Abstract
To understand how extant viruses interact with their hosts, we need a historical framework of their evolutionary association. Akin to retrovirus or hepadnavirus viral fossils present in eukaryotic genomes, bracoviruses are integrated in braconid wasp genomes and are transmitted by Mendelian inheritance. However, unlike viral genomic fossils, they have retained functional machineries homologous to those of large dsDNA viruses pathogenic to arthropods. Using a phylogenomic approach, we resolved the relationships between bracoviruses and their closest free relatives: baculoviruses and nudiviruses. The phylogeny showed that bracoviruses are nested within the nudivirus clade. Bracoviruses establish a bridge between the virus and animal worlds. Their inclusion in a virus phylogeny allowed us to relate free viruses to fossils. The ages of the wasps were used to calibrate the virus phylogeny. Bayesian analyses revealed that insect dsDNA viruses first evolved at ∼310 Mya in the Paleozoic Era during the Carboniferous Period with the first insects. Furthermore the virus diversification time frame during the Mesozoic Era appears linked to the diversification of insect orders; baculoviruses that infect larvae evolved at the same period as holometabolous insects. These results imply ancient coevolution by resource tracking between several insect dsDNA virus families and their hosts, dating back to 310 Mya.
Keywords: Baculoviridae, Polydnaviridae, virus age estimate, time to most recent common ancestor, paleovirology
All organisms experience a wide range of associations with both closely and distantly related species. With time, organisms coevolve, leading to various degrees of reciprocal interactions ranging from local coadaptation to cospeciation (1). Parasites and their hosts, being involved in durable and intimate interactions best characterized as a continuum between antagonism and mutualism (2), are well suited as subjects for studying coevolution. Replicating exclusively in host cells, viruses in particular share obligate interactions with their hosts, implicating long-term coevolution (3). Establishing a historical framework of virus/host evolutionary associations is fundamental to understanding how extant viruses interact with their hosts. Few attempts, focusing on single mammalian or plant virus families and none spanning several virus families, have been made to set the macroevolutionary history of virus/host relationships (4–7). To dig deeper, we need to estimate the divergence times of virus families. However, viral geologic fossils required in molecular dating are uncommon and impossible to assign to extant taxa even at the family level (8). Current molecular dating approaches of viruses use viral fossils, as would be present in eukaryotic genomes as calibration anchors (9, 10). However, because of their nonfunctional nature, these fossils degrade with time and genomic traces of ancient viral integrations are eventually lost, making it so far impossible to reconstruct the macroevolutionary history of viruses beyond 100 Mya (5).
Although insects are taxonomically the most diverse group of potential eukaryotic host species (11), as yet few reports have focused on insect virus macroevolution (12). Here we explore the paleontology of several insect virus families in relation to their insect hosts, based on the uniqueness of obligatory symbiotic viruses of braconid parasitoid wasps. We focus on large circular dsDNA viruses, including the nudiviruses, the genus Bracovirus, and the families Baculoviridae and Hytrosaviridae, all of which are exclusively pathogenic to arthropods, harbor rod-shaped enveloped nucleocapsids, and replicate in the nucleus (13–16). Previous phylogenetic studies clearly show that the nudiviruses and baculoviruses are sister groups (17, 18). The Hytrosaviridae are outside, although how distantly related remains uncertain. Whole-proteome clustering analysis placed them at the base of the DNA virus tree (18). However, based on gene content, they are thought to form a monophyletic group with the Baculoviridae, nudiviruses, and Nimaviridae (19). Within the Polydnaviridae, bracoviruses are associated with braconid wasps of the microgastroid complex (20). Parasitoid wasps have domesticated these viruses for transferring virulence genes (21–23), interfering with the immune response and development of their lepidopteran hosts (24, 25). The long-debated genuine viral nature of bracoviruses has recently been demonstrated with the discovery in wasp genomes of genes involved in bracovirus particle production (26–28). These genes, related to nudivirus and baculovirus core genes (27), establish a bridge between the phylogeny of free dsDNA viruses and geologic animal fossil data, thus allowing the exploration of deep evolutionary timescales.
Here, we performed phylogenomic analyses based on 19 bracovirus genes. Furthermore, we estimated the divergence times of the insect dsDNA virus families based on the age estimations of parasitoid wasp lineages (29). This work provides unique insights into the coevolutionary interactions between viruses and arthropods dating since the Paleozoic Era.
Results
Insect dsDNA Virus Phylogeny.
Analyses were conducted on 19 bracovirus genes for which homologs have been found in free insect dsDNA viruses (Table S1). Phylogenetic congruence analyses on unrooted trees showed that there was no conflicting signal between the different genes (SI Text, Fig. S1, and Table S2). The phylogenomic analyses are therefore based on a concatenated alignment of 19 genes (7,238 aa), representing all of the data available for three integrated symbiotic bracoviruses, aligned to homologs of 10 free pathogenic viruses from the nudiviruses, the Baculoviridae, and the Hytrosaviridae as outgroups.
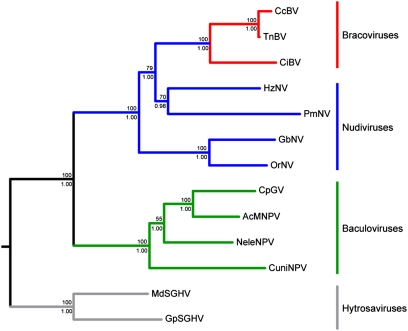
Bayesian inference and maximum likelihood phylogenetic analyses gave identical highly supported trees, differing only in branch lengths (Fig. 1). Interrelationships within and between the baculovirus and nudivirus clades are in accordance with, and better supported than, phylogenies based on diverse datasets (12, 17–19). Consistent with their chromosomal inheritance, the relationships within bracoviruses mirror the phylogeny of the wasps (20, 29).
Fig. 1.
Insect dsDNA virus phylogeny obtained from maximum likelihood and Bayesian inference analyses of the combined dataset. Support for nodes above branches indicates maximum likelihood nonparametric bootstraps (1,000 replicates) and below branches indicates Bayesian inference posterior probabilities. Red, blue, and green branches indicate bracovirus, nudivirus, and baculovirus lineages, respectively. Gray branches indicate hytrosavirus (salivary gland hypertrophy virus) lineage used as outgroup.
The most notable result from this tree is the paraphyly of the nudivirus clade, which includes the bracoviruses. Thus, this phylogeny demonstrates that the bracovirus lineage derives from within the nudiviruses and shares a common ancestor with the Heliothis zea nudivirus (HzNV)–Penaeus monodon nudivirus (PmNV) clade.
Molecular Dating.
Molecular dating requires both a phylogenetic species tree and calibration anchors. The dual identity of bracoviruses as integrated wasp symbionts and viruses allowed us to relate geologic fossils to the insect dsDNA virus phylogeny. Bracoviruses are solely inherited vertically in the chromosomes of their associated braconid wasp. Each bracovirus and its wasps together act as a single genomic entity sharing the same evolutionary history. So their speciation events are contemporaneous (20). Thus, to calibrate the dsDNA virus tree, we used the age, phylogenetically estimated based on fossil amber calibration, of the wasps belonging to the bracovirus-bearing microgastroid complex (29). The time to the most recent common ancestor (TMRCA) of Chelonus inanitus bracovirus (CiBV) and Cotesia congregata bracovirus (CcBV) was set to 103.38 ± 4.41 Mya, corresponding to C. inanitus (Cheloninae) and C. congregata (Microgastrinae) lineage divergence times. Similarly, the TMRCA of CcBV and Toxoneuron nigriceps bracovirus (TnBV) was set at ∼87 ± 5 Mya, the divergence times between the Microgastrinae and Cardiochilinae (Fig. S2). Because bracoviruses are now included in the insect dsDNA virus phylogeny, a Bayesian inference approach, using an autocorrelated lognormal relaxed clock and calibration priors based on the above age estimates, was performed to estimate the divergence times of the nudivirus and baculovirus dsDNA virus lineages.
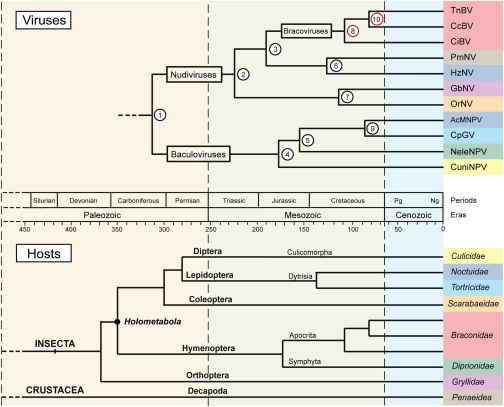
Age estimates and 95% highest posterior density (HPD) intervals were obtained for each node (virus tree in Fig. 2; Table 1). The TMRCA of nudiviruses and baculoviruses (node 1) is estimated at ∼310 Mya (Carboniferous, late Paleozoic Era). The TMRCA of the nudivirus (node 2) and the baculovirus (node 4) lineages are estimated at ∼222 Mya (Triassic, early Mesozoic Era) and ∼178 Mya (Jurassic, middle Mesozoic Era), respectively. Furthermore, the results outline that the diversification of the nudivirus and baculovirus lineages stretches back to the Mesozoic Era and occurred primarily during the Jurassic and the Cretaceous Periods.
Fig. 2.
Evolutionary timeline of insect dsDNA virus and their associated host species. Nodes 8 and 10 were used to calibrate the insect dsDNA virus phylogeny, and the host tree is drawn from the literature (SI Text). Age estimates and their 95% HPD intervals for the corresponding nodes are shown in Table 1. Colors associate each virus to its respective host family.
Table 1.
Age estimates and 95% HPD intervals of the nodes indicated on Fig. 2
| Node | Time, Mya | 95% HPD, Mya |
| 1 | 312 | 127–569 |
| 2 | 222 | 107–392 |
| 3 | 190 | 101–340 |
| 4 | 178 | 40–368 |
| 5 | 153 | 31–321 |
| 6 | 128 | 11–265 |
| 7 | 116 | 21–235 |
| 8 | 104 | 95–112 |
| 9 | 86 | 10–189 |
| 10 | 83 | 74–93 |
To relate the evolution of the insect dsDNA viruses to their hosts, the host species tree (Fig. 2) was drawn from the literature (SI Text). This tree shows that the insect and crustacean lineages associated with the dsDNA viruses of this study appeared in the middle Paleozoic Era and the diversification of insect orders occurred during the second half of the Paleozoic Era. Diversification within insect orders occurred mainly during the Mesozoic Era after the Permian crisis. Within the insect lineage, the appearance of holometabolous insects at ∼350 Mya is also highlighted on the host tree.
Discussion
Previous studies on braconid wasps have established the monophyly of the lineage bearing bracoviruses, the microgastroid complex (20, 29), suggesting the integration of an ancestral virus in the genome of the most recent common ancestor of these wasps. Recently, homologous viral machineries, related to those of nudiviruses, were discovered to produce distantly related bracoviruses (27). The first objective of this study was to resolve the phylogenetic relationships between the bracoviruses and their free virus relatives, the nudiviruses and baculoviruses. Our phylogenomic analyses clearly show the paraphyly of the nudiviruses with the inclusion of the bracoviruses (Fig. 1), thus closing the demonstration that bracoviruses derive from nudiviruses. The phylogenetic tree is also in accordance with recent studies indicating that the baculoviruses and nudiviruses are closely related but form distinct lineages and as such should be classified in distinct virus families (16, 17, 19, 30).
Interestingly, bracoviruses share a common ancestor with the HzNV-PmNV clade (Fig. 1), for which the highest genome content similarities (six unique genes) have been found (27). HzNV is able to develop particular infection strategies such as having a tropism for gonadal cells (31) and the establishment of latent infections (32). Both of these mechanisms could have been instrumental for the evolution of bracoviruses. The nudiviral ancestor of the bracoviruses could have used this particular infection strategy to establish a persistent latent infection in hymenopteran gonad cells, leading to its integration in the wasp chromosome.
The parasitoid wasps obligatorily depend on the symbiotic bracoviruses for their parasitic success. These two entities share the same genetic inheritance and undergo synchronized speciation, which led us to assume that the TMRCAs of both wasp and bracovirus lineages are contemporaneous. The additional peculiarity of bracoviruses is their phylogenetic relationships with other free dsDNA viruses. This link between the animal and the virus worlds allowed the estimation of baculovirus and nudivirus divergence times for inferring their macroevolutionary history.
Our dataset is suitable for molecular dating approaches on a large timescale because it fills two important criteria relative to low evolutionary rates. First, unlike RNA viruses, large dsDNA virus genomes have relatively slow evolutionary rates (33–35), approaching those of bacteria and lower unicellular eukaryotes (36). They are therefore appropriate subjects for molecular dating, as shown in the case of hepadnaviruses, for which long-term evolution has been inferred from integrated genomic fossils (9). Similarly, we used the bracovirus sequences integrated in the wasp genomes as calibration anchors. The second criterion relates to the dataset itself. Unlike for the hepadnavirus example, here the bracovirus sequences are still functionally active. The gene set is involved in the viral machinery, produces the viral particles, and is conserved in baculoviruses, nudiviruses, and bracoviruses. The functional homology even remains observable at the level of viral particle morphology because bracovirus and nudivirus nucleocapsids are very similar (37). Overall, this observation points to relatively slow evolutionary rates on a large timescale, which we assume to be similar between the free viruses and the eukaryotic viral symbionts. Given these assumptions, we inferred the age estimates of the dsDNA virus phylogeny from the eukaryotic calibration points.
The results show that the baculoviruses and nudiviruses are rooted at 310 Mya, deep in the Paleozoic Era in the Carboniferous Period, suggesting that the ancestors of these viruses had already infected the first insects, which appeared during the Devonian and Carboniferous Periods. Currently, this is the oldest age estimate to have been determined for any group of viruses, going far beyond the 19 Ma estimated for avian hepadnaviruses (DNA viruses) (9) or the 100 Ma established for mammalian retroviruses (RNA viruses) (5).
In the late Carboniferous Period, when the nudivirus and baculovirus families diverged, insects were also diversifying into new orders (Fig. 2). The phylogenies of both the baculovirus and nudivirus lineages are clearly not the mirror images of that of their hosts. This lack of cophylogeny shows that the diversification of both virus families and host orders occurred independently. On the macroevolutionary timescale, this finding suggests that these viruses may have colonized new insect hosts and also crustaceans many times during their evolution. However, the time frame of nudivirus and baculovirus diversification, starting at 222 Mya and 178 Mya, respectively (Table 1), does not appear to be totally independent of that of their hosts. Both virus families diversified during the Mesozoic Era, after the major Permian extinction, which is contemporary with the major diversification of insect orders and families (38).
Remarkably, at the late Carboniferous Period, the baculovirus lineage appeared shortly after the appearance of holometabolous insects, which are insects with larval stages undergoing complete metamorphosis (Fig. 2). These insects form a monophyletic clade, including (among others) the orders Diptera, Lepidoptera, Coleoptera, and Hymenoptera (39). Extant baculoviruses have only been characterized in holometabolous insect hosts from the orders Diptera, Hymenoptera, and Lepidoptera. Baculovirus phylogenetic studies have shown the ancient coevolution between baculoviruses and insects of these three orders (12), although the lack of dating analyses did not allow inference of how long the relationship could have lasted. Our data suggest that the baculovirus ancestor was already specialized to infect the new susceptible host niche that represents the larval stage of holometabolous insects. Pathological observations strongly support this idea because baculovirus infections typically occur when insect larvae ingest viral particles (40, 41).
In contrast to baculoviruses, nudiviruses appear to be a more generalist group of viruses. Although the pathological data are sparse, they are known to affect adult as well as larval stages with more varied infection strategies. The lack of environmentally resistant occlusion bodies in nudiviruses has been proposed to lead to the evolution of close association with their hosts, often involving viral persistence or latency (42). Nudiviruses have a broad host spectrum (Fig. 2), encompassing many insect orders but also crustaceans like PmNV (17, 19). The derived phylogenetic position of PmNV may imply a large host shift between insects and crustaceans, which could date as far back as 125 Mya.
Relating the large-scale evolutionary history of insect dsDNA viruses to that of their hosts showed that the ancestors of the baculovirus and nudivirus lineages infected the already-evolved ancestors of their extant hosts, suggesting a coevolutionary scenario of colonization by resource tracking whereby the resources for these viruses are the insects (43). When new insect species evolved, providing in part an evolutionarily modified form of the ecological niche required for the virus replication cycle, virus individuals, adapted to the ancestral host, colonized the new host. This ecological speciation mechanism eventually produced new virus species. On the macroevolutionary timescale, with the extinction of many host and virus phylogenetic lineages, cophylogenetic divergence should not be expected. However, closely related virus species should infect closely related insects, as shown in baculoviruses, for which particular phylogenetic lineages are specialized to infect particular insect orders (12). When more data become available for nudiviruses, similar lineage specializations are likely to be revealed. This coevolutionary process does not exclude large host shifts nor that dsDNA viruses evolve by codivergence with their hosts on the microevolutionary timescale. Indeed, most dsDNA viruses have a strong specificity toward their hosts and therefore have only a small probability of infecting a new host species during any short interval (44). The insect dsDNA virus phylogeny bears witness to the ancestral and intimate coevolutionary relationships between viruses and their insect hosts and shows that the expansion of their host range has been driven by punctuated host-shift events through the ages. This result is in line with recent findings on the evolution of foamy retroviruses, which have coevolved with mammals for over 100 Ma (5), suggesting that common macroevolutionary scenarios affect viruses regardless of their replication modes or origin.
Our analysis is the largest timescale macroevolutionary scenario, supported with the oldest age estimates, reported for any extant virus families. Our findings broaden the general scope of paleovirology studies (45) and anchor the timeline of virus macroevolution to test hypotheses on the role of viruses in the origin of cellular life. Ultimately, it should be possible to extend the molecular dating to the whole phylogeny of large dsDNA virus families (18). Similar combined phylogenomic approaches could prove useful for dating free-living unicellular organisms related to macroorganism symbionts.
Materials and Methods
Sequence Data.
Amino acid sequences of CiBV, CcBV, and TnBV nudivirus-related genes were retrieved from Bézier et al. (27). BLASTp analyses (46) were used to identify homologs (E-value <0.01) from nudivirus and baculovirus genomes. When available, homologous sequences from hytrosaviruses (salivary gland hypertrophy virus) were used as outgroups.
Four species were used for the nudivirus family. Among these species, three nudivirus genomes have been fully sequenced [HzNV (47), Gryllus bimaculatus nudivirus (GbNV) (48), and Oryctes rhinoceros nudivirus (OrNV) (16)] and one has been partially sequenced (PmNV). Four species representative of each genus of the baculovirus family were chosen: Culex nigripalpus nucleopolyhedrovirus (CuniNPV) (49), Neodiprion lecontei nucleopolyhedrovirus (NeleNPV) (50), Autographa californica multiple nucleopolyhedrovirus (AcMNPV) (51), and Cydia pomonella granulovirus (CpGV) (52). Two species represent the hytrosavirus family: Musca domestica salivary gland hypertrophy virus (MdSGHV) (53) and Glossina pallidipes salivary gland hypertrophy virus (GpSGHV) (54).
The dataset comprises 19 nudivirus-related genes: 13 belong to the 20 core genes conserved by all nudiviruses and baculoviruses, 1 gene (odv-e66) is only found in some nudiviruses and exclusively in baculoviruses infecting Lepidoptera, and 5 genes are found only in nudiviruses (Table S1). Multiple amino acid alignments were obtained for each of the 19 genes with the program MAFFT (55), then they were trimmed with the program Gblocks (56) to only keep conserved domains before concatenation.
Phylogenomic Analyses.
Phylogenomic analyses of the concatenated multiple amino acid alignment were performed by using both maximum likelihood and Bayesian inference. The appropriate substitution model and model parameters for maximum likelihood analysis were selected with ProtTest (57) and the Akaike information criterion (Blosum62 with the +I, +G, and +F parameters). Maximum likelihood analysis was performed with RAxML (58), and support for node in ML tree was obtained from 1,000 bootstrap iterations.
Mixed-model Bayesian phylogenomic analysis was performed with MrBayes (59). ProtTest and Bayesian information criteria were used to select appropriate substitution models and model parameters for each gene (Table S3). MrBayes analyses were run across four Monte Carlo Markov chains (MCMC) for 1 million generations, sampling every 500 generations. The consensus tree was obtained after a burn-in of 500 generations, and the value of average SD of split frequencies was used as a proof of stationarity if this value was under 0.01.
Molecular Dating Analysis.
To test whether the combined dataset evolved in a clocklike fashion, we compared the lnL of an unconstrained tree versus a tree constrained to evolve clockwise with the AAML program from the PAML package (60). Divergence times were then estimated by using a Bayesian inference approach implemented in BEAST (61). The analysis was performed on the combined dataset, preliminarily converted into codon-based sequence, and without outgroup because of the uncertainty concerning the sister-group relationships between the hytrosaviruses and the ingroup (16, 18). We assumed an uncorrelated lognormal relaxed clock and free mean substitution rates. TMRCAs of braconid wasp lineages bearing bracoviruses were used as priors to calibrate the insect dsDNA phylogeny under a normal distribution and a Yule process prior. Six independent MCMCs were run for 100 million generations sampling every 1,000 generations, using a burn-in of 1,000. The results of the six MCMCs were combined with the log-combiner program of the BEAST package, and age estimates were analyzed (effective sample size >200) with the Tracer program (62).
Supplementary Material
Acknowledgments
We thank the reviewers for their critical comments. This work was supported by European Research Council Grant 205206 GENOVIR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in EMBL (www.ebi.ac.uk/) (accession nos. FN557481–FN557487).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105580108/-/DCSupplemental.
References
- 1.Brooks D, McLennan D. Phylogeny, Ecology, and Behavior: A Research Program in Comparative Biology. Chicago: Univ of Chicago Press; 1991. pp. 189–204. [Google Scholar]
- 2.Combes C. Parasitism: The Ecology and Evolution of Intimate Interaction. Chicago: Univ of Chicago Press; 2001. [Google Scholar]
- 3.Lovisolo O, Hull R, Rösler O. Coevolution of viruses with hosts and vectors and possible paleontology. Adv Virus Res. 2003;62:325–379. doi: 10.1016/s0065-3527(03)62006-3. [DOI] [PubMed] [Google Scholar]
- 4.Pagán I, Holmes EC. Long-term evolution of the Luteoviridae: Time scale and mode of virus speciation. J Virol. 2010;84:6177–6187. doi: 10.1128/JVI.02160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katzourakis A, Gifford RJ, Tristem M, Gilbert MTP, Pybus OG. Macroevolution of complex retroviruses. Science. 2009;325:1512. doi: 10.1126/science.1174149. [DOI] [PubMed] [Google Scholar]
- 6.Fargette D, et al. Diversification of rice yellow mottle virus and related viruses spans the history of agriculture from the Neolithic to the present. PLoS Pathog. 2008;4:e1000125. doi: 10.1371/journal.ppat.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rector A, et al. Ancient papillomavirus-host co-speciation in Felidae. Genome Biol. 2007;8:R57. doi: 10.1186/gb-2007-8-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poinar G, Jr, Poinar R. Fossil evidence of insect pathogens. J Invertebr Pathol. 2005;89:243–250. doi: 10.1016/j.jip.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert C, Feschotte C. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol. 2010;8:e1000495. doi: 10.1371/journal.pbio.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.May RM. How many species are there on Earth? Science. 1988;241:1441–1449. doi: 10.1126/science.241.4872.1441. [DOI] [PubMed] [Google Scholar]
- 12.Herniou EA, Olszewski JA, O'Reilly DR, Cory JS. Ancient coevolution of baculoviruses and their insect hosts. J Virol. 2004;78:3244–3251. doi: 10.1128/JVI.78.7.3244-3251.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herniou EA, et al. Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. 2011. Baculoviridae. in press. [Google Scholar]
- 14.Lietze V-U, Abd-Alla AMM, Vreysen MJB, Geden CJ, Boucias DG. Salivary gland hypertrophy viruses: A novel group of insect pathogenic viruses. Annu Rev Entomol. 2011;56:63–80. doi: 10.1146/annurev-ento-120709-144841. [DOI] [PubMed] [Google Scholar]
- 15.Drezen J-M, et al. Polydnavirus genome: Integrated vs. free virus. J Insect Physiol. 2003;49:407–417. doi: 10.1016/s0022-1910(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Bininda-Emonds ORP, van Oers MM, Vlak JM, Jehle JA. The genome of Oryctes rhinoceros nudivirus provides novel insight into the evolution of nuclear arthropod-specific large circular double-stranded DNA viruses. Virus Genes. 2011;42:444–456. doi: 10.1007/s11262-011-0589-5. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Burand J, Jehle J. Nudivirus genomics: Diversity and classification. Virol Sin. 2007;22:128–136. [Google Scholar]
- 18.Wu GA, Jun S-R, Sims GE, Kim S-H. Whole-proteome phylogeny of large dsDNA virus families by an alignment-free method. Proc Natl Acad Sci USA. 2009;106:12826–12831. doi: 10.1073/pnas.0905115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Jehle JA. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: New insights on an old topic. J Invertebr Pathol. 2009;101:187–193. doi: 10.1016/j.jip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Whitfield JB. Estimating the age of the polydnavirus/braconid wasp symbiosis. Proc Natl Acad Sci USA. 2002;99:7508–7513. doi: 10.1073/pnas.112067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beckage NE, Gelman DB. Wasp parasitoid disruption of host development: Implications for new biologically based strategies for insect control. Annu Rev Entomol. 2004;49:299–330. doi: 10.1146/annurev.ento.49.061802.123324. [DOI] [PubMed] [Google Scholar]
- 22.Espagne E, et al. Genome sequence of a polydnavirus: Insights into symbiotic virus evolution. Science. 2004;306:286–289. doi: 10.1126/science.1103066. [DOI] [PubMed] [Google Scholar]
- 23.Espagne E, et al. A virus essential for insect host-parasite interactions encodes cystatins. J Virol. 2005;79:9765–9776. doi: 10.1128/JVI.79.15.9765-9776.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupuy C, Huguet E, Drezen J-M. Unfolding the evolutionary story of polydnaviruses. Virus Res. 2006;117:81–89. doi: 10.1016/j.virusres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Webb BA, et al. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology. 2006;347:160–174. doi: 10.1016/j.virol.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Bézier A, et al. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science. 2009;323:926–930. doi: 10.1126/science.1166788. [DOI] [PubMed] [Google Scholar]
- 27.Bézier A, Herbinière J, Lanzrein B, Drezen J-M. Polydnavirus hidden face: The genes producing virus particles of parasitic wasps. J Invertebr Pathol. 2009;101:194–203. doi: 10.1016/j.jip.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Wetterwald C, et al. Identification of bracovirus particle proteins and analysis of their transcript levels at the stage of virion formation. J Gen Virol. 2010;91:2610–2619. doi: 10.1099/vir.0.022699-0. [DOI] [PubMed] [Google Scholar]
- 29.Murphy N, Banks JC, Whitfield JB, Austin AD. Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol Phylogenet Evol. 2008;47:378–395. doi: 10.1016/j.ympev.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 30.Jehle J. In: Insect Virology. Asagari S, Johnson K, editors. Norfolk, UK: Caister Academic Press; 2010. pp. 153–170. [Google Scholar]
- 31.Burand JP, Rallis CP, Tan W. Horizontal transmission of Hz-2V by virus infected Helicoverpa zea moths. J Invertebr Pathol. 2004;85:128–131. doi: 10.1016/j.jip.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Wu Y-L, et al. The early gene hhi1 reactivates Heliothis zea nudivirus 1 in latently infected cells. J Virol. 2010;84:1057–1065. doi: 10.1128/JVI.01548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: Patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 34.Firth C, et al. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol. 2010;27:2038–2051. doi: 10.1093/molbev/msq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gago S, Elena SF, Flores R, Sanjuán R. Extremely high mutation rate of a hammerhead viroid. Science. 2009;323:1308. doi: 10.1126/science.1169202. [DOI] [PubMed] [Google Scholar]
- 37.Stoltz DB, Whitfield JB. Virology. Making nice with viruses. Science. 2009;323:884–885. doi: 10.1126/science.1169808. [DOI] [PubMed] [Google Scholar]
- 38.Grimaldi D, Engel M. Evolution of the Insects. Cambridge, UK: Cambridge Univ Press; 2005. pp. 119–147. [Google Scholar]
- 39.Savard J, et al. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of holometabolous insects. Genome Res. 2006;16:1334–1338. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slack J, Arif BM. The baculoviruses occlusion-derived virus: Virion structure and function. Adv Virus Res. 2007;69:99–165. doi: 10.1016/S0065-3527(06)69003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohrmann GF. Baculovirus Molecular Biology. 2nd Ed. Bethesda, MD: National Center for Biotechnology Information; 2011. [PubMed] [Google Scholar]
- 42.Burand J. In: The Insect Viruses. Miller LK, Ball LA, editors. New York: Plenum; 1998. pp. 69–90. [Google Scholar]
- 43.Brooks D, McLennan D. The Nature of Diversity: An Evolutionary Voyage of Discovery. Chicago: Univ of Chicago Press; 2002. pp. 465–476. [Google Scholar]
- 44.Villarreal LP, Defilippis VR, Gottlieb KA. Acute and persistent viral life strategies and their relationship to emerging diseases. Virology. 2000;272:1–6. doi: 10.1006/viro.2000.0381. [DOI] [PubMed] [Google Scholar]
- 45.Emerman M, Malik HS. Paleovirology—modern consequences of ancient viruses. PLoS Biol. 2010;8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng C-H, et al. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J Virol. 2002;76:9024–9034. doi: 10.1128/JVI.76.18.9024-9034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Kleespies RG, Huger AM, Jehle JA. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J Virol. 2007;81:5395–5406. doi: 10.1128/JVI.02781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afonso CL, et al. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J Virol. 2001;75:11157–11165. doi: 10.1128/JVI.75.22.11157-11165.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lauzon HAM, et al. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J Virol. 2004;78:7023–7035. doi: 10.1128/JVI.78.13.7023-7035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 52.Luque T, Finch R, Crook N, O'Reilly DR, Winstanley D. The complete sequence of the Cydia pomonella granulovirus genome. J Gen Virol. 2001;82:2531–2547. doi: 10.1099/0022-1317-82-10-2531. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Maruniak A, Maruniak JE, Farmerie W, Boucias DG. Sequence analysis of a non-classified, non-occluded DNA virus that causes salivary gland hypertrophy of Musca domestica, MdSGHV. Virology. 2008;377:184–196. doi: 10.1016/j.virol.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abd-Alla AMM, et al. Genome analysis of a Glossina pallidipes salivary gland hypertrophy virus reveals a novel, large, double-stranded circular DNA virus. J Virol. 2008;82:4595–4611. doi: 10.1128/JVI.02588-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Misawa K, Kuma K-i, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 57.Abascal F, Zardoya R, Posada D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 58.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 59.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 60.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 61.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rambault A, Drummond A. Tracer v1.4: MCMC trace analyses tool. 2007. Available from http://beast.bio.ed.ac.uk/Tracer.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.