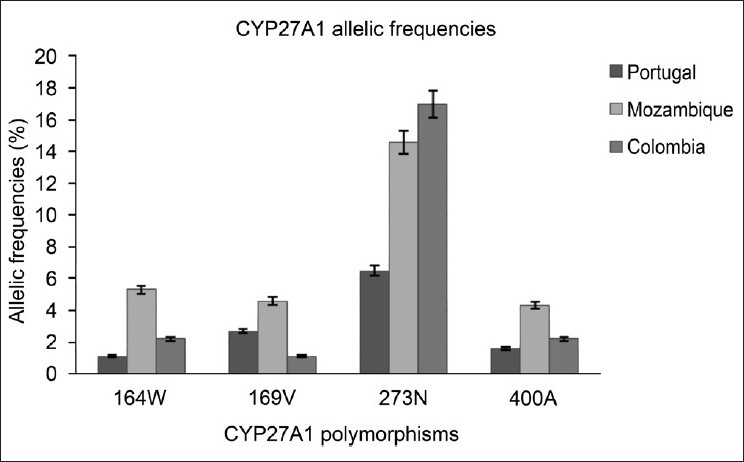
For CYP27A1, the variants R164W, A169V and V400A had a low allelic frequency for the populations studied. The variant D273N was significantly more prevalent in Mozambican (14.6%; P < 0.001) and Colombian (17%; P < 0.001) individuals than in the Portuguese (1.6%) population.
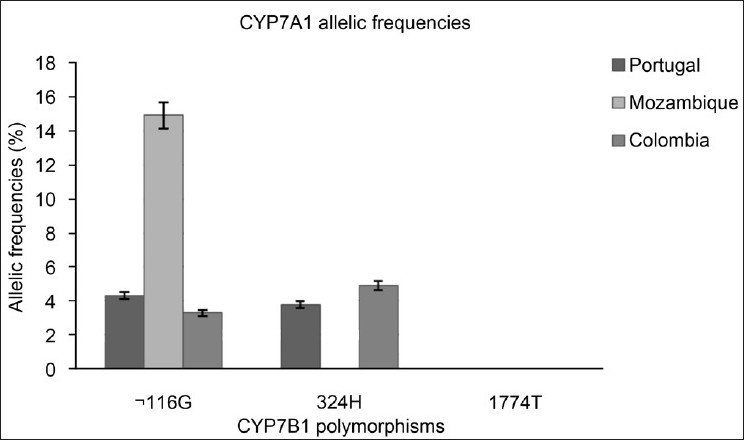
For CYP7B1, the variant –116C>G had a low frequency in Portuguese (4.3%) and Colombian (3.3%) individuals, but had a higher frequency among Mozambicans (14.9%; P < 0.001). The variant R324H had low frequency between Portuguese (3.8%) and Colombian (4.9%) populations, but was not found in Mozambican (0%; P < 0.001) individuals.
None of the genetic variants showed any significant deviation from Hardy–Weinberg equilibrium.
Discussion
Drug metabolizing enzymes play a key role in cholesterol metabolism and may be important in the metabolism of some lipid-lowering drugs such as some statins. Although influx and efflux transporters are important for the disposition and efficacy of many drugs, CYP enzymes are important contributors for inter-individual variation in drug response.
As the response to a large range of drugs is highly variable, pharmacogenetic diversity may in part contribute to this variability, being important in the context of therapeutic efficacy and safety. An evaluation of SNP frequencies among different populations with variable ethnic background will certainly be useful to characterize the changes in drug pharmacokinetics and predict differential susceptibility to possible adverse effects.
Except for the polymorphism of –203A>C (CYP7A1) and some variations of CYP27A1 found in some patients with cerebrotendinous xanthomatosis, there is not much information about CYP7A1, CYP27A1 and CYP7B1 polymorphisms. However, these enzymes are extremely important in bile acid synthesis and their accumulation in the organism has harmful consequences for individual health. So, it is important not only to know the genetic background of populations from different origins, but also to determine the functional consequences of these genetic variants for the enzyme activity.
We analyzed the frequencies of 12 SNPs in three CYP enzymes in individuals from Portugal, Mozambique and Colombia. From these SNPs, CYP7A1 N233S and CYP7B1 1774C>T were not detected in any of the three populations in the study.
The variant N233S of the CYP7A1 is localized in exon 3 and CYP7B1 1774C>T is localized in exon 6, 3’-downstream region. The absence of these SNPs in the populations analyzed may indicate that these mutations are localized in conservative regions of the gene and their substitution may lead to non-functional proteins.
The allelic frequency for CYP7A1 –203A>C among Caucasians ranges from 36.7 to 48%.[22–27] The estimate of 41.8% found in Portugal is in the range for a European/Caucasian population. The allelic frequency obtained for Mozambicans (52.6%) is lower than that reported by Nakamoto et al. for an African (58.3%) or an Afro-American (58%) population, and the frequency for Colombians (40.7%) is significantly higher than that for a Mexican-American (24%) population.[27] This discrepancy may be caused by differences in sample sizes between these two studies, but the most probable cause is the origin of the samples. Africa has different ethnic groups in accordance with the geographic localization. The African subjects analyzed by Nakamoto et al. were from Yoruba of Ibadan, Nigeria, localized in Guinea Golf, West coast, while the individuals analyzed in this study were from Mozambique, localized in East coast of Africa, which can explain the allelic frequencies observed. The high frequency determined in Colombians is consistent with the European (Spanish) ancestry of a wide percentage of the population.
CYP7A1 –203A>C was associated with LDL-cholesterol[12] and in patients with hypercholesterolemia, –203A>C was associated with LDL-lowering response,[24] as was observed after treatment with statin therapy.[28] Therefore, this SNP contributes to the efficiency of statin treatment. Recently, this SNP was also positively associated with subclinical atherosclerosis in healthy postmenopausal women[29] and it was considered a genetic risk factor for gallbladder carcinoma, but with a modest contribution in the susceptibility to gallstone disease.[30]
The allelic frequencies obtained for CYP7A1 –496C>T were high but similar between the three populations studied. The results obtained for Portugal (36.9%) and for Mozambique (50%) are in agreement with the previous reports for a Caucasian (36.7%) and African (55%) population, respectively.[27]
The allelic frequency of CYP7A1 –346C>T was high for the three populations studied, but significantly lower for the population from Mozambique (13.9%; P < 0.001).
The allelic frequency obtained for CYP7B1 –116C>G (4.3%) in Portuguese is in agreement with the frequencies obtained by Jakobsson et al. for a Swedish population (4.04%).[31] In the population of Mozambique, the allelic frequency (14.9%) was significantly higher than that observed for the populations of Portugal and Colombia (3.3%). However, reports on the populations from these geographic points are not found in the literature for comparison.
The genetic variants –203C>A, –346C>T and –496C>T are localized in the promoter region of the CYP7A1 gene, and therefore, it is much likely that these polymorphisms have functional significance. In fact, studies on the promoter region of CYP7A1 revealed several cell-specific enhancer elements[32] and in another study, the deletion of this region in HepG2 cells resulted in a 2.5-fold increase of basal promoter activity and this segment bound a negative regulatory protein of HepG2 cells.[33] These results suggest that polymorphisms in CYP7A1 promoter region may modulate transcriptional activity and consequently the rate of cholesterol catabolism and its flux through the body.
CYP7B1 –116C>G is localized in the promoter region of the gene and it was found to be associated with an altered promoter activity.[31] Besides, this polymorphism was considered ethnic-dependent since significant differences in allele frequencies were observed for populations with distinct geographic localization,[31] which is in agreement with our observations.
To the best of our knowledge, this is the first description of several polymorphisms analyzed in this study, namely CYP7A1 (–346C>T, N233S, G347S), CYP27A1 (R164W, A169V, D273N, V400A), CYP7B1 (R324H, 1774C>T).
CYP7A1 G347S and CYP27A1 R164W, A169V and V400A were rare, with similar allelic distribution among populations.
The polymorphism G347S is localized in exon 4 of CYP7A1. Gly (G) is an important amino acid for proper folding of the protein and is known to be a helix breaker and folding blocker of proteins. Hence, the substitution of a glycine in the protein can lead to conformational changes of CYP7A1.
The allelic frequency of CYP27 D273N for the Portuguese population (6.5%) was significantly lower than that observed for Mozambique (14.6%) and Colombia (17%) populations and it represents the only polymorphism in this study, with a distribution significantly different from the Portuguese population.
The variants R164W and A169V are localized in exon 3, while the variants D273N and V400A are localized in exon 4 of CYP27A1. The variants R164W and A169V appear to be located in C-helix α, which is involved in conformational structure of the protein.[34] Arg residues (R) play important roles in proper folding of proteins as they can form salt bridges with Asp (D) and Glu (E) residues to stabilize the tertiary structure of proteins and, as these polymorphisms are near to conservative amino acids, with important functions in folding and stabilization of the conformational structure of the protein, these genetic variations may be important in predicting the correct folding of CYP27A1. In fact, more than 70% of the mutations reported in cerebrotendinous xanthomatosis are Arg, Gly or Pro polymorphisms, suggesting that these patients have conformational CYP27A1 changes.
D273N appears to be located on the third substrate recognition site and V400A in the fifth substrate recognition site sheet β1–4, which is involved in heme binding.[34] These sites are very important for the functionality of CYP27A1, and although the polymorphisms do not lead to substitutions of very different amino acids, the low frequency found for these SNPs suggests that they are localized in semi-conservative regions.
Alterations in protein amino acids can lead to complete loss of its folding and, consequently, its structure, which can have drastic effects such as the elimination of enzyme active sites, creation of new binding sites or general dysfunction.
The allelic frequency for CYP7B1 R324H was similar between Portuguese (3.8%) and Colombian (3.3%) populations, but significantly different from Mozambican (P < 0.001) population, where the polymorphism was not found.
The genetic variant R324H is localized in exon 4 of CYP7B1. The low allelic frequency observed for this SNP may be explained by the fact that Arg (R) is involved in proper folding of proteins and can form salt bridges with Asp and Glu residues to stabilize the tertiary structure of the protein.
While polymorphisms in the promoter region can modify the degree of gene expression, polymorphisms in the coding region may have a direct effect on enzymatic activity, altering drug pharmacokinetics.
Considering Africans, Asians and Europeans as major population groups, it is becoming more documented that there is a clear genetic variation among these populations, with the main reason attributed to the early evolutionary history of human origins in Africa and subsequent expansion throughout the world.[35] The location of African population analyzed in this study, Mozambique, was a Portuguese colony in the late 15th century, and some admixture between Mozambicans and Portuguese was expected. However, studies performed in the Mozambican population only detected, in the male gene pool, a small fraction of lineages from European origin.[36] Therefore, although the Iberian Peninsula was under Muslim influence for centuries, DNA analysis shows that that the Portuguese population is closely related to other populations of Western Europe.[35,37] According to Dupanloup et al., an earlier European-wide study had pointed to a small North African and Arab element in modern day Iberian ancestry when compared to the pre-Islamic ancestral basis, and the Gibraltar Strait seems to have functioned much more as a genetic barrier than a bridge.[35] The genetic background of the Colombian population is considered a mixture of Spanish and native individuals, with prevalence of European ancestry of today's population.[38] Our results suggest genetic similarities of all polymorphisms studied (except for CYP27A1 D273N) between Portuguese and Colombian populations, but major differences in SNP frequencies between Portuguese/Colombian and Mozambican populations. These results demonstrate that analyses of populations from different ethnic backgrounds are essential for obtaining adequate results in genotype/phenotype correlation studies and to predict differential susceptibility to possible adverse effects.
Pharmacogenetics constitutes the most important part of future molecular medicine. For this, molecular diagnostics along with molecular specified drug therapy will allow a safer and more efficient treatment.
We consider CYP7A1, CYP27A1 and CYP7B1 as good candidates for pharmacogenetic analysis of cholesterol-lowering therapy since these CYP enzymes play key roles in cholesterol homeostasis, and genetic variations in these genes most likely have a great impact on total body lipid metabolism. The characterization of variants in these genes among individuals from distinct geographic backgrounds will allow the development of new, more effective and safer drugs for each person. An evaluation of SNP frequencies among different populations with variable ethnic background will certainly be useful to characterize the changes in drug pharmacokinetics and predict differential susceptibility to possible adverse effects.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Roots I, Laschinski G, Arjomand-Nahad F, Kirchheiner J, Schwartz D, Brockmöller J, et al. Genotype and phenotype relationship in drug metabolism. Ernst Schering Res Found Workshop. 2009;59:81–100. doi: 10.1007/978-3-540-49529-1_6. [DOI] [PubMed] [Google Scholar]
- 2.Sarkis A, Roman RJ. Role of cytochrome P450 metabolites of arachidonic acid in hypertension. Curr Drug Metabol. 2004;5:245–56. doi: 10.2174/1389200043335603. [DOI] [PubMed] [Google Scholar]
- 3.Siest G, Ferrari L, Accaoui MJ, Batt AM, Visvikis S. Pharmacogenomics of drugs affecting the cardiovascular system. Clin Chem Lab Med. 2003;41:590–9. doi: 10.1515/CCLM.2003.089. [DOI] [PubMed] [Google Scholar]
- 4.Schmitz G, Langerman T. Pharmacogenetics of cholesterol lowering therapy. Vasc Pharmacol. 2006;44:75–89. doi: 10.1016/j.vph.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz G, Langerman T. Transcriptional regulatory networks in lipid metabolism control ABCA1 expression. Biochimica Biophysica Acta. 2005;1735:1–9. doi: 10.1016/j.bbalip.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz G, Langerman T, Heimer S. Role of ABCG1 and other ABCG family in lipid metabolism. J Lipid Res. 2001;42:1513–20. [PubMed] [Google Scholar]
- 7.Chiang JY. Regulation of bile acid synthesis: Pathways, nuclear receptors, and mechanisms. J Hepatol. 2004;40:539–51. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–15. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 9.Cohen JC, Cali JJ, Jelinek DF, Mehrabian M, Sparkes RS, Lusis AJ, et al. Cloning of the human cholesterol 7-alpha-hydroxylase gene (CYP7) and localization to chromosome 8q11-q12. Genomics. 1992;14:153–61. doi: 10.1016/s0888-7543(05)80298-8. [DOI] [PubMed] [Google Scholar]
- 10.Paumgartner G, Sauerbruch T. Gallstones: Pathogenesis. Lancet. 1991;338:1117–21. doi: 10.1016/0140-6736(91)91972-w. [DOI] [PubMed] [Google Scholar]
- 11.Angelin B, Hershon KS, Brunzell JD. Bile acid metabolism in hereditary forms of hypertriglyceridemia: Evidence for an increased synthesis rate in monogenic familial hypertriglyceridemia. Proc Nat Acad Sci. 1987;84:5434–8. doi: 10.1073/pnas.84.15.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Freeman DJ, Grundy SM, Levine DM, Guerra R, Cohen JC. Linkage between cholesterol 7-alpha-hydroxylase and high plasma low-density lipoprotein cholesterol concentrations. J Clin Invest. 1998;101:1283–91. doi: 10.1172/JCI1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tontonoz P, Mangeslsdorf J. Liver X Signaling pathways in cardiovascular disease. Mol Endocrinology. 2003;17:985–93. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson PA, Crosby AH, Turner C, Patel H, Wood NW, Schapira AH, et al. A clinical and genetic study of SPG5A linked to autosomal recessive hereditary spastic paraplegia. Neurology. 2003;61:235–8. doi: 10.1212/01.wnl.0000069920.42968.8d. [DOI] [PubMed] [Google Scholar]
- 15.Tsaousidou MK, Ouahchi K, Warner TT, Yang Y, Simpson MA, Laing NG, et al. Sequence alterations within CYP7B1 implicate defective cholesterol homeostasis in motor-neuron degeneration. Am J Hum Genet. 2008;82:510–5. doi: 10.1016/j.ajhg.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Setchell KD, Schwarz M, O’Connell NC, Lund EG, Davis DL, Lathe R, et al. Identification of a new inborn error in bile acid synthesis: Mutation of the oxysterol 7-alpha-hydroxylase gene causes severe neonatal liver disease. J Clin Invest. 1998;102:1690–703. doi: 10.1172/JCI2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Chen W, Chiang JY. PXR induces CYP27A1 and regulates cholesterol metabolism in the intestine. J Lip Res. 2007;48:372–84. doi: 10.1194/jlr.M600282-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Guyant-Marechal L, Verrips A, Girard C, Wevers RA, Zijlstra F, Sistermans E, et al. Unusual cerebrotendinous xanthomatosis with fronto-temporal dementia phenotype. Am J Med Genet. 2005;139A:114–7. doi: 10.1002/ajmg.a.30797. [DOI] [PubMed] [Google Scholar]
- 19.Hylemon P, Zhou H, Pandak W, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lip Res. 2009;36:263–7. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavaco I, Reis R, Gil P, Ribeiro V. CYP3A4*1B and NAT2*14 alleles in a native African population. Clin Chem Lab Med. 2003;41:606–9. doi: 10.1515/CCLM.2003.091. [DOI] [PubMed] [Google Scholar]
- 21.Cavaco I, Gil J, Gil-Berglund E, Ribeiro V. CYP3A4 and MDR1 alleles in a Portuguese population. Clin Chem Lab Med. 2003;41:1345–50. doi: 10.1515/CCLM.2003.206. [DOI] [PubMed] [Google Scholar]
- 22.Hofman M, Weggemans R, Zock P, Schouten E, Katan M, Princen H. CYP7A1 A-278C polymorphism affects the response of plasma lipids after dietary cholesterol or cafestol interventions in humans. J Nut. 2004;134:2200–4. doi: 10.1093/jn/134.9.2200. [DOI] [PubMed] [Google Scholar]
- 23.Hofman M, Princen H, Zwiderman A, Jukema J. Genetic variation in the rate-limiting enzyme in cholesterol catabolism (cholesterol 7α-hydroxylase) influences the progression of atherosclerosis and risk of new clinical events. Clin Sci. 2005;108:530–45. doi: 10.1042/CS20040339. [DOI] [PubMed] [Google Scholar]
- 24.Kajinami K, Brousseau ME, Ordovas J, Schaefer E. Interactions between common genetic polymorphisms in ABCG5/G8 and CYP7A1 on LDL cholesterol-lowering response to atorvastatin. Atherosclerosis. 2004;175:287–93. doi: 10.1016/j.atherosclerosis.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Kajinami K, Brousseau ME, Ordovas J, Schaefer E. A promoter polymorphism in cholesterol 7α-hydroxylase interacts with apolipoprotein E genotype in the LDL-lowering response to atorvastatin. Atherosclerosis. 2005;180:407–15. doi: 10.1016/j.atherosclerosis.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Juzyszyn Z, Kurzawski M, Lener A, Modrzejewski A, Pawlik A, Drozdzik M. Cholesterol 7alpha-hydroxylase (CYP7A1)c.-278A>C promoter polymorphism in gallstone disease patients. Gene Test. 2008;12:97–100. doi: 10.1089/gte.2007.0067. [DOI] [PubMed] [Google Scholar]
- 27.Nakamoto K, Wang S, Jenison R, Guo G, Klaassen C, Wan Y, et al. Linkage disequilibrium blocks, haplotype structure, and htSNPs of human CYP7A1 gene. BMC Genetics. 2006;7:1–12. doi: 10.1186/1471-2156-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kajinami K, Brousseau ME, Nartsupha C, Ordovas JM, Schaefer EJ. ATP binding cassette transporter G5 and G8 genotypes and plasma lipoprotein levels before and after treatment with atorvastatin. J Lipid Res. 2004;45:653–6. doi: 10.1194/jlr.M300278-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Lambrinoudaki IV, Kaparos GI, Vlachou SA, Stamatelopoulos KS, Georgiopoulos GA, Sergentanis TN, et al. CYP A-204C polymorphism is associated with subclinical atherosclerosis in postmenopausal women. Menopause. 2008;15:1163–8. doi: 10.1097/gme.0b013e31817727c3. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava A, Pandey SN, Choudhuri G, Mittal B. Role of genetic variant A-204C of cholesterol 7alpha-hydroxylase (CYP7A1) in susceptibility to gallbladder cancer. Mol Genet Metab. 2008;94:83–9. doi: 10.1016/j.ymgme.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Jakobsson J, Karypidis H, Johansson JE, Roh HK, Rane A, Ekström L. A functional C-G polymorphism in the CYP7B1 promoter region and its different distribution in Oriental and Caucasians. Pharmacogenomics J. 2004;4:245–50. doi: 10.1038/sj.tpj.6500236. [DOI] [PubMed] [Google Scholar]
- 32.Molowa DT, Chen WS, Cimis GM, Tan CP. Transcriptional regulation of the human cholesterol 7 alpha-hydroxylase gene. Biochemistry. 1992;31:2539–44. doi: 10.1021/bi00124a014. [DOI] [PubMed] [Google Scholar]
- 33.Cooper AD, Chen J, Botelho-Yetkinler MJ. Characterization of hepatic-specific regulatory elements in the promoter region of the human cholesterol 7-hydroxylase gene. J Biol Chem. 1997;272:3444–52. [PubMed] [Google Scholar]
- 34.Sawata N, Sakaki T, Kitanaka S, Kato S, Inouye K. Structure-function analysis of CYP27B1 and CYP27A1. Eur J Biochem. 2001;268:6607–15. doi: 10.1046/j.0014-2956.2001.02615.x. [DOI] [PubMed] [Google Scholar]
- 35.Dupanloup I, Bertorelle G, Chikhi L, Barbujani G. Estimating the Impact of Prehistoric Admixture on the Genome of Europeans. Mol Biol Evol. 2004;21:1361–72. doi: 10.1093/molbev/msh135. [DOI] [PubMed] [Google Scholar]
- 36.Pereira L, Gusmão L, Alves C, Amorim A, Prata M. Bantu and European Y-lineages in Sub-Saharan Africa. Ann Hum Genet. 2002;66:369–78. doi: 10.1017/S0003480002001306. [DOI] [PubMed] [Google Scholar]
- 37.Novembre J, Johnson T, Bryc K, Kutalik Z, Adam R, Adam I, et al. Genes mirror geography within Europe. Nature. 2008;456:98. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bedoya G, Montoya P, Garcia J, Soto I, Bourgeois S, Carvajal L, et al. Admixture dynamics in Hispanics: A shift in the nuclear genetic ancestry of a South American population isolate. Proc Natl Acad Sci USA. 2006;103:7234–9. doi: 10.1073/pnas.0508716103. [DOI] [PMC free article] [PubMed] [Google Scholar]


