Abstract
The zinc-finger antiviral protein (ZAP) was originally identified as a host factor that inhibits the replication of Moloney murine leukemia virus. Here we report that ZAP inhibits HIV-1 infection by promoting the degradation of specific viral mRNAs. Overexpression of ZAP rendered cells resistant to HIV-1 infection in a ZAP expression level-dependent manner, whereas depletion of endogenous ZAP enhanced HIV-1 infection. Both human and rat ZAP inhibited the propagation of replication-competent HIV-1. ZAP specifically targeted the multiply spliced but not unspliced or singly spliced HIV-1 mRNAs for degradation. We provide evidence indicating that ZAP selectively recruits cellular poly(A)-specific ribonuclease (PARN) to shorten the poly(A) tail of target viral mRNA and recruits the RNA exosome to degrade the RNA body from the 3′ end. In addition, ZAP recruits cellular decapping complex through its cofactor RNA helicase p72 to initiate degradation of the target viral mRNA from the 5′ end. Depletion of each of these mRNA degradation enzymes reduced ZAP's activity. Our results indicate that ZAP inhibits HIV-1 by recruiting both the 5′ and 3′ mRNA degradation machinery to specifically promote the degradation of multiply spliced HIV-1 mRNAs.
Keywords: retrovirus, host restriction factor, RNA degradation
The infection of cells by HIV type 1 (HIV-1) can be restricted by host factors through a variety of mechanisms (for review, see ref. 1). APOBEC3G and some of its family members restrict HIV-1 infection by deaminating the cytosine in the minus-strand DNA generated during reverse transcription, eventually leading to the destruction of the viral genome (2, 3). However, APOBEC3G can only restrict the replication of Vif-minus HIV-1 but not wild-type virus (2). The Vif protein of HIV-1 bridges the interaction between APOBEC3G and an ubiquitin E3 ligase complex and triggers the polyubiquitination and degradation of APOBEC3G (4, 5). Trim5α and its derivative TRIMCyp restrict HIV-1 infection at early steps of the viral life cycle in a species-specific manner (6, 7). Trim5α mediates the degradation of the capsid (CA) protein of HIV-1 through its ubiquitin E3 ligase activity (8). There is also evidence showing that Trim5α induces rapid uncoating of the HIV-1 capsid, resulting in failure to form a functional preintegration complex (6, 9). Differences in the activities of Trim5α proteins from different species against HIV-1 result from the differences in their abilities to interact with HIV-1 CA (9, 10). Tetherin is a recently identified restriction factor against HIV-1, which inhibits the release of HIV-1 particles from host cells by linking virus particles to each other and “tethering” them to the plasma membrane (11, 12). The anti-HIV activity of tetherin is antagonized by the Vpu protein of HIV-1 (11, 13).
The zinc-finger antiviral protein (ZAP) was initially recovered as a host factor that inhibits the replication of Moloney murine leukemia virus (MLV) (14). In addition to MLV, ZAP inhibits the replication of certain alphaviruses and filoviruses (15, 16). ZAP is not a universal antiviral factor; some viruses replicate normally in ZAP-expressing cells (15). Analysis of viral nucleic acids to determine the step at which ZAP inhibits the replication of MLV has shown that the formation of the integrated provirus is normal, but the level of viral mRNA in the cytoplasm is significantly reduced (14). Further studies have demonstrated that ZAP binds directly to viral mRNA and recruits the RNA exosome to degrade target RNA (17–19). The DEAD-box RNA helicase p72 directly interacts with ZAP as a cofactor and is required for optimal function of ZAP (20).
Mammalian cells have evolved a highly organized and complex mRNA degradation system (21). With the exception of histone mRNA, mature mammalian mRNA bears two protective elements, a 5′ m7G cap and a poly(A) tail. In general mRNA decay, degradation is initiated by shortening of the poly(A) tail by deadenylase, poly(A)-specific ribonuclease (PARN), the CCR4–NOT complex, or the Pan2–Pan3 complex (21). The deadenylated RNA body is degraded 3′–5′ by a 3′–5′ exoribonuclease complex, the exosome (22, 23). The cap structure is removed by the decapping complex Dcp1a–Dcp2 (24), followed by 5′–3′ degradation of the RNA body by 5′–3′ exoribonuclease Xrn1 (25). The relationship between the 3′–5′ and 5′–3′ degradation pathways is not very clear. In transacting factor-mediated specific mRNA decay, for example, AU-rich element (ARE)-mediated mRNA decay and nonsense-mediated mRNA decay, this mRNA degradation machinery is also used to degrade inherently short-lived mRNAs or aberrant mRNAs (26–28).
In this report we show that ZAP inhibits HIV-1 infection by recruiting the cellular mRNA degradation machinery to degrade specific viral mRNAs. ZAP recruits deadenylase PARN and the exosome to degrade target mRNAs from the 3′ end. We further provide evidence that ZAP also recruits the 5′–3′ mRNA degradation machinery to degrade the target viral mRNAs.
Results
Expression of ZAP Inhibits HIV-1 Infection.
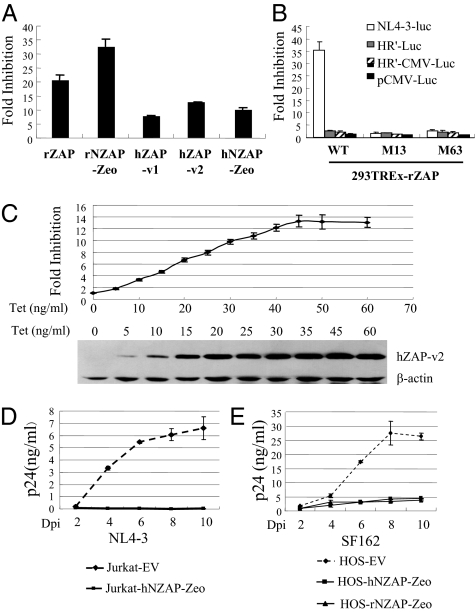
To probe the antiviral activity of ZAP against HIV-1, various forms of ZAP (Fig. S1) were tested against vesicular stomatitis virus G protein (VSV-G)–pseudotyped HIV-1 vector NL4-3-luc, which contains most of the sequence of the HIV-1 genome (29) (Fig. S2). The N-terminal domain of rat ZAP (rZAP) fused with the zeocin resistance gene (rNZAP-Zeo) has been reported to display antiviral activity similar to that of full-length rZAP on MLV (14). There are two forms of human ZAP (hZAP) protein arising from alternative splicing, which differ only in the C-terminal domain (30) (Fig. S1). Full-length rZAP, rNZAP-Zeo, both forms of hZAP (hZAP-v1 and hZAP-v2), and the N-terminal domain of hZAP fused with the zeocin resistance gene (hNZAP-Zeo) were individually expressed in HEK293 cells in a tetracycline-inducible manner (Fig. S1) and assayed for their antiviral activities. All of the ZAP proteins displayed significant inhibitory activities against NL4-3-luc (Fig. 1A). To confirm the specificity of the inhibitory effect of ZAP against NL4-3-luc, we tested two ZAP mutants previously reported to fail in inhibiting MLV infection (31). The antiviral activities of the proteins were also tested against HR′-Luc and HR′-CMV-Luc vectors, in which a large fraction of the HIV-1 genome is deleted (32) (Fig. S2). Wild-type ZAP displayed significant antiviral activity against NL4-3-luc, but its activity against HR′-Luc or HR′-CMV-Luc was very modest (Fig. 1B). As expected, the ZAP mutants failed to inhibit any of the HIV-1 vectors (Fig. 1B).
Fig. 1.
Expression of ZAP inhibits HIV-1 infection. (A) 293TREx cells expressing the indicated ZAP proteins were infected with VSV-G–pseudotyped NL4-3-luc. At 3 h after infection, cells were mock treated or treated with 1 μg/mL tetracycline to induce ZAP expression. The cells were lysed, and luciferase activities were measured at 48 h after infection. Fold inhibition was calculated as the ratio of the luciferase activity in mock-treated cells to that in tetracycline-treated cells. Data presented are means ± SD of three independent experiments. (B) 293TREx-rZAP, 293TREx-rZAP-Zm13, and 293TREx-rZAP-Zm63 cells were respectively infected with VSV-G–pseudotyped NL4-3-luc, HR′-Luc, and HR′-CMV-Luc or transfected with pCMV-Luc. At 3 h after infection or 6 h after transfection, cells were mock treated or treated with 1 μg/mL tetracycline. Cells were lysed, and luciferase activities were measured at 48 h after infection or after transfection. Fold inhibition was calculated as the ratio of the luciferase activity in the mock-treated cells to that in tetracycline-treated cells. Data presented are means ± SD of three independent experiments. (C) 293TREx-hZAP-v2 cells were infected with VSV-G–pseudotyped NL4-3-luc. At 3 h after infection, cells were mock treated or treated with tetracycline at the indicated concentrations. Cells were lysed at 48 h after infection, luciferase activities were measured, and fold inhibition was calculated (Upper). Data presented are means ± SD of three independent measurements. The expression levels of hZAP-v2 were measured by Western blotting (Lower). (D) Jurkat cells transduced with empty vector (EV) or retroviral vector expressing hNZAP-Zeo were pooled and infected with HIV-1 strain NL4-3 (1 ng p24/mL) in duplicate. At the indicated time points supernatants were collected, and p24 levels were measured. Data presented are means ± SE of two independent measurements. (E) HOS-CD4-CCR5 cells transduced with empty vector (EV) or retroviral vector expressing hNZAP-Zeo or rNZAP-Zeo were pooled and infected with HIV-1 strain SF162 (20 ng p24/mL) in duplicate. At the indicated time points supernatants were collected, and p24 levels were measured. Data are means ± SE of two independent measurements. Dpi, days postinfection.
To test whether the antiviral activity of ZAP is expression level-dependent, 293TREx-hZAP-v2 cells were treated with increasing concentrations of tetracycline to induce hZAP-v2 expression and assayed for their activity to inhibit the expression of NL4-3-luc. Indeed, the antiviral activity of hZAP-v2 correlated well with its expression level (Fig. 1C).
To test whether ZAP is able to inhibit the propagation of replication-competent HIV-1, hNZAP-Zeo was expressed in Jurkat cells. The cells were infected with NL4-3 virus, and viral replication was monitored by assaying HIV-1 p24 protein concentration in the supernatant over successive days. Viral replication was easily detected in control cells (Fig. 1D). However, viral replication was profoundly blocked in cells expressing hNZAP-Zeo (Fig. 1D). rNZAP-Zeo and hNZAP-Zeo were also expressed in HOS-CD4-CCR5 cells, a human osteosarcoma cell line engineered to express HIV-1 receptors and commonly used as HIV-1 reporter cells (33), and cells were infected with HIV-1 strain SF162. Both rNZAP-Zeo and hNZAP-Zeo blocked viral replication (Fig. 1E). These results establish that ZAP inhibits the replication of live HIV-1.
ZAP Prevents the Accumulation of nef-luc mRNA from NL4-3-luc.
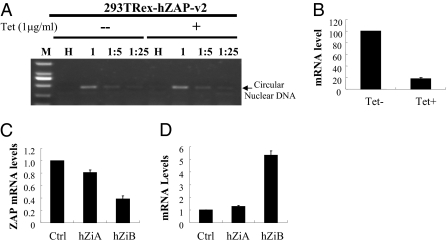
rZAP has been shown to inhibit MLV infection by promoting viral mRNA degradation in the cytoplasm without affecting the formation and nuclear entry of viral DNA (14). To test whether hZAP inhibits HIV-1 infection by the same mechanism, 293TREx-hZAP-v2 cells were infected with VSV-G–pseudotyped NL4-3-luc, and the levels of nuclear circular viral DNA in the absence and presence of hZAP were compared. As expected, no difference was detected in the levels of nuclear circular viral DNA with and without hZAP expression (Fig. 2A), indicating that expression of hZAP does not affect the formation and nuclear entry of viral DNA.
Fig. 2.
Expression of hZAP reduces the level of nef-luc mRNA. (A) 293TREx-hZAP-v2 cells were infected with VSV-G–pseudotyped NL4-3-luc at the indicated dilutions. At 3 h after infection, cells were mock treated or treated with 1 μg/mL tetracycline. At 24 h after infection cells were lysed, and Hirt DNA was extracted. LTR–LTR junction of nuclear circular viral DNA was detected by PCR. M, molecular weight marker; HI, heat-inactivated NL4-3-luc. (B) 293TREx-hZAP-v2 cells were infected with VSV-G–pseudotyped NL4-3-luc. At 3 h after infection, cells were mock treated (Tet-) or treated (Tet+) with 1 μg/mL tetracycline. At 48 h after infection, cytoplasmic RNA was extracted and subjected to real-time PCR analysis to measure the levels of nef-luc and gapdh mRNAs. The relative level of nef-luc mRNA was normalized to that of gapdh mRNA. The nef-luc mRNA level in mock-treated cells was set as 100. Data presented are means ± SD from three independent experiments. (C) The endogenous hZAP mRNA levels in Jurkat cells stably expressing control shRNA (Ctrl) or shRNAs against hZAP (hZiA and hZiB) were measured by real-time PCR. The relative ZAP mRNA level in Jurkat-Ctrl cells was set as 1. Data presented are means ± SD of three independent experiments. (D) Cells were infected with VSV-G–pseudotyped NL4-3-luc. At 48 h after infection, nef-luc mRNA levels were measured by real-time PCR. The relative nef-luc mRNA level in Jurkat-Ctrl cells was set as 1. Data presented are means ± SD of three independent experiments.
In NL4-3-luc, the luciferase reporter is inserted into the coding sequence of Nef (Fig. S1). To analyze whether ZAP targets HIV-1 mRNA for degradation, we compared nef-luc mRNA levels before and after the induction of hZAP expression. Indeed, upon expression of ZAP, the nef-luc mRNA levels were reduced by approximately fivefold (Fig. 2B).
To test whether endogenous ZAP is active against HIV-1 infection, Jurkat cells stably expressing control shRNA (Jurkat-Ctrl) or shRNAs against hZAP (Jurkat-hZiA and Jurkat-hZiB) were generated. The expression levels of hZAP in Jurkat-hZiA and Jurkat-hZiB cells were reduced by approximately 20% and 60%, respectively (Fig. 2C). The cells were infected with VSV-G–pseudotyped NL4-3-luc. In Jurkat-hZiA and Jurkat-hZiB cells, approximately 1.5-fold and fivefold increases in the mRNA levels were observed, respectively, compared with that in control cells (Fig. 2D). Similar results were also observed in HEK293 cells (Fig. S3) and HOS cells (Fig. S4). These results show that down-regulation of endogenous hZAP increases nef-luc mRNA levels.
ZAP Specifically Targets Multiply Spliced HIV-1 mRNAs.
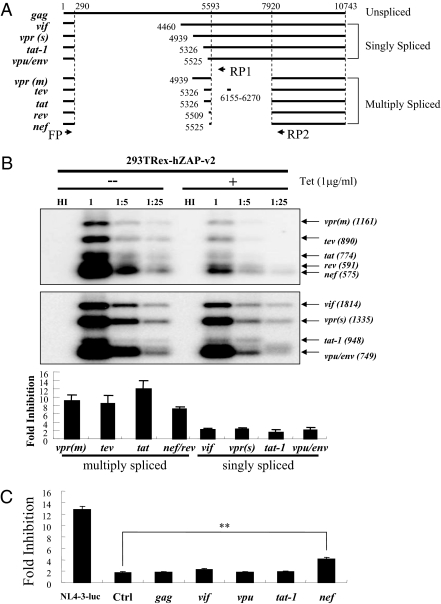
HIV-1 precursor RNA is known to be spliced into multiple mRNA species (34) (Fig. 3A). To analyze whether other HIV-1 mRNAs are also targeted by ZAP for degradation, HIV-1 mRNAs levels in cells infected with NL4-3-luc were measured by RT-PCR followed by Southern blotting using primers that specifically amplify all singly or multiply spliced HIV-1 mRNAs (Fig. 3A). Upon hZAP expression, levels of multiply spliced HIV-1 mRNAs were all reduced significantly (Fig. 3B, Upper). In contrast, levels of singly spliced mRNAs were only modestly affected (Fig. 3B). To substantiate these results, 293TREx-hZAP-v2 cells were infected with NL4-3-luc, and two individual clones harboring the provirus were picked and expanded. The relative mRNA levels of gag, vif, and nef-luc were measured by real-time PCR. ZAP expression significantly reduced the level of nef-luc mRNA but not that of gag or vif (Fig. S5). We noted that the fold inhibition in these two clones was lower than that observed in the above experiment. A possible explanation for this is that newly synthesized multiply spliced HIV-1 mRNAs are more sensitive to ZAP for reasons that are yet to be determined.
Fig. 3.
(A) Schematic representation of HIV-1 mRNAs expressed from NL4-3-luc. Primers FP and RP1 were used for PCR amplification of cDNAs from singly spliced mRNAs. Primers FP and RP2 were used for PCR amplification of cDNAs from multiply spliced mRNAs. The primers are represented by arrows. (B) 293TREx-hZAP-v2 cells were infected with VSV-G–pseudotyped NL4-3-luc at the indicated dilutions. At 3 h after infection, the cells were mock treated or treated with 1 μg/mL tetracycline. At 48 h after infection, cytoplasmic RNA was extracted. The levels of the mRNAs indicated were analyzed by RT-PCR followed by Southern blotting and quantified by phosphorimaging. Fold inhibition was calculated as the ratio of the mRNA level in mock-treated cells to that in tetracycline-treated cells. Data presented are means ± SE of the fold inhibition values from infections with undiluted or fivefold diluted virus. (C) 293TREx-hZAP-v2 cells were infected with VSV-G–pseudotyped NL4-3-luc, HR′-CMV-Luc virus (Ctrl), or HR′-CMV-Luc virus containing the 5′ UTR from the indicated HIV-1 mRNAs. At 3 h after infection, cells were mock treated or treated with 1 μg/mL tetracycline. Cells were lysed, and luciferase activities were measured at 48 h after infection. Fold inhibition was calculated as the ratio of the luciferase activity in mock-treated cells to that in tetracycline-treated cells. Data presented are means ± SD of three independent experiments. **P < 0.01.
Multiply spliced mRNAs differ from singly spliced mRNAs at the second splicing junction (Fig. 3A). The above results suggest that the target of ZAP is the 5′ UTR of HIV-1 nef mRNA, which contains the second splicing junction. To substantiate this notion, the sequences corresponding to the 5′ UTR of unspliced gag, singly spliced vif, vpu, and tat-1, and multiply spliced nef were cloned into HR′-CMV-Luc upstream of the luciferase coding sequence. The vectors were analyzed for their sensitivities to hZAP. Indeed, nef 5′ UTR rendered HR′-CMV-Luc sensitive to ZAP, whereas the other 5′ UTRs failed to do so (Fig. 3C).
3′–5′ mRNA Degradation Enzymes Are Required for ZAP-Mediated Viral mRNA Degradation.
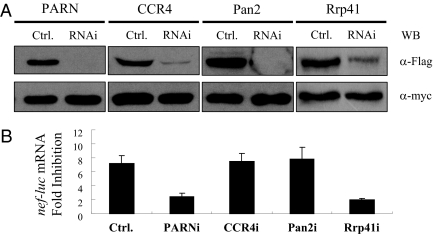
rZAP has been shown to recruit the RNA-processing exosome to degrade target mRNA (18). To test whether hZAP degrades HIV-1 mRNAs through the RNA exosome, we first tested whether the exosome is required for hZAP to degrade nef-luc mRNA. The ability of the shRNA directed against exosome component hRrp41 (Rrp41i) to down-regulate the expression of hRrp41 was validated (Fig. 4A). As expected, depletion of hRrp41 reduced the activity of hZAP in the degradation of nef-luc mRNA (Fig. 4B). Depletion of hRrp42 also reduced the antiviral activity of hZAP (Fig. S6).
Fig. 4.
3′–5′ mRNA degradation enzymes are required for ZAP-mediated viral mRNA degradation. (A) Plasmids expressing the Flag-tagged proteins and shRNAs indicated were cotransfected into HEK 293 cells, together with a plasmid expressing myc-tagged GFP to control transfection efficiency and sample handling. At 48 h after transfection, protein expression levels were measured by Western blotting. (B) 293TREx-hZAP-v2 cells were transfected twice with the indicated shRNA-expressing plasmids. Cells were infected with VSV-G–pseudotyped NL4-3-luc for 3 h and then mock treated or treated with 1 μg/mL tetracycline to induce ZAP expression. At 48 h after infection, cytoplasmic RNA was extracted and subjected to real-time PCR analysis to measure the mRNA levels of nef-luc and gapdh. Levels of nef-luc mRNA were normalized to those of gapdh. Fold inhibition was calculated as the ratio of the normalized nef-luc mRNA level in mock-treated cells to that in tetracycline-treated cells. Data presented are means ± SD from three independent experiments.
HIV-1 mRNAs are protected by poly(A) tails at their 3′ ends. For the exosome to degrade target mRNA, the poly(A) tail needs to be removed first. In mammalian cells there are three known deadenylases: PARN, the CCR4–NOT complex, and the Pan2–Pan3 complex. Each deadenylase was individually down-regulated by shRNA and examined for the effect on ZAP-mediated nef-luc mRNA degradation in 293TREx-hZAP-v2 cells. Compared with control shRNA, shRNAs against PARN, CCR4, and Pan2 all reduced the expression levels of target proteins to 10% or less (Fig. 4A). Down-regulation of PARN significantly reduced ZAP's activity, whereas down-regulation of CCR4 or Pan2 had little effect (Fig. 4B). These results establish that deadenylation by PARN and subsequent 3′–5′ mRNA degradation by the exosome are required for ZAP-mediated viral mRNA degradation.
5′–3′ mRNA Degradation Enzymes Are Required for Optimal Function of ZAP.
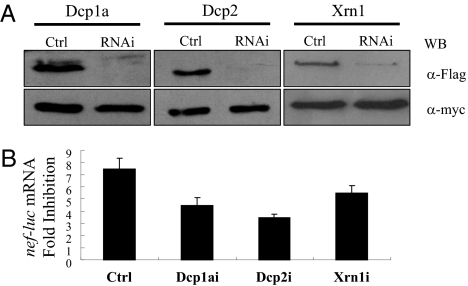
We next analyzed whether 5′–3′ mRNA degradation machinery is involved in ZAP-mediated viral mRNA degradation. The abilities of the shRNAs directed against the decapping enzymes Dcp1a and Dcp2 and 5′–3′ exoribonulease Xrn1 to down-regulate the expression of the target proteins were confirmed (Fig. 5A). Down-regulation of endogenous Dcp1a, Dcp2, and Xrn1 in 293TREx-hZAP-v2 cells all reduced the activity of hZAP (Fig. 5B). These results establish that 5′–3′ mRNA degradation machinery is involved in hZAP-mediated degradation of viral mRNA.
Fig. 5.
5′–3′ mRNA degradation enzymes are required for ZAP-mediated viral mRNA degradation. Experiments were performed as described in the legend to Fig. 4. (A) Validation of the abilities of the shRNAs indicated to down-regulate the expression of the proteins indicated. (B) Down-regulation of Dcp1a, Dcp2, and Xrn1 reduced ZAP's activity in the degradation of nef-luc mRNA. Data presented are means ± SD from three independent experiments.
Interactions Between ZAP and mRNA Degradation Enzymes.
The above results indicate that both 3′–5′ and 5′–3′ mRNA degradation enzymes participate in ZAP-mediated viral mRNA degradation. We next set out to examine the interactions between ZAP and these enzymes.
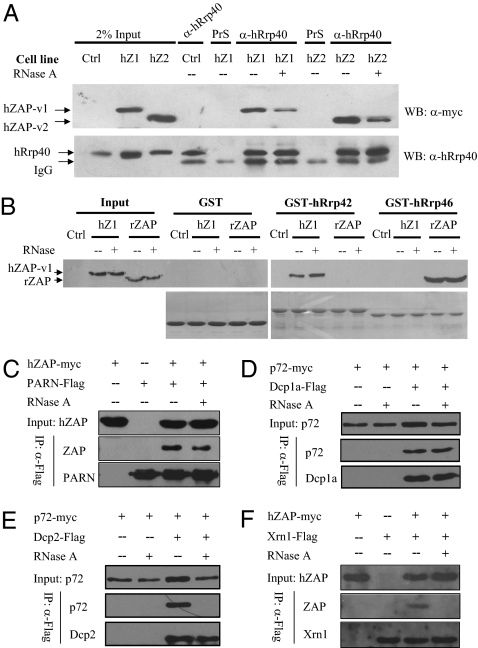
We previously reported that rZAP interacts with the exosome through directly binding to the exosome component Rrp46 (18). Here, we first analyzed the interaction between hZAP and the exosome. As expected, immunoprecipitation of the endogenous exosome component hRrp40 coprecipitated hZAP-v1 and hZAP-v2 (Fig. 6A). To test how hZAP interacts with the exosome, pull-down assays were performed using bacterially expressed human exosome components. Surprisingly, unlike rZAP, hZAP did not interact with hRrp46 (Fig. 6B). Other exosome components were tested for their interactions with hZAP, and hRrp42 was found to bind hZAP (Fig. 6B). Similar experiments were also performed using rat exosome components. In line with the above results, hZAP interacted with rat Rrp42 (rRrp42), whereas rZAP interacted with rat Rrp46 (rRrp46) (Fig. S7).
Fig. 6.
Interactions between ZAP and mRNA degradation enzymes. (A) 293TREx-hZAP-v1 and 293TREx-hZAP-v2 cells were treated with tetracycline to induce hZAP expression. Cells were lysed in lysis buffer with (+) or without (−) RNase A (50 μg/mL). Proteins were immunoprecipitated with rabbit anti-hRrp40 antibody and Western blotted with anti-myc antibody (Upper) or anti-hRrp40p antibody (Lower). (B) The bacterially expressed GST proteins indicated were immobilized onto glutathione-Sepharose 4B resin and incubated with lysates of cells expressing rZAP or hZAP-v1 in the presence of RNase A at 4 °C for 4 h. Resins were washed and boiled in sample loading buffer. Proteins were resolved by SDS/PAGE and detected by Western blotting with anti-myc antibody (Upper) or by Coomassie blue staining (Lower). Input, total cell lysates; PrS, rabbit preimmune serum; Ctrl, 293TREx cells; hZ1, 293TREx-hZAP-v1 cells; hZ2, 293TREx-hZAP-v2 cells; rZAP, 293TREx-rZAP cells. (C–F) Plasmids expressing the indicated proteins were cotransfected into HEK293T cells. At 48 h after transfection, cells were lysed in lysis buffer with (+) or without (−) RNase A (50 μg/mL). Proteins were immunoprecipitated with anti-Flag antibody and Western blotted with anti-myc antibody or anti-Flag antibody.
Coimmunoprecipitation assays were used to analyze the interaction between hZAP and PARN. RNase A was added to the cell lysate to test whether the interaction was caused by nonspecific RNA tethering. Immunoprecipitation of PARN coprecipitated hZAP in the absence or presence of RNase A, indicating that hZAP interacts with PARN in an RNA-independent manner (Fig. 6C). Similar assays were also performed to test interactions between ZAP and 5′–3′ mRNA degradation enzymes. ZAP interacted with Dcp1a and Dcp2 only in the absence of RNase A (Fig. S8), suggesting that the interaction was not direct. Our previous results have demonstrated that RNA helicase p72 directly interacts with ZAP and is required for optimal functioning of ZAP (20). The possibility exists that ZAP might recruit the decapping complex through p72. To test this idea, we analyzed the interaction between p72 and Dcp1a or Dcp2. Both Dcp1a and Dcp2 interacted with p72 in the absence of RNase A (Fig. 6 D and E). However, Dcp1a but not Dcp2 interacted with p72 in the presence of RNase A (Fig. 6 D and E). These results suggest that ZAP recruits the decapping complex Dcp1a/Dcp2 through p72 to remove the cap structure. Only RNA-dependent interactions between ZAP and XrnI were detected (Fig. 6F).
Discussion
In this report we show that overexpression of ZAP inhibits HIV-1 infection in a ZAP expression level-dependent manner by preventing the accumulation of multiply spliced HIV-1 mRNAs (Figs. 1–3). Down-regulation of endogenous ZAP enhances nef-luc mRNA levels (Fig. 2 C and D). Both human and rat ZAP inhibit the propagation of replication-competent HIV-1 (Fig. 1 D and E), suggesting that ZAP is neither species-specific nor antagonized by HIV-1 proteins.
ZAP dramatically reduced the levels of multiply spliced HIV-1 mRNAs, but its effects on unspliced and singly spliced mRNAs were modest (Fig. 3B and Fig. S5). Because all of the sequences in multiply spliced mRNAs are also present in singly spliced mRNAs, these results suggest that the sequence that arises as a result of the second splicing is important for ZAP recognition of the target RNA. This is supported by the observation that cloning of the 5′ UTR of nef rendered HR′-CMV-luc sensitive to ZAP, but the 5′ UTR of tat-1 failed to do so (Fig. 3C). Tat protein is required for efficient transcription of the unspliced mRNA (35). In ZAP-expressing cells the level of tat mRNA derived from multiple splicing was dramatically reduced (Fig. 3B). However, the level of tat-1 mRNA derived from single splicing was only modestly affected (Fig. 3B). The residual expression of Tat seemed to be sufficient to maintain transcription of viral mRNA at a level not much lower than that without ZAP. The modest reduction in unspliced and singly spliced mRNA levels could be accounted for by the reduction in the expression of Tat protein. Collectively these results indicate that ZAP directly targets multiply spliced mRNAs but not singly spliced mRNAs for degradation.
Both rZAP and hZAP interacted directly with the exosome, but through binding to different components of the exosome (Fig. 6B and Fig. S7). The six core components of the human exosome form a barrel-like structure, with hRrp42 and hRrp46 located opposite to each other (22, 23). It is conceivable that the ability of ZAP to deliver target RNA to the exosome should not be affected by whether it binds to hRrp42 or to hRrp46. This may explain why rZAP and hZAP display similar antiviral activities.
In addition to the 3′–5′ mRNA degradation machinery, ZAP recruits the decapping enzyme through its cofactor p72. ZAP did not directly interact with 5′–3′ exoribonuclease XrnI (Fig. 6F). Whether ZAP interacts with XrnI through a yet to be identified factor remains to be determined. Once the cap structure is removed from the mRNA, the RNA body is exposed and becomes accessible to XrnI (21), possibly removing the absolute requirement for ZAP to recruit XrnI, although recruitment of XrnI to the target mRNA might expedite the degradation process.
We noticed that ZAP inhibited the expression of Nef-luc protein by approximately 13-fold (Fig. 1A) but reduced the level of nef-luc mRNA by only approximately fivefold (Fig. 2B). This discrepancy suggests that ZAP may also repress the translation of target viral mRNA. How ZAP inhibits translation of target viral mRNA remains to be elucidated.
The short form of ZAP (hZAP-v2) was recently reported to stimulate the RIG-I signaling pathway (36). To analyze whether ZAP inhibits HIV-1 infection through the RIG-I pathway, RIG-I was depleted by RNAi in 293TREx-hZAP-v2 cells. Down-regulation of RIG-I did not affect the antiviral activity of hZAP-v2 against NL4-3-luc (Fig. S9), indicating that the antiviral activity of hZAP against HIV-1 reported here is independent of the RIG-I pathway.
On the basis of the results presented previously and in this study, we propose a working model for ZAP-mediated mRNA degradation. ZAP binds directly to its target mRNA, recruits PARN to remove the poly(A) tail, and further recruits the exosome to degrade the RNA body from the exposed 3′ end. ZAP also recruits the decapping enzyme through its cofactor p72 to remove the cap structure of the target mRNA. The RNA body is further degraded by XrnI from the exposed 5′ end.
In conclusion, we have shown here that ZAP inhibits HIV-1 replication by selectively targeting multiply spliced mRNAs for degradation. Whether ZAP inhibits HIV-1 infection and how HIV-1 evades ZAP to establish effective infection in vivo await further investigation.
Materials and Methods
DNA construction and cell culture are described in SI Materials and Methods.
Real-Time PCR.
The mRNA levels of hZAP, Gag, Vif, and RIG-I were measured by SYBR Green real-time PCR in Rotor-gene 6000 (Corbett Life Science) using the following program: (i) 50 °C 2 min, 1 cycle; (ii) 95 °C 10 min, 1 cycle; (iii) 95 °C 15 s; 60 °C 30 s; 72 °C 30 s, 40 cycles; (iv) 72 °C 10 min, 1 cycle. The sequences of the primers were as follows:
qhZAP FP: 5′-CCACATCTTCTAGGGTGGATGA-3′
qhZAP RP: 5′-CGTCCAGGTTTTACCAATAAGCA-3′
qGag FP: 5′-GTGTGGAAAATCTCTAGCAGTGG-3′
qGag RP: 5′-CGCTCTCGCACCCATCTC-3′
qVif FP: 5′-GGCGACTGGGACAGC-3′
qVif RP: 5′-CACACAATCATCACCTGCC-3′
qRIG-I FP: CCTACCTACATCCTGAGCTACAT
qRIG-I RP: TCTAGGGCATCCAAAAAGCCA.
nef-luc mRNA levels were measured by Taqman real-time PCR in Rotor-gene 6000 (Corbett Life Science) with the following program: (i) 50 °C 2 min; (ii) 95 °C 10 min; (iii) 95 °C 15 s, 60 °C 1 min, 40 cycles; (iv) 30 °C 1 min. TaqMan GAPDH Control Reagents (human) (ABI) were used for gapdh mRNA real-time PCR. The Taqman probe and primers for nef-luc mRNA were as follows:
qnef-luc probe: 5′-FAM-AAGCAACCCACCTCCC-NFQ-3′
qnef-luc FP: 5′-ACAGTCAGACTCATCAAGCTTCTCT-3′
qnef-luc RP: 5′-CGGGTCCCCTCGGGATT-3′.
Measurement of Viral mRNAs by RT-PCR.
Cytoplasmic RNA was extracted using an RNeasy Kit (Qiagen) according to the manufacturer's instructions. Four micrograms of total cytoplasmic RNA was used for RT by MLV reverse transcriptase using random primers in a 20-μL system. One microliter of the RT product was amplified in a 50-μL PCR.
The cDNAs from singly spliced HIV-1 mRNAs were amplified using primers FP and RP1. The cDNAs from the multiply spliced HIV-1 mRNAs were amplified using primers FP and RP2. The PCR conditions used were 94 °C for 30 s, 45 °C for 30 s, and 72 °C for 90 s for 16 cycles. To confirm the identities of the PCR products, the cDNAs of vif, vpu, tat, rev, and nef were PCR-amplified for 46 cycles under the same conditions, and the PCR products were recovered and sequenced.
PCR products were resolved on a 1.2% agarose gel, followed by Southern blotting. To detect the RT-PCR products from the singly spliced mRNAs, a fragment of the HIV-1 genome PCR-amplified from pNL4-3-luc using primers HIV6078F and HIV6621R was used as a template for the probes. To detect RT-PCR products from multiply spliced mRNAs, a fragment of the HIV-1 genome PCR-amplified from pNL4-3-luc using primers HIV8396F and HIV8776R was used as a template for the probes. The quantities of the PCR products were measured by phosphorimaging. The sequences of the primers were as follows:
FP: 5′-AGTAAAGCCAGAGGAGATCTCTCG-3′
RP1: 5′-ATTGGTATTAGTATCATTCTTCAAATC-3′
RP2: 5′-GCCACCCATCTTATAGCAAAATCC-3′
HIV6078F: 5′-GTAGCAATAGTAGCATTAGTAGTAGC-3′
HIV6621R: 5′-ATTGGTATTAGTATCATTCTTCAAATC-3′
HIV8396F: 5′-ACCCGACAGGCCCGAAGGAATAG-3′
HIV8776R: 5′-GCCACCCATCTTATAGCAAAATCC-3′.
Statistical Analysis.
The mean values ± SD or mean values ± SE were calculated from at least three independent experiments unless otherwise indicated, and P values were calculated using the Student t test.
Supplementary Material
Acknowledgments
We thank Dr. Shan Cen, Nidhanapati K. Raghavendra, and Xiaoyu Li for technical support. This work was supported in part by National Science Foundation Grants 30530020 and 81028011, Ministry of Science and Technology 973 Program Grant 2006CB504302, and Ministry of Health of China Grant 2009ZX09501-029 (to G.G.); and by National Science Foundation Grant 30800053 and Ministry of Health of China Grant 2008ZX10001-002 (to G.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101676108/-/DCSupplemental.
References
- 1.Wolf D, Goff SP. Host restriction factors blocking retroviral replication. Annu Rev Genet. 2008;42:143–163. doi: 10.1146/annurev.genet.42.110807.091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 5.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 6.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 7.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 8.Chatterji U, et al. Trim5alpha accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J Biol Chem. 2006;281:37025–37033. doi: 10.1074/jbc.M606066200. [DOI] [PubMed] [Google Scholar]
- 9.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 12.Perez-Caballero D, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 15.Bick MJ, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller S, et al. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol. 2007;81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol. 2004;78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA. 2007;104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y, Gao G. ZAP-mediated mRNA degradation. RNA Biol. 2008;5:65–67. doi: 10.4161/rna.5.2.6044. [DOI] [PubMed] [Google Scholar]
- 20.Chen G, Guo X, Lv F, Xu Y, Gao G. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc Natl Acad Sci USA. 2008;105:4352–4357. doi: 10.1073/pnas.0712276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 23.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 24.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens A. 5′-exoribonuclease 1: Xrn1. Methods Enzymol. 2001;342:251–259. doi: 10.1016/s0076-6879(01)42549-3. [DOI] [PubMed] [Google Scholar]
- 26.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gherzi R, et al. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 29.Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 30.Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Lv F, Gao G. Mutagenesis analysis of the zinc-finger antiviral protein. Retrovirology. 2010;7:19. doi: 10.1186/1742-4690-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 33.Deng H, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 34.Mary C, et al. Quantitative and discriminative detection of individual HIV-1 mRNA subspecies by an RNAse mapping assay. J Virol Methods. 1994;49:9–23. doi: 10.1016/0166-0934(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 35.Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 36.Hayakawa S, et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol. 2011;12:37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.