Abstract
Phosphatidylserine (PS) is a relatively minor constituent of biological membranes. Despite its low abundance, PS in the plasma membrane (PM) plays key roles in various phenomena such as the coagulation cascade, clearance of apoptotic cells, and recruitment of signaling molecules. PS also localizes in endocytic organelles, but how this relates to its cellular functions remains unknown. Here we report that PS is essential for retrograde membrane traffic at recycling endosomes (REs). PS was most concentrated in REs among intracellular organelles, and evectin-2 (evt-2), a protein of previously unknown function, was targeted to REs by the binding of its pleckstrin homology (PH) domain to PS. X-ray analysis supported the specificity of the binding of PS to the PH domain. Depletion of evt-2 or masking of intracellular PS suppressed membrane traffic from REs to the Golgi. These findings uncover the molecular basis that controls the RE-to-Golgi transport and identify a unique PH domain that specifically recognizes PS but not polyphosphoinositides.
Keywords: cholera toxin, endocytosis
Phosphatidylserine (PS) is an anionic phospholipid class in eukaryotic biomembranes. PS is highly enriched in the plasma membrane (PM) and plays key roles in various physiological processes such as the coagulation cascade, recruitment and activation of signaling molecules, and clearance of apoptotic cells (1). These functions of PS are known to be executed by a number of proteins that have PS-recognition modules, such as a gamma-carboxyglutamic acid domain in prothrombin (2), a C2 domain in protein kinase C (3), a discoidin-type C2 domain in lactadherin (4), and a kinase associated-1 domain in MARK/PAR1 kinases (5). PS is also found in endocytic organelles (4, 6), where its functions are largely unknown.
Proteins newly synthesized at the endoplasmic reticulum (ER) that are destined for secretion or for residence within organelles move through the Golgi to their final destinations (7). This membrane outflow is counteracted by retrograde membrane flow that originates from either the PM or the endosomal system (8). The retrograde membrane traffic to the Golgi is used by several Golgi proteins to maintain their predominant Golgi localization, such as mannose 6-phosphate receptors (acid-hydrolase receptors), furin (a transmembrane enzyme), TGN38/46 (trans-Golgi resident proteins), and Wntless (a sorting receptor for Wnt) (9, 10). Intriguingly, some protein toxins produced by bacteria and plants, e.g., cholera toxin, Shiga toxin, and ricin, highjack this retrograde traffic to reach the cytosol, where they exert their toxicity (11, 12). The retrograde membrane traffic from the PM to the Golgi is known to pass through early endosomes (EEs)/recycling endosomes (REs) (10, 13). The molecular mechanism underlying this traffic has just begun to be elucidated (8, 9).
Here we report that PS is essential for retrograde membrane traffic at REs. PS was most concentrated in REs among intracellular organelles, and evectin-2 (evt-2), a protein of previously unknown function, was targeted to REs by the binding of its pleckstrin homology (PH) domain to PS. Depletion of evt-2 or masking of intracellular PS suppressed membrane traffic from REs to the Golgi. These findings uncover the molecular basis that controls the retrograde transport through REs, and identify a PH domain that specifically recognizes PS.
Results and Discussion
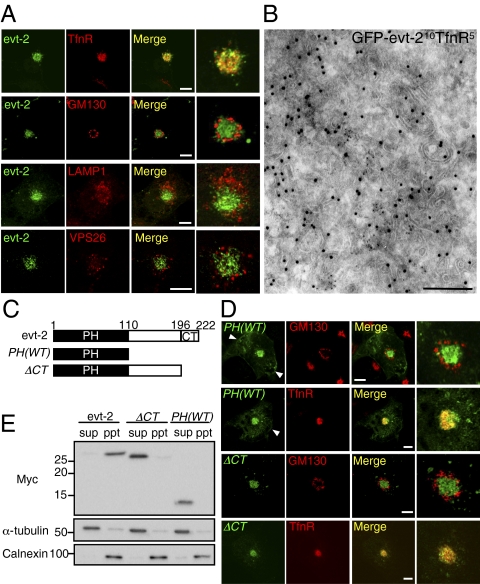
Evectin-1 (evt-1) and -2 are identified as post-Golgi proteins of unknown function (14) that have a PH domain that typically binds polyphosphoinositides (15). Evt-2 is expressed in a broad range of tissues, whereas evt-1 is expressed specifically in the nervous system. We first examined the subcellular localization of evt-2 using COS-1 cells in which organelles are spatially well separated (16, 17) (Fig. S1). Evt-2 colocalized with an RE marker, transferrin receptor (TfnR), but not with a Golgi marker (GM130), a lysosomal marker (LAMP1), or an early endosomal marker (VPS26) (Fig. 1A, for quantification see Fig. S2A). By immunoelectron microscopy using ultrathin cryosections, GFP–evt-2 (labeled with 10 nm gold particles) was specifically detected in tubulovesicular structures, and TfnR (labeled with 5 nm gold particles) was detected in some of them (Fig. 1B and Fig. S3). We thus concluded that evt-2 is localized predominantly to REs. Evt-2 also colocalized with the RE marker TfnR in HeLa cells (Fig. S4A).
Fig. 1.
A PH-domain–containing protein, evt-2, localizes to REs. (A) Evt-2 tagged with Myc was transiently expressed in COS-1 cells. The cells were then fixed and stained for Myc, TfnR, GM130, LAMP1, and VPS26. Magnified images around the Golgi/REs region are shown in the Right column. (Scale bars, 10 μm.) (B) Electron micrograph of an ultrathin cryosection of COS-1 cells. Cells expressing GFP–evt-2 were labeled with antibodies against GFP (10 nm in diameter) and TfnR (5 nm in diameter). (Scale bar, 250 nm.) (C) Domain structures of evt-2 and its truncation mutants. Myc-epitope was added to all of the constructs at the N terminus. (D) PH(WT) and ΔCT were transiently expressed. The cells were then fixed and stained for Myc, GM130, and TfnR. Arrowheads indicate PM. (Scale bars, 10 μm.) (E) Evt-2, ΔCT, and PH(WT) were transiently expressed in COS-1 cells for 24 h. Cell lysates were spun at 100,000 g for 60 min at 4 °C, and the resultant supernatant (sup) and pellet (ppt) were immunoblotted with anti-Myc antibody. α-Tubulin and calnexin were immunoblotted as a control.
Evt-2 has an N-terminal PH domain and a C-terminal hydrophobic region (CT) (Fig. 1C). PH(WT), a Myc-tagged evt-2 PH domain, was targeted to REs and to the PM to some extent (Fig. 1D). The PH domain is thus sufficient for evt-2 targeting to REs. ΔCT was exclusively localized to REs as evt-2, suggesting that the domain between PH and CT constrains the RE localization of evt-2. Evt-2 was recovered in the pellet after ultracentrifugation of cell lysate, whereas truncation mutants (ΔCT and PH(WT)) were found in the supernatant (Fig. 1E), showing that CT is required for evt-2 association with membranes.
The human proteome has ∼300 proteins with PH domains. About 10% of these proteins bind specifically to phosphatidylinositol phosphates (PIPs) through their PH domains, whereas the ligands of the rest of these proteins remain unclear (15). To determine the lipid specificity of evt-2 PH, we measured the binding of several negatively charged lipids on liposomes to recombinant evt-2 PH. Unexpectedly, PS bound to the PH, but phosphatidic acid, phosphatidylinositol, sulfatide, and all PIPs did not (Fig. 2 A and B). Lys20 is highly conserved in other PH domains (18). Evt-2 PH (K20E) lost the ability to bind to PS.
Fig. 2.
Evt-2 PH binds to PS. (A and B) His-tagged evt-2 PH (WT or K20E) was mixed with liposomes harboring the indicated negatively charged lipid (20% mol/mol of total lipids). After 15 min, the mixture was spun at 100,000 g for 30 min, and the resultant supernatant (S) and pellet (P) were subjected to SDS-PAGE. The gels were stained with Coomassie blue (A). The intensities of individual bands were quantitated with NIH ImageJ. The values of “P (bound)/S (free)” are shown in a bar graph (B). Data represent mean values ± SD of three independent experiments. PE, phosphatidylethanolamine; PC, phosphatidylcholine; PA, phosphatidic acid; PI, phosphatidylinositol. (C) Confocal images of wild-type yeast cells and cho1Δ cells expressing GFP-tagged 2 × PH (a tandem fusion of evt-2 PH) or GFP–Lact-C2. (Scale bars, 10 μm.) (D) COS-1 cells were transfected with GFP–Lact-C2, then fixed, and stained for GM130 or TfnR. Arrowheads indicate PM. (Scale bars, 10 μm.)
A Saccharomyces cerevisiae mutant (cho1Δ) deficient in PS (19, 20) is used to analyze protein binding to PS in vivo (4, 5). We expressed evt-2 PH in wild-type yeast and found that it was not associated with the PM. Because a tandem fusion of lipid-binding modules, such as the FYVE domain of EEA1 and Hrs, greatly enhances the lipid-binding affinity of the FYVE domain (21), we generated a tandem evt-2 PH (2 × PH) and expressed it in both the wild-type and cho1Δ mutant. 2 × PH was observed predominantly on the PM of the wild-type yeast, whereas it was cytosolic in the PS-deficient mutant (Fig. 2C). The C2 domain of lactadherin (Lact-C2), a specific probe for PS (4), was used as a positive control. These findings showed that evt-2 PH recognizes PS in vivo.
Several intracellular organelles possess unique phospholipids such as PIPs, although the specific phospholipids in REs are not well characterized (22). We therefore expressed a variety of phospholipid probes in cells to see their distribution. None of the PIP probes stained REs (Fig. S5), but Lact-C2 predominantly stained REs and the PM (Fig. 2D, for quantification see Fig. S2B), revealing that REs are most enriched with PS among intracellular organelles. The enrichment of PS in REs was also confirmed in HeLa cells (Fig. S4B). Given the ligand specificity of the evt-2 PH, the binding of evt-2 PH to PS is likely to be involved in evt-2 localization to REs.
We were able to grow crystals of human evt-2 PH (a recombinant 110-amino acid domain) in complex with O-phospho-l-serine, the head group of PS, and analyzed them by X-ray crystallography at 1.0 Å resolution. The data collection and refinement statistics are summarized in Table S1. The overall structure was similar to the standard PH domain fold, with seven β strands forming two orthogonal antiparallel β sheets and two α helices containing the major C-terminal α helix (Fig. 3A). O-phospho-l-serine binds to the positively charged pocket made by three basic residues (Arg11, Arg18, and Lys20) and the backbone nitrogen atoms of three residues (Thr14, Ile15, and Leu16) of the β1/β2 loop (Fig. 3 B and C and Table S2). Arg11 and Arg18 each make two salt bridges with the l-serine oxygen atoms and the phosphate oxygen of the ligand, respectively (Fig. 3B and Table S2). In addition, Lys20 makes salt bridges with both moieties of the ligand. The nitrogen atom of O-phospho-l-serine forms a salt bridge with the side chain of Glu44 in one of the two conformers in the crystal (Fig. S6A).
Fig. 3.
High-resolution structure of evt-2 PH bound to phosphoserine. (A) Overall structure of human evt-2 PH in complex with O-phospho-l-serine, consisting of seven β strands and two helices. The amino and carboxy termini are denoted in red as N and C, respectively. O-phospho-l-serine is shown as a stick model. (B) Stereoview of the O-phospho-l-serine binding site of evt-2 PH. Interacting residues are shown as stick models. A σa-weighted Fo-Fc omit map (3.0 σ level; in gray mesh) is superposed on O-phospho-l-serine. Hydrogen bonds and salt bridges are shown as red broken lines. (C) Charge distribution surface model of evt-2 PH in complex with O-phospho-l-serine (stick model). The surface is colored according to the electrostatic potential of the residues (blue, positive; red, negative). Only one of the double conformers of Glu44 is shown.
We next examined whether the amino acid residues involved in the ligand binding in the crystal are essential for evt-2 localization to REs. The full-length point mutant K20E, in which Lys20 was changed to Glu, lost RE localization and showed puncta around the Golgi (Fig. 4A, for quantification see Fig. S2A). These puncta did not colocalize with TfnR (REs), but colocalized in part with VPS26 (EEs) and CD63 (late endosomes,LEs). The PH point mutant of Arg11, Arg18, or Lys20 (PH(R11E), PH(R18E), and PH(K20E)) did not show any specific membrane localization (Fig. 4B), suggesting that these residues bind to the head group of PS in vivo. All of the residues involved in the direct interaction of the ligand (Table S2) were conserved in the evt-2 PH of other species (Fig. 4C), further implicating these residues in the recognition of PS.
Fig. 4.
The amino acid residues involved in the ligand binding in the crystal are essential for evt-2 localization to REs. (A) The full-length point mutant K20E, in which Lys20 was changed to Glu, was transiently expressed in COS-1 cells. The cells were then fixed and stained for Myc, GM130, TfnR, VPS26, and CD63. Arrowheads indicate puncta where K20E and endosomal markers are juxtaposed or colocalized. (Scale bars, 10 μm.) (B) The PH point mutants PH(K20E), PH(R11E), and PH(R18E), in which Lys20, Arg11, or Arg18 was changed to Glu, were transiently expressed. The cells were then fixed and stained for Myc and GM130. (Scale bars, 10 μm.) (C) Alignment of vertebrate evt-2 PH domains. The secondary structure elements of human evt-2 PH are shown above the sequence. Conserved residues are boxed in white on a black background. Similar residues are boxed in black with a white background. Asterisks indicate the residues directly involved in the interaction of the ligand.
Two membrane trafficking pathways pass through REs: one is a recycling pathway and the other is a retrograde pathway that links the PM to the Golgi/ER (10). We examined whether evt-2 is involved in these pathways. Evt-2 knockdown did not cause any gross change in Tfn recycling to the PM (Fig. S7 A and B). Cholera toxin travels from the PM through endosomes (EEs/REs) to the Golgi/ER, and is finally translocated into the cytosol (11). Alexa 594-labeled cholera toxin B subunit (CTxB) was pulsed for 5 min and chased (Fig. 5A). After a 5-min uptake, CTxB accumulated around the Golgi, colocalizing in part with VPS26, showing that it reached EEs (Fig. S8A). After a 15-min pulse/chase, CTxB accumulated at the cell center, colocalizing with Tfn, showing it reached REs (Fig. S8B). After a 60–90 min pulse/chase, CTxB showed a good colocalization with GM130. These results showed the sequential retrograde transport of CTxB from the PM to the Golgi through EEs and then REs. In cells depleted of evt-2 by using siRNA 1 (Fig. 5B), CTxB transport to REs proceeded normally after a 15-min pulse/chase, but CTxB transport from REs to the Golgi was significantly impaired after a 60- or 90-min pulse/chase. The other siRNA oligos 2 and 3 also impaired CTxB traffic to the Golgi (Fig. S7C, for quantification see D). Thus, evt-2 is required for retrograde transport of CTxB from REs to the Golgi.
Fig. 5.
Evt-2 and PS regulate retrograde transport through REs. (A) COS-1 cells were pulsed for 5 min at 37 °C with Alexa 594-CTxB and chased. Cells were then fixed at the indicated times from the beginning of the pulse, and stained for GM130. (Scale bars, 10 μm.) (B) Cells were treated with evt-2 siRNA 1 or control siRNA for 48 h. The pulse/chase of CTxB was performed as described in A. (Scale bars, 10 μm.) (C) Cells were treated with evt-2 siRNA 1 or mock treated for 48 h. Cells were then incubated with 35S and CTxB-GS at 37 °C. CTxB-GS was immunoprecipitated and analyzed by autoradiography (Upper). A parallel experiment on Vero cells was carried out as control. The Lower band (II) represents the sulfated, but nonglycosylated CTxB. The Upper band (I) is N glycosylated. Thus, CTxB traveled from the PM to the Golgi and a significant fraction of the Golgi-modified protein moved to the ER. In cells depleted of evt-2, the intensity of 35S-labeling of CTxB at both 1 and 3 h was reduced by 62–68%. In contrast, transport of CTxB beyond the Golgi to the ER was not affected (about 10% of the sulfated CTxB was glycosylated in both conditions). (Lower) Immunoblot (WB) of samples using an antibody against CTxB as loading control for autoradiogram. Top band (III) shows CTxB-GS. Bands labeled IV are degradation products. (D) Cells were treated with either evt-2 siRNA 1 or transfection reagent only for 48 h. Following addition of 1 nM cholera toxin, intracellular cAMP levels were measured at 50-min time point: evt-2 siRNA-treated cells (gray column), control cells (transfection reagent only, black column), and the cells without cholera toxin addition (white column). Data represent mean values of three independent experiments. (E) Cells treated with either evt-2 siRNA 1 or control siRNA were fixed and costained for GM130/TGN46 or GP73/GM130. Magnified images around the Golgi/REs region in one of the cells for each condition are shown in the Right column. (Scale bars, 10 μm.) (F) Cells were cotransfected with evt-2 siRNA 1 and siRNA-resistant mouse evt-2 constructs tagged with Myc (WT or K20E). After 72 h, cells were fixed and costained for Myc/TGN46. Arrowheads indicate cells expressing evt-2, and asterisks indicate nonexpressing cells. (Scale bars, 10 μm.) (G) Transient expression of GFP–Lact-C2. Cells were pulsed for 5 min with Alexa 594-CTxB, chased, fixed at 60 min after the beginning of the pulse, and stained for GM130. The arrowhead indicates a cell expressing GFP–Lact-C2, and the asterisk indicates a nonexpressing cell. (Scale bar, 10 μm.)
The effect of evt-2 knockdown on retrograde transport was also examined by biochemical assays using a mutant CTxB (CTxB-GS) harboring tyrosine sulfation and N-glycosylation sites to monitor its arrival at the Golgi or ER (23), and using cholera toxin to measure the increase of intracellular cAMP that occurs when the toxin arrives in the cytosol. Both assays confirmed the impaired retrograde traffic of cholera toxin upon depletion of evt-2 (Fig. 5 C and D and Fig. S7E).
Evt-2 depletion did not significantly alter the subcellular localizations of a Golgi marker (GM130), a lysosomal marker (LAMP2), LE markers [CD63 and cation-independent mannose 6-phosphate receptor (CI-MPR)], or an RE marker (TfnR) (Fig. S7F). However, Golgi proteins TGN46 and GP73 no longer localized to the Golgi, but localized on punctate structures upon evt-2 depletion (Fig. 5E, for quantification see Fig. S2C). These two proteins are known to circulate between the Golgi and the PM through endosomes (10, 24). When retrograde transport from REs to the Golgi is impaired by depletion of evt-2, TGN46 and GP73 may be shunted to circulate among PM, EEs, and REs, resulting in their puncta localization.
Next, we performed knockdown/rescue experiments. Cells were depleted of evt-2 with siRNA 1 and transfected with an siRNA-resistant evt-2 construct (a mouse evt-2 WT or K20E mutant defective in PS binding). Golgi localization of TGN46 was restored in the cells transfected with mouse evt-2 WT (Fig. 5F). In contrast, TGN46 remained scattered throughout the cytosol in the cells transfected with evt-2 K20E mutant. Therefore, the binding of evt-2 PH to PS is essential for evt-2 function in endosomal membrane trafficking.
We further examined how blocking PS would affect endosomal membrane transport. Lact-C2 was overexpressed to mask PS in the cytosolic leaflet of REs. CTxB transport to the Golgi was severely delayed and accumulated in REs in the cells transfected with Lact-C2 (Fig. 5G, for quantification see Fig. S2D), showing that exposure of PS in the cytosolic surface of REs is required for CTxB transport from REs to the Golgi.
Recent studies in yeast and mammalian cells have established that the retromer complex at EEs has a critical function in retrograde membrane traffic to the Golgi (8, 25). In this study, besides EEs, we showed that REs are involved in this membrane traffic for certain cargo proteins. Retrograde and recycling traffickings diverge at REs, with a previously uncharacterized PH-domain–containing protein evt-2, which is essential and specific for retrograde traffic. We found that GP73, which circulates between the Golgi and the PM through endosomes, binds to evt-2 (Fig. S9). Together with the fact that evt-2 depletion abolished GP73 localization at the Golgi (Fig. 5E), we assume that evt-2 functions as a sorting device at REs to recruit specific cargo molecules that follow retrograde transport to the Golgi. We and other groups have shown that REs also play an obligatory role in the exocytic pathway for various secretory cargos (17, 26, 27). REs, therefore, can handle three membrane trafficking pathways: the recycling pathway (EEs → REs → PM), the exocytic pathway (the Golgi → REs → PM), and the retrograde pathway (EEs → REs → the Golgi).
The PH domain in evt-2 specifically binds PS, but not PIPs. What is the structural basis underlying the specificity of evt-2 PH to PS? All high-affinity, stereospecific PH domains for PIPs share a similar binding site (15). The β1/β2 loop functions as a platform for the interaction with the head group of the ligand. This loop lines a deep binding pocket and contains the sequence motif KXn(K/R)XR (where X is any amino acid), in which the basic side chains participate in most phosphate-group interactions. The corresponding motif in evt-2 PH is R11XnK20XN, where italics indicate changes, and of note, the two nitrogen atoms (Nη1 and Nη2) of Arg11 make two salt bridges neatly with the carboxyl group of O-phospho-l-serine, which is absent in PIPs. A search for 3D structures homologous to human evt-2 PH using the DALI server (28) yielded the DAPP1/PHISH PH domain in complex with Ins(1,3,4,5)P4 (Protein Data Bank code, 1FAO) as the best match (29). A comparison of the structures of the two complexes suggests that some evt-2 PH amino acids would clash with the head group of Ins(1,3,4,5)P4 (Fig. S6 B and C). Thr14, Ile15, and Leu16 of evt-2 PH are close to the ligand, and the ligand-binding site is much smaller than that of DAPP1 PH. Therefore, the tightness of the ligand-binding site might account for the specificity of evt-2 PH for PS. Several simultaneous recognitions were identified in evt-2 PH (Fig. 3B and Table S2). Among them, Lys20 and Ile15 appear particularly important because they recognize both the l-serine and phosphate regions of the ligand. Simultaneous recognition of multiple regions of a ligand by interacting residues might enhance the binding affinity and specificity.
In this study, we showed that PS recognition by the PH domain of evt-2 is essential for endosomal membrane transport from the PM to the Golgi. The data presented here provide compelling evidence that intracellular PS has a critical role in membrane traffic and uncover the molecular basis that controls the RE-to-Golgi transport.
Materials and Methods
Cell Culture and Transfection.
COS-1 cells were cultured at 37 °C with 5% CO2 in DMEM containing 10% heat-inactivated FCS. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Structure Determination.
The complex structure of human evt-2 PH with O-phospho-l-serine was determined by the molecular replacement method at 1.0 Å resolution using the data collected at beamline AR-NW12A of the Photon Factory. The crystal belongs to space group P21, with a = 31.7 Å, b = 48.4 Å, c = 64.3 Å, and β = 92.2°. The coordinates and structure factors of the human evt-2 PH structure have been deposited in the Protein Data Bank with the accession code 3AJ4.
Additional materials and methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
A special thanks to Wendy Hamman for help with tissue culture and transfection conditions. This work was supported by the Core Research for Evolutional Science and Technology, Japan Science and Technology Agency (H.A. and T.T.), the Program for Promotion of Basic and Applied Research for Innovations in Bio-Oriented Industry (H.A.), the 21st Century Center of Excellence Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.T.), Grants-in-aid for Scientific Research (20370045 to H.A. and 18050019 to T.T.), and a Senri Life Science Foundation Grant (to T.T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3AJ4).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109101108/-/DCSupplemental.
References
- 1.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 2.Huang M, et al. Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins. Nat Struct Biol. 2003;10:751–756. doi: 10.1038/nsb971. [DOI] [PubMed] [Google Scholar]
- 3.Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gómez-Fernández JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 5.Moravcevic K, et al. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gagescu R, et al. The recycling endosome of Madin-Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell. 2000;11:2775–2791. doi: 10.1091/mbc.11.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellman I, Warren G. The road taken: Past and future foundations of membrane traffic. Cell. 2000;100:99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- 8.Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 9.Johannes L, Popoff V. Tracing the retrograde route in protein trafficking. Cell. 2008;135:1175–1187. doi: 10.1016/j.cell.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 11.Lencer WI, Tsai B. The intracellular voyage of cholera toxin: Going retro. Trends Biochem Sci. 2003;28:639–645. doi: 10.1016/j.tibs.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Sandvig K, van Deurs B. Membrane traffic exploited by protein toxins. Annu Rev Cell Dev Biol. 2002;18:1–24. doi: 10.1146/annurev.cellbio.18.011502.142107. [DOI] [PubMed] [Google Scholar]
- 13.Mallard F, et al. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of shiga toxin B-fragment transport. J Cell Biol. 1998;143:973–990. doi: 10.1083/jcb.143.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krappa R, Nguyen A, Burrola P, Deretic D, Lemke G. Evectins: Vesicular proteins that carry a pleckstrin homology domain and localize to post-Golgi membranes. Proc Natl Acad Sci USA. 1999;96:4633–4638. doi: 10.1073/pnas.96.8.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 16.Misaki R, Nakagawa T, Fukuda M, Taniguchi N, Taguchi T. Spatial segregation of degradation- and recycling-trafficking pathways in COS-1 cells. Biochem Biophys Res Commun. 2007;360:580–585. doi: 10.1016/j.bbrc.2007.06.101. [DOI] [PubMed] [Google Scholar]
- 17.Misaki R, et al. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. J Cell Biol. 2010;191:23–29. doi: 10.1083/jcb.200911143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowler S, et al. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkinson KD, et al. Yeast mutants auxotrophic for choline or ethanolamine. J Bacteriol. 1980;141:558–564. doi: 10.1128/jb.141.2.558-564.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hikiji T, Miura K, Kiyono K, Shibuya I, Ohta A. Disruption of the CHO1 gene encoding phosphatidylserine synthase in Saccharomyces cerevisiae. J Biochem. 1988;104:894–900. doi: 10.1093/oxfordjournals.jbchem.a122579. [DOI] [PubMed] [Google Scholar]
- 21.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 23.Fujinaga Y, et al. Gangliosides that associate with lipid rafts mediate transport of cholera and related toxins from the plasma membrane to endoplasmic reticulm. Mol Biol Cell. 2003;14:4783–4793. doi: 10.1091/mbc.E03-06-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puri S, Bachert C, Fimmel CJ, Linstedt AD. Cycling of early Golgi proteins via the cell surface and endosomes upon lumenal pH disruption. Traffic. 2002;3:641–653. doi: 10.1034/j.1600-0854.2002.30906.x. [DOI] [PubMed] [Google Scholar]
- 25.Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ang AL, et al. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J Cell Biol. 2004;167:531–543. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray RZ, Kay JG, Sangermani DG, Stow JL. A role for the phagosome in cytokine secretion. Science. 2005;310:1492–1495. doi: 10.1126/science.1120225. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson KM, et al. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.