Abstract
When grafting or wounding disconnects stem tissues, new tissues are generated to restore the lost connection. In this study, the molecular mechanism of such healing was elucidated in injured stems of Arabidopsis. Soon after the inflorescence stems were incised, the pith cells started to divide. This process was strongly inhibited by the elimination of cauline leaves, shoot apices, or lateral buds that reduced the indole-3-acetic acid supply. Microarray and quantitative RT-PCR analyses revealed that genes related to cell division, phytohormones, and transcription factors were expressed because of incision. Among them, two plant-specific transcription factor genes, ANAC071 and RAP2.6L, were abundantly expressed. ANAC071 was expressed at 1–3 d after cutting exclusively in the upper region of the cut gap, with concomitant accumulation of indole-3-acetic acid. In contrast, RAP2.6L was expressed at 1 d after cutting exclusively in the lower region, with concomitant deprivation of indole-3-acetic acid. The expression of ANAC071 and RAP2.6L were also promoted by ethylene and jasmonic acid, respectively. In transformants suppressing the function of RAP2.6L or ANAC071, the division of pith cells was inhibited. Furthermore, the ethylene signaling-defective ein2 mutant showed incomplete healing. Hence, plant-specific transcription factors differentially expressed around the cut position were essential for tissue reunion of Arabidopsis wounded flowering stems and were under opposite control by polar-transported auxin, with modification by the ethylene and jasmonic acid wound-inducible hormones.
Keywords: regeneration, meristem, stress
Functionally specialized leaves, flowers, and roots of vascular plants are integrated via stems to enable plant responses to changing environments (1). Plant stems provide essential mechanical support of the plant body and deliver nutrients as well as physiological chemical information through vascular bundles (1). Auxin is required to establish the vertical axis and is produced in shoot apices and transported to roots through stele parenchyma or procambial cells with auxin-specific transporters (2). Biotic or biophysical plant environmental stresses often result in wounding that may disintegrate and endanger plant tissues. Cellular responses to wounding involve reconstruction of damaged tissues and also activation of the synthesis of defense-related proteins, including basic pathogenesis-related proteins, as well as wound hormones such as ethylene and jasmonic acid (JA) (3). During reconstruction of damaged tissues, vascular and/or other cells are transformed so as to physiologically connect the existing tissues (4, 5). Such activities have long been observed in grafting techniques, which are advantageous for agriculture and horticulture (6) and also for basic research on systemic physiological events, such as flower induction (7) and hormone actions (8, 9). Low grafting efficiency and tissue incompatibility are often encountered, necessitating an understanding of the healing mechanism underlying grafting. In previous studies on grafting as well as repairing of wounded tissues, much effort has focused on understanding the regeneration of vascular elements, demonstrating the involvement of phytohormones such as auxin and cytokinin, and several NAC (NAM, ATAF1,2 and CUC2) transcription factors (TFs) (4, 10).
Processes reuniting ground tissues such as the cortex and pith in injured regions comprise cell division and successive cell differentiation, although the underlying molecular mechanisms have been less studied than vascular regeneration. Active pectin biosynthesis occurs in cucumber and tomato hypocotyls, and gibberellin (GA), likely produced in and exported from cotyledons, is used for cell division during cortical tissue reunion (11–13). Such regeneration ability has been studied in terms of totipotency in tissue culture, suggesting that TFs are required for regulating gene expression related to regeneration (14, 15). However, the TFs and accompanying molecular processes involved in reuniting ground tissues remain to be determined.
The research reported here aimed to understand the molecular events during the tissue-reunion process in plant stems. To this end, a tissue-reunion inducible system was established by incising Arabidopsis inflorescence stems. The incised tissues were investigated by microarray analysis followed by expression/functional analysis of the up-regulated genes. Two plant-specific TF genes, ANAC071 and RAP2.6L, with localized expression in the upper and lower regions of the cut gap, respectively, were required for the division of pith cells in the reunion process. Differential controls of these genes by auxin and the wound-related ethylene and JA hormones are also described.
Results
Histological Time Course of Cell Division During Wounding of the Arabidopsis Inflorescence Stems.
Arabidopsis inflorescence stems were used 7–10 d after bolting. The internodes between the first cauline leaves and rosette leaves, which were no longer capable of cell division and elongation, were incised to half-diameter depth with a microsurgical knife (Fig. S1 A and B).
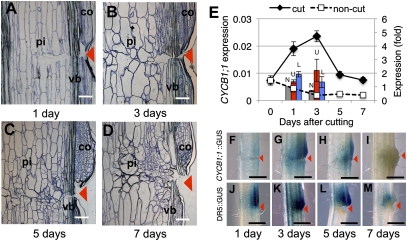
The pith cells around the cut site then began to randomly divide and elongate intrusively toward the cut surface (Fig. 1 A–D). To obtain an estimate of cell division activity in the wounded stems, quantitative RT-PCR (qRT-PCR) analyses were performed using mitotic cyclin-encoding cyclin B1;1 (At4g37490), a mitosis marker gene expressed only around the G2/M transition (16) (Fig. 1E). The cyclin gene was clearly up-regulated in the wounded region 3 d after cutting, where cell division was initiated in both pith and cortical cells neighboring the wounded regions (Fig. 1B and E), as was previously observed in wounded cucumber hypocotyls (11). Spatiotemporal histochemical changes in mitotic activity in response to tissue reunion were pursued using the cyclin B1;1 promoter (designated as pcyclin B)::GUS transgenic plants (Fig. 1 F–I). The GUS activity culminated 3 d after cutting (Fig. 1G), in agreement with both qRT-PCR analysis (Fig. 1E) and morphological observations (Fig. 1 A–D). The GUS activity was localized in the pith, vascular tissues, and cortex in both the upper and lower regions of the cut gap, although the intensity was much higher in the upper region (Fig. 1G).
Fig. 1.
Process of tissue reunion in the wounded flowering stem of Arabidopsis. (A–D) Light micrographs of the tissue-reunion process. Arrowheads indicate position of the cut. pi, pith; co, cortex; vb, vascular bundle. (E) Expression of Cyclin B1;1. Each value is the mean ± SE (n = 3). Bars show comparative expression between upper (U; red bar) or lower regions (L; blue bar) of cut stem. The noncut (N; gray bar) control was arbitrarily set to 1, and the mean ± SE is shown (n = 4). (F–I) pCyclin B::GUS plants. (J–M) DR5::GUS plants. Photographs were taken at 1 d (A, F, and J), 3 d (B, G, and K), 5 d (C, H, and L), and 7 d (D, I, and M) after cutting. (Scale bars, 100 μm in A–D, 1 mm in F–M.)
Roles of Auxin in Tissue Recovery in Incised Arabidopsis Stems.
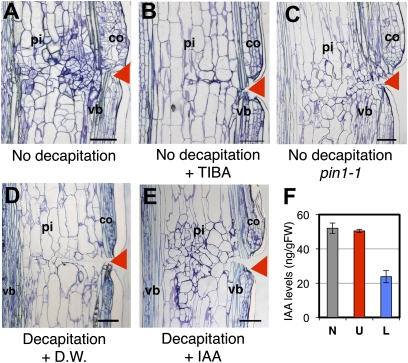
The level of auxin was histochemically analyzed using artificial auxin-responsive promoter (DR5)::GUS transgenic plants (Fig. 1 J–M). Strong GUS activity occurred at 1–5 d after cutting, with higher intensity in the upper region (Fig. 1 J–L), indicating that auxin accumulation may have occurred in the upper region immediately after cutting, lasting for 5 d and diminishing, and then tissue reunion completed within almost 7 d (Fig. 1 L and M). Auxin is derived from either cauline leaves, shoot apices, or lateral buds. Removal of both the shoot apex and lateral buds or only the cauline leaves resulted in minor inhibitory effects (Fig. S1 E and F), but simultaneous removal of all three severely retarded cell division and pcyclin B::GUS activity in the pith tissue (Fig. S1 D and H). Involvement of auxin in this phenomenon was substantiated by the fact that 10−3 M indole-3-acetic acid (IAA) application to the top of the decapitated stem nullified the inhibitory effect of decapitation (Fig. 2E). Furthermore, the healing processes in the wound tissue were also inhibited by the application of 10−3 M 2,3,5-triiodobenzoic acid (TIBA), an inhibitor for polar auxin transport, to the upper part of the cut gap (Fig. 2B). Similar effects were also observed in the polar auxin transport mutant pin1-1 (17) (Fig. 2C). In contrast, cortical cell division was activated without regard to the removal of the auxin-delivering tissues (Fig. S1 C–J).
Fig. 2.
Roles of auxin on tissue reunion in Arabidopsis wounded stems. (A and B) Control wild type without decapitation. (C) pin1-1. (D and E) Decapitated wild type. Lanolin paste containing distilled water (D.W.) (A and D), 10−3 M TIBA (B), or 10−3 M IAA (E) was applied. Arrowheads indicate position of cut. pi, pith; co, cortex; vb, vascular bundle. Images were taken 7 d after stem cutting. (Scale bars in A–E, 100 μm.) (F) Quantification of endogenous IAA in upper (U; red bar), lower (L; blue bar), or noncut region (N; gray bar). Mean ± SE is shown (n = 3).
Finally, the auxin level in the wound tissues was simultaneously measured by evaluating the expression of IAA5, an early auxin-inducible gene. Compared with the control, the IAA level was not altered in the upper region but was decreased by 50% in the lower region 1 d after cutting (Fig. 2F). Simultaneously, the IAA5 gene was strongly up-regulated exclusively in the upper region and then strongly reduced, possibly by negative feedback (Fig. S2A).
Microarray Analysis of the Tissue-Reunion Process and Gene Expression Profiles of ANAC071 and RAP2.6L.
Explants shorter than 5 mm with or without nicks were collected from the same internodes 1, 3, and 5 d after cutting and subjected to an oligonucleotide-based microarray to determine what genes initiate and regulate the tissue-reunion process (Fig. S3 and Dataset S1). Table S1 shows selected genes with expression patterns that correlate with tissue reunion.
The microarray analysis led us to focus on the expression of TFs that were highly up-regulated at an early stage. Two plant-specific TF genes were selected, ANAC071 (Arabidopsis NAC domain containing protein 71; At4g17980) encoding NAC domain-TF, and RAP2.6L (At5g13330) encoding an ERF/AP2 -TF (Fig. 3 A and B and Table S1). ANAC071 belongs to the NAC domain-TF family known to include 105 predicted NAC proteins in Arabidopsis (18), which are involved in the formation of organ primordia and other biological functions, including defense responses (19). RAP2.6L belongs to the APETALA2 (AP2)/ERBP family, which is one of the large families of TFs in Arabidopsis involved in many different developmental processes and environmental response events (20, 21). The family is composed of 144 members in Arabidopsis and has been divided into five subfamilies: the AP-2 subfamily, RAV subfamily, DREB (A) subfamily, ERF (ethylene response factor; B) subfamily, and others (22).
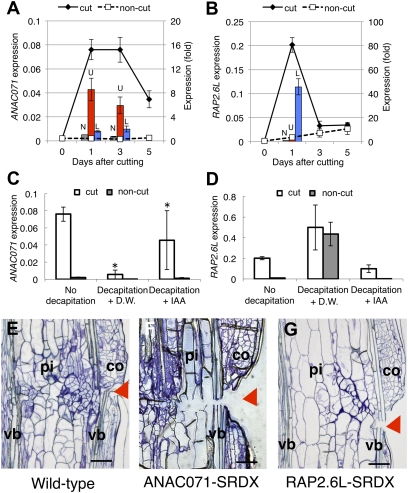
Fig. 3.
Gene expression of ANAC071 and RAP2.6L TFs and phenotypes of gene-suppressing transformants. (A and B) Gene expression of ANAC071 (A) and RAP2.6L (B). For time-course analysis, the mean ± SE is shown (n = 3). Bars show comparative expression between upper (U; red bar) or lower region (L; blue bar) of wounded stem. The noncut (N; gray bar) control was arbitrarily set to 1, and mean ± SE is shown (n = 4). (C and D) Effects of decapitation and IAA application on the expression of ANAC071 (C) or RAP2.6L (D). Mean ± SE is shown (n = 3). *P < 0.05 (Fisher's test). (E–G) Representative phenotype of SRDX transgenic plants. (E) Wild type. (F) ANAC071-SRDX. (G) RAP2.6L-SRDX. Images were taken 7 d after cutting. Arrowheads indicate position of cut. pi, pith; co, cortex; vb, vascular bundle. (Scale bars in E–G, 100 μm.)
The ANAC071 transcript was highly expressed 1–3 d after cutting, and the transcript was very intense in the upper cut region (Fig. 3A). Intensive expression of ANAC071 in the upper region was also observed in pANAC071::GUS transgenic plants, and the promoter activity was predominantly observed in pith and vascular tissue (Fig. S2B). The transcript level was drastically reduced after decapitation but was restored by IAA application (Fig. 3C).
The RAP2.6L transcript was transiently highly expressed 1 d after cutting, with exclusive expression in the lower region (Fig. 3B). The RAP2.6L transcript was scarce in the noncut stem (Fig. 3B). Decapitation strongly enhanced the level of the RAP2.6L transcript in the cut and noncut stems, and this effect was nullified by IAA application (Fig. 3D).
Histological Analysis of Transformants Defective in ANAC071 and RAP2.6L.
In general, plant TFs constitute a large family that shares highly conserved DNA-binding domains, resulting in much redundancy. Although gene-suppressing transformants are difficult to obtain for such genes (23), this can be overcome by using chimeric repressor silencing technology (CRES-T) (24). To determine the roles of ANAC071 and RAP2.6L in cell division, the gene-suppressing transformants of these genes, ANAC071-SRDX and RAP2.6L-SRDX, were prepared using the CRES-T method (Fig. S4 A and B). In ANAC071-SRDX, cell division was strongly inhibited but cell elongation was enhanced around the lesion area, resulting in incomplete tissue reunion (Fig. 3 E and F and Fig. S4 C and D). In RAP2.6L-SRDX, moderate inhibition was observed (Fig. 3 E and G and Fig. S4 E and F). Transformants with the plant expression vector alone showed normal reunion.
Involvement of Ethylene in Tissue Reunion and Gene Expression of RAP2.6L and ANAC071.
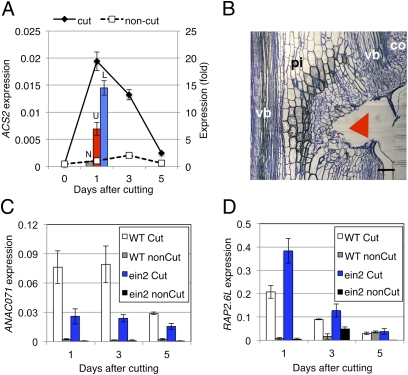
qRT-PCR analyses revealed that stem cutting promoted the expression of the aminocyclopropane carboxylic acid (ACC) synthase (ACS) gene, ACS2 (At1g01480), shortly (1 d) after cutting in both the lower and upper regions (Fig. 4A). Likewise, nearly equal promoter activity was observed in the upper and lower regions in the pACS2::GUS transgenic plants 1 d after cutting (Fig. S5A). ACS2 is one of the ACC synthases encoded by a multigene family in many plant species and produced in response to various environmental stimuli (25). Decapitation suppressed ACS2 transcript expression, although such suppression was not restored by exogenous IAA (Fig. S6). It is likely that the ACS2 gene is spatially regulated by either IAA or injury.
Fig. 4.
Involvement of ethylene in tissue reunion and expression of RAP2.6L and ANAC071. (A) Expression of ACS2. For time-course analysis, the mean ± SE is shown (n = 3). Bars show comparative expression between upper (U; red bar) and lower region (L; blue bar) of cut stem. The noncut (N; gray bar) control was arbitrarily set to 1, and the mean ± SE is shown (n = 4). (B) Light micrograph of ein2 cut stem. Images were taken 7 d after cutting. Arrowheads indicate position of cut. pi, pith; co, cortex; vb, vascular bundle. (Scale bar, 100 μm.) The image is a composite to form the complete figure. (C and D) Relative expression of ANAC071 (C) and RAP2.6L (D) in cut stem (white bar) or noncut stem (gray bar) of wild type, and cut stem (blue bar) or noncut stem (black bar) of ein2. Mean ± SD (n = 2) is shown.
To determine whether ethylene was involved in tissue reunion, the ethylene-insensitive mutant ein2 was investigated. Five days after cutting the ein2 stem, cell division was observed only in the cortex neighboring the cut and not in the pith, unlike in the wild-type stem (Fig. S5 B–D). Complete reunion was not observed up to 7 d after cutting the ein2 stem (Fig. 4B). ANAC071 expression in the ein2 cut was significantly lower than in the wild type (Fig. 4C), indicating that ANAC071 was up-regulated by ethylene. In contrast, RAP2.6L expression level was approximately two times higher in the ein2 than in wild type 1 d after cutting (Fig. 4D), indicating that RAP2.6L was down-regulated by ethylene.
Regulation of RAP2.6L by JA.
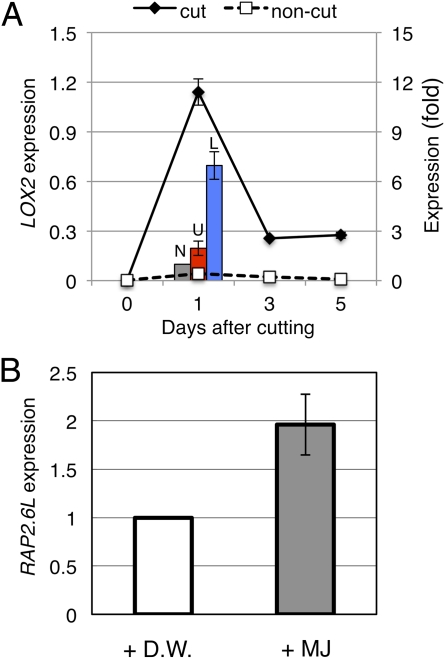
Microarray and qRT-PCR analyses showed that some JA biosynthesis genes were up-regulated during the tissue-reunion process. LOX2 [encoding lipoxygenase 2 (At3g45140), a JA biosynthesis enzyme] was selected because this gene was exclusively expressed in wounded tissue (Fig. 5A) and is known to be wound-inducible (26). LOX2 expression peaked 1 d after stem cutting, with higher expression in the lower cut region (Fig. 5A) and a superimposable expression profile with RAP2.6L (Fig. 3B). RAP2.6L was up-regulated by JA methyl ester upon administration to the intact flowering stem (Fig. 5B), in accord with the AtGenExpress microarray database (27).
Fig. 5.
Regulation of RAP2.6L expression by JA. (A) Gene expression of LOX2. For time-course analysis, the mean ± SE is shown (n = 3). Bars show comparative expression between upper (U; red bar) and lower region (L: blue bar) of cut stem. The noncut stem control (N; gray bar) was arbitrarily set to 1, and the mean ± SE is shown (n = 4). (B) Effects of methyl jasmonate (MJ) application on the expression of RAP2.6L in the intact flowering stem. The distilled water (D.W.) control was arbitrarily set to 1, and the mean ± SE is shown (n = 3).
Discussion
Recovery of Wounded Inflorescence Stems in Arabidopsis Is Different from That of Cucumber and Tomato Hypocotyls.
As found in our earlier work (11), injured cucumber or tomato hypocotyls never healed when cotyledons were removed. This was due to GA depletion caused by removal of the GA-supplying cotyledons (11, 12). Exogenous GA, but not IAA, can replace the cotyledons. Hypocotyls are embryonic organs consisting of the epidermis, cortex, endodermis, and vascular tissues, but lacking a pith. Its growth requires import of nutrients and hormones from cotyledons because of its juvenility. In cucumber and tomato hypocotyls, injury recovery is accompanied by cell division and elongation mainly in the cortical cells, which characteristically have pectin-rich thick cell walls.
Before the present study, no involvement of GA was found in wound healing because the wounded stems of the GA-deficient Arabidopsis mutant ga3ox1/ga3ox2 recovered with no difficulty. The tissue-reunion mechanism likely changes as developmental stages proceed. Arabidopsis inflorescence stem internodes analyzed in this study had no dividing cells and consisted of the epidermis, cortex, endodermis, vascular tissues, and a pith occupying much of the stem center. In the stems, cell proliferation-based reunion of the wounded area proceeded mainly in the pith, in contrast to the events in hypocotyls (Fig. 1). Involvement of auxin in healing the injured stems was substantiated by the fact that auxin-deprived stems, because of TIBA treatment or pin1-1 mutation, were not capable of wound healing. Interestingly, cortical cell division occurred in both the upper and lower cut stem regions and was not affected by decapitation, auxin transport inhibition (Fig. 2), or GA biosynthesis inhibition, or in GA-deficient mutants.
Upper Cut Region of Wounded Inflorescence Stems Was Healed by ANAC071.
Microarray analysis followed by qRT-PCR of wounded stems revealed two TF genes, ANAC071and RAP2.6L, that were strongly up-regulated immediately after artificial wounding. Furthermore, ANAC071and RAP2.6L were spatially selectively expressed in the upper and lower cut regions, respectively (Fig. 3 A and B). The requirement of these TFs for pith cell division in tissue reunion was indicated by analyzing TF gene-suppressing transformants (Fig. 3 E–G and Fig. S4).
ANAC071 was most likely induced by auxin because auxin accumulated in the upper cut region, and ANAC071 was not present in the auxin-poor lower region. Furthermore, this finding is supported by a previous report that ANAC071-TF and related NAM (no apical meristem)-like TFs are up-regulated in auxin-rich callus induction medium (28, 29). Furthermore, auxin and microRNAs may regulate the spatial patterns of the transcription of some NAC family genes, including CUC1 and CUC 2 (30). A group of microRNAs was also predicted to target the Populus homolog of ANAC071 (31). The biological functions of NAC-TFs remain to be determined, but several NAC-TFs may be involved in shoot meristem formation, organ boundary specification, and secondary wall thickening (10, 32, 33), together with abscisic acid-mediated stress responses against drought and high salinity (18, 19, 34).
Ethylene, a wound-inducible hormone, was also an important factor activating ANAC071 because ANAC071-TF expression was significantly reduced in the ein2 mutant (Fig. 4C), which had unusual reunion morphology (Fig. 4B). ACC, the direct precursor of ethylene, is synthesized by ACSs in response to biotic or abiotic stimuli, and the ACSs are rate-limiting enzymes in ethylene production. Although numerous ACS genes are known, ACS2 was specifically up-regulated in both the upper and lower cut regions in this study (Fig. 4A). Decapitation decreased ACS2 expression in the Arabidopsis flowering stem, although this negative effect was not fully compensated after IAA application (Fig. S6A). Taken together, the results suggest that auxin activated ANAC071 and ethylene enhanced ANAC071 expression in the auxin-rich upper cut region, inducing pith cell division during tissue reunion (Fig. S7).
Lower Cut Region of Wounded Inflorescence Stems Was Healed by RAP2.6L-TF.
The RAP2.6L gene is thought to regulate genes involved in meristem maintenance during shoot regeneration, because T-DNA knockdown of RAP2.6L reduced the frequency of shoot development from root explants and hampered the expression of shoot meristem-specific genes (28, 29). In the present study, RAP2.6L gene expression appeared in the lower cut region, which had a lower IAA level due to blocked polar auxin transport. It is likely that RAP2.6L was up-regulated in response to wounding in a region below the cut where auxin concentrations were low. Furthermore, RAP2.6L was not expressed in the IAA-rich upper cut region, indicating that it was down-regulated by IAA. In support of this conclusion, Che et al. (28, 29) clearly showed that RAP2.6L is up-regulated in cytokinin-rich, shoot-generating media for Arabidopsis root explants but not in auxin-rich, root-inducing media.
LOX2 was also thought to be involved in injury healing because it was up-regulated in the lower cut region immediately after cutting (Fig. 5A). LOX2 is a lipoxygenase participating in JA synthesis that converts linolenic acid to its hydroperoxide. The plant hormone JA is a key regulator of plant responses to environmental stresses and biotic challenges (3). RAP2.6L-TF expression was enhanced upon application of the JA derivative methyl jasmonate to the flowering stems (Fig. 5B). However, decapitation with or without IAA application did not affect the expression of LOX2 during the reunion process (Fig. S6B). It is most likely that this gene is activated by wounding independently of IAA. However, it is not known why LOX2 was not expressed in the upper cut region. On the other hand, RAP2.6L seems to be down-regulated by ethylene because its expression was increased in the ein2 mutant (Fig. 4D). In conclusion, in the lower cut region, RAP2.6L was activated because of auxin depletion, and this activation process was positively regulated by JA but negatively regulated by ethylene, resulting in the net incremental increase in RAP2.6L expression (Fig. S7), facilitating pith cell division during tissue reunion.
Materials and Methods
Arabidopsis thaliana seeds were germinated and grown in artificial soil under continuous white fluorescent light. After 7–10 d of bolting, stem internodes between the first or second cauline leaves and rosette leaves were cut through half of their diameter with a microsurgical knife (Fig. S1 A and B), and the plant was then grown for an additional 14 d. Detailed experimental procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Hiroshi Ezura (University of Tsukuba, Japan) and Dr. Eiji Nambara (University of Toronto, Canada) for the kind gifts of plant materials, Ms. Miho Shimizu (University of Tsukuba) for technical assistance, and Drs. Masao Tasaka and Miyo Terao-Morita (Nara Institute of Science and Technology, Japan) for valuable advice on auxin. This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences and a Grant-in-Aid for Scientific Research on Priority Areas (21027004 to S.S.) and by a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists (04J11879 to M.A.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110443108/-/DCSupplemental.
References
- 1.Satoh S. Organic substances in xylem sap delivered to above-ground organs by the roots. J Plant Res. 2006;119:179–187. doi: 10.1007/s10265-005-0257-8. [DOI] [PubMed] [Google Scholar]
- 2.Kramer EM, Bennett MJ. Auxin transport: A field in flux. Trends Plant Sci. 2006;11:382–386. doi: 10.1016/j.tplants.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 3.León J, Rojo E, Sánchez-Serrano JJ. Wound signalling in plants. J Exp Bot. 2001;52:1–9. doi: 10.1093/jexbot/52.354.1. [DOI] [PubMed] [Google Scholar]
- 4.Sachs T. Integrating cellular and organismic aspects of vascular differentiation. Plant Cell Physiol. 2000;41:649–656. doi: 10.1093/pcp/41.6.649. [DOI] [PubMed] [Google Scholar]
- 5.Flaishman MA, Loginovsky K, Lev-Yadun S. Regenerative xylem in inflorescence stems of Arabidopsis thaliana. J Plant Growth Regul. 2003;22:253–258. [Google Scholar]
- 6.Davis AR, et al. Cucurbit grafting. Crit Rev Plant Sci. 2008;27:50–74. [Google Scholar]
- 7.Kobayashi Y, Weigel D. Move on up, it's time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- 8.Katsumi M, Foard DE, Phinney BO. Evidence for the translocation of gibberellin A3 and gibberellin like substances in graft between normal, dwarf1 and dwarf5 seedling of Zea mays L. Plant Cell Physiol. 1983;24:379–388. [Google Scholar]
- 9.Notaguchi M, Daimon Y, Abe M, Araki T. Graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Signal Behav. 2009;4:123–125. doi: 10.4161/psb.4.2.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi M, Kubo M, Fukuda H, Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008;55:652–664. doi: 10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- 11.Asahina M, et al. Gibberellin produced in the cotyledon is required for cell division during tissue reunion in the cortex of cut cucumber and tomato hypocotyls. Plant Physiol. 2002;129:201–210. doi: 10.1104/pp.010886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asahina M, et al. Effects of the removal of cotyledons on endogenous gibberellin levels in hypocotyls of young cucumber and tomato seedlings. Plant Biotechnol. 2007;24:99–106. [Google Scholar]
- 13.Asahina M, Gocho Y, Kamada H, Satoh S. Involvement of inorganic elements in tissue reunion in the hypocotyl cortex of Cucumis sativus. J Plant Res. 2006;119:337–342. doi: 10.1007/s10265-006-0278-y. [DOI] [PubMed] [Google Scholar]
- 14.Zimmerman JL. Somatic embryogenesis: A model for early development in higher plants. Plant Cell. 1993;5:1411–1423. doi: 10.1105/tpc.5.10.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD. Organ regeneration does not require a functional stem cell niche in plants. Nature. 2009;457:1150–1153. doi: 10.1038/nature07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 17.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 18.Ooka H, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 19.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Sakuma Y, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 21.Iwase A, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol. 2011;21:508–514. doi: 10.1016/j.cub.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- 23.Riechmann JL, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 24.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 25.Tsuchisaka A, Theologis A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 2004;136:2982–3000. doi: 10.1104/pp.104.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suza WP, Staswick PE. The role of JAR1 in Jasmonoyl-L: -isoleucine production during Arabidopsis wound response. Planta. 2008;227:1221–1232. doi: 10.1007/s00425-008-0694-4. [DOI] [PubMed] [Google Scholar]
- 27.Goda H, et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 28.Che P, Lall S, Howell SH. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta. 2007;226:1183–1194. doi: 10.1007/s00425-007-0565-4. [DOI] [PubMed] [Google Scholar]
- 29.Che P, Lall S, Nettleton D, Howell SH. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 2006;141:620–637. doi: 10.1104/pp.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aida M, Tasaka M. Genetic control of shoot organ boundaries. Curr Opin Plant Biol. 2006;9:72–77. doi: 10.1016/j.pbi.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Barakat A, Wall PK, Diloreto S, Depamphilis CW, Carlson JE. Conservation and divergence of microRNAs in Populus. BMC Genomics. 2007;8:481. doi: 10.1186/1471-2164-8-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsuda N, et al. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen MK, et al. The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem J. 2010;426:183–196. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.