Abstract
Mouse ES cells use a mitochondrial threonine dehydrogenase (TDH) enzyme to catabolize threonine into glycine and acetyl-CoA. Measurements of mRNA abundance have given evidence that ES cells express upwards of 1,000-fold higher levels of TDH mRNA than any of seven other mouse tissues tested. When cell culture medium is deprived of threonine, ES cells rapidly discontinue DNA synthesis, arrest cell division, and eventually die. Such studies led to the conclusion that mouse ES cells exist in a threonine-dependent metabolic state. Proceeding with the assumption that the active TDH enzyme should be essential for the growth and viability of mouse ES cells, we performed a drug screen in search of specific inhibitors of the purified TDH enzyme. Such efforts led to the discovery of a class of quinazolinecarboxamide (Qc) compounds that inhibit the ability of the TDH enzyme to catabolize threonine into glycine and acetyl-CoA. Administration of Qc inhibitors of TDH to mouse ES cells impeded cell growth and resulted in the induction of autophagy. By contrast, the same chemicals failed to affect the growth of HeLa cells at concentrations 300-fold higher than that required to kill mouse ES cells. It was likewise observed that the Qc class of TDH inhibitors failed to affect the growth or viability of ES cell-derived embryoid body cells known to have extinguished TDH expression. These studies demonstrate how it is possible to kill a specific mammalian cell type on the basis of its specialized metabolic state.
Keywords: specialized metabolism, threonine catabolism, embryonic stem cells
The diverse spectrum of life forms found in nature makes use of a variety of specialized metabolic pathways to support growth and reproduction. Over the past century, these often unanticipated specializations in metabolism have enabled scientists to pharmacologically poison specific organisms or cell types to the benefit of human health. One of the first examples of the utility of this strategy was the sulfonamide class of antibiotics. Discovered in the 1930s, sulfonamides act by inhibiting the bacterial enzyme dihydropteroate synthetase (DHPS) (1, 2). The DHPS enzyme is needed by bacteria to synthesize folic acid derivatives, which are required for the de novo synthesis of nucleotides. Mammals do not encode DHPS enzymes and instead obtain folates from their diet. For this reason sulfonamides are well tolerated by humans. A second example of an antibiotic that works by exploiting metabolic specialization is metronidazole. Metronidazole becomes toxic only after being reduced in the absence of oxygen; hence this chemical toxin selectively targets anaerobic microorganisms (3). Many drugs that block the growth of eukaryotic pathogens also work by attacking points of vulnerability built around specialized differences in metabolism. These include the azole, echinocandin, and allylamine classes of antifungal drugs (4–6), as well as the antiparasitics thiabendazole and diethylcarbazamine (7, 8).
Human cancer cells have also been targeted by virtue of their specialized metabolic state. Because most somatic cells are quiescent and have ceased DNA replication, a variety of chemotherapeutic agents has been used to inhibit nucleotide synthesis in a wide range of cancer cells. Antifolates, purine and pyrimidine antagonists, and ribonucleotide reductase inhibitors all act by interfering with nucleotide biosynthesis (9–11). Whereas these drugs affect many types of cancer, the asparaginase enzyme has been used to specifically inhibit the growth of acute lymphoblastic leukemia (ALL) cells (12). Having turned off expression of the asparagine synthetase gene, ALL cells are unusually sensitive to asparaginase-mediated deprivation of asparagine (13). In addition to these existing therapies, longstanding evidence suggests that many tumor cells rely heavily on glycolysis, instead of respiration, for the majority of their energy production (14). Specialized inhibitors of glycolysis are currently the subject of intense scrutiny as putative anticancer agents (15).
We have previously shown that mouse ES cells exist in a metabolic state that is highly dependent upon the amino acid threonine (16). It was concluded that threonine dependence stems from the activity of the mitochondrial enzyme threonine dehydrogenase (TDH), which is expressed at a much higher level in ES cells than in any other mammalian tissue or cell type examined to date. TDH catabolizes threonine to produce glycine and acetyl-CoA, which provide fuel for nucleotide synthesis and ATP generation, respectively (17). Thus, flux through the TDH pathway was proposed to be required to sustain the exceptionally rapid cell division rate of mouse ES cells (16). In the present study we have used unbiased chemical screening to identify specific and potent inhibitors of the TDH enzyme. We hereby demonstrate that chemical inhibition of TDH enzyme activity in ES cells results in a form of cellular autophagy that is insufficient to maintain viability. Importantly, TDH-specific chemical inhibitors displayed no toxicity to somatic cells that do not express the TDH enzyme. These results confirm the importance of the threonine-dependent metabolic state for mouse ES cell viability and further demonstrate that an understanding of metabolic specialization can be used to successfully impede the viability of a particular mammalian cell type.
Results
Identification of Selective Chemical Inhibitors of the TDH Enzyme.
To initiate biochemical analysis of threonine catabolism, we expressed and purified the mouse TDH enzyme. Although the full-length protein was insoluble and contained in bacterial inclusion bodies, deletion of 39 amino acids from the amino terminus resulted in the production of milligram quantities of soluble, highly purified TDH protein. This 39 amino acid stretch corresponds to a predicted mitochondrial signal sequence, which is likely to be cleaved after the protein's translocation into the mitochondrion (18). It is therefore assumed that the purified recombinant protein closely resembles the TDH enzyme active in the mitochondria of mouse ES cells.
Incubation of the purified TDH enzyme with threonine and NAD+ resulted in a dose-dependent, saturable increase in absorbance at 340 nm, indicative of formation of reduced coenzyme (NADH). This activity was dependent on the presence of both threonine and NAD+ in the reaction mixture. To confirm that the activity was not caused by the presence of small amounts of copurifying bacterial TDH, we further purified the recombinant mouse TDH enzyme using anion exchange and gel filtration chromatography. The threonine-dependent, NAD+-reducing activity fractionated with the major band on the gel, which was recognized by a TDH-specific antibody. To determine kinetic parameters for the TDH enzyme, we constructed Lineweaver-Burk double reciprocal plots for both substrates by measuring reaction velocity as a function of threonine and NAD+ concentrations. The Michaelis constants (Km) for NAD+ and threonine were 180 μM and 14 mM, respectively, and the turnover number (kcat) was ≈60,000 s−1. These values are in close agreement with kinetic constants reported for other TDH enzymes of eukaryotic origin (19, 20). We conclude that the purified mouse TDH enzyme is capable of catalyzing the same chemical reaction as has been demonstrated for TDH orthologs from diverse metazoan and microbial species (19–22).
If the growth of mouse ES cells is dependent on TDH-mediated threonine catabolism, then selective inhibition of the TDH enzyme should result in reduced proliferative capacity of ES cells. To begin to address this hypothesis, we screened the University of Texas Southwestern Medical Center (UTSWMC) chemical library for TDH inhibitors using an in vitro TDH assay. The high throughput screen (HTS) was performed in 364-well plates, and assays were initiated with 5 μL of a mixture of both substrates at 10 mM and incubated for 30 min at room temperature (Methods). Of the ≈200,000 compounds screened, ≈1,000 were observed to inhibit TDH activity with a z-score >3 relative to the 1% DMSO control. The inhibitory action of these molecules was then confirmed by repeating the same TDH absorbance assay in triplicate, and dose-dependent inhibition was determined using compound concentrations of 1, 3, and 10 μM. To identify and eliminate generic dehydrogenase inhibitors, we performed a similar absorbance assay using hydroxysteroid dehydrogenase (HSDH), the enzyme most closely related to mouse TDH by primary sequence homology. Any compounds that inhibited HSDH were deemed nonspecific dehydrogenase inhibitors and eliminated from further study.
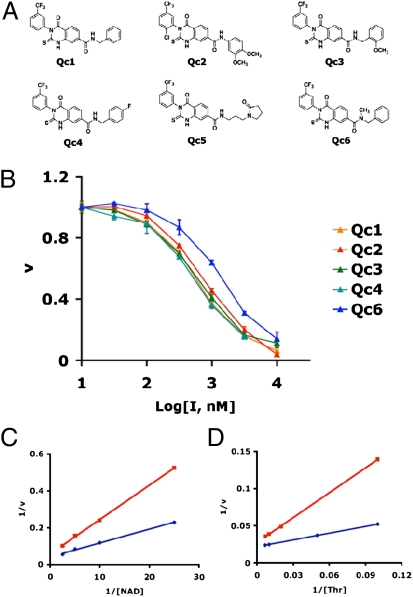
A cluster of six closely related quinazolinecarboxamide (Qc) compounds emerged from the HTS screen and were chosen for further study (Fig. 1A). To test for specificity against TDH, we examined whether these compounds were able to inhibit alcohol dehydrogenase, lactate dehydrogenase, or glucose-6-phosphate dehydrogenase. None of the six Qc small molecules showed any inhibitory activity against the other dehydrogenase enzymes when tested at a concentration of 10 μM. To determine IC50s of the Qc compounds against TDH, we titrated the inhibitors from 10 nM to 10 μM and measured TDH activity (Fig. 1B). The IC50 for all of the Qc molecules tested was ≈0.5 μM. To establish the mechanism of TDH inhibition, we constructed Lineweaver-Burk plots for both substrates in the presence and absence of inhibitor Qc1 (Fig. 1 C and D). We determined that both Km and Vmax were altered upon addition of Qc1, a pattern known as mixed noncompetitive inhibition. We also tested the reversibility of the Qc1 inhibitor by incubating purified TDH enzyme with varying concentrations of inhibitor for 30 min at room temperature and then dialyzing the mixture overnight. After dialysis, TDH activity was comparable to the DMSO control at all concentrations of inhibitor tested (up to 10 μM, or 20-fold greater than the IC50). We conclude that TDH inhibition by the Qc1 compound is fully reversible.
Fig. 1.
Chemical structures and potency of TDH inhibitors. (A) Structures of the six best hits from the TDH inhibitor screen. The compounds contained a Qc scaffold with various peripheral modifications. (B) IC50 values were determined by titrating the compounds from 10 nM to 10 μM and measuring TDH activity (Methods). The approximate IC50 for all six compounds was 0.5 μM. (C and D) Lineweaver-Burk analysis of enzyme inhibition. TDH activity was assayed in the absence and presence of the Qc inhibitor at the NAD+ and threonine concentrations shown. Blue curves depict data obtained in the absence of inhibitor, and red curves depict data obtained in the presence of inhibitor. Both Vmax (y intercept) and Km (x intercept) were altered in the presence of inhibitor, indicative of mixed noncompetitive inhibition.
Qc Class of TDH Inhibitors Selectively Kills Mouse ES Cells.
Previous work has shown that threonine deprivation severely impedes mouse ES cell proliferation (16). To determine whether the observed growth impediment results from selective disruption of the TDH metabolic pathway, we cultured ES cells in complete culture medium containing TDH inhibitors. To identify any off-target toxicity resulting from this treatment, we performed parallel experiments using 3T3 fibroblasts and human cancer (HeLa) cells, which do not express the TDH enzyme. When tested at 10 μM, all six of the Qc compounds severely inhibited the proliferation of ES cells but not fibroblasts or cancer cells. We likewise observed that the Qc compounds had no discernible effect on the growth or viability of embryoid body cells, which extinguish TDH expression during the course of differentiation (16). Whereas the Qc class of TDH inhibitors killed ES cells with an EC50 of ≈3 μM, the EC50 against the other cell types tested was ≈1 mM. Thus, the Qc compounds have a 300-fold greater level of toxicity for ES cells vs. other mammalian cell types, strongly arguing that these molecules are killing ES cells through specific inhibition of the TDH enzyme.
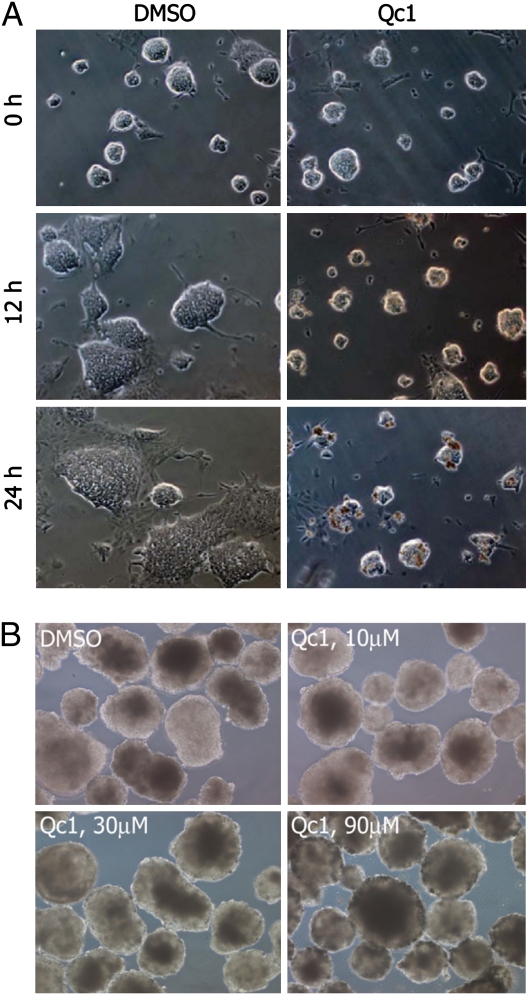
To more closely examine the effects of TDH inhibition on ES cell growth, we incubated ES cell colonies grown in glass chamber slides with the Qc1 compound and monitored colony morphology using phase contrast microscopy (Fig. 2A). Colonies treated with TDH inhibitor appeared normal but failed to proliferate, whereas those treated with vehicle (DMSO) grew rapidly in size. After 24 h of TDH inhibition, groups of dead cells became apparent at the surface of colonies, and longer incubation resulted in the majority of cells appearing as collections of opaque cells with indistinct boundaries. By contrast, when embryoid bodies were exposed for 24 h to 10-μM, 30-μM, or 90-μM levels of the Qc1 inhibitor of TDH, no impediment was observed with respect to either cell growth or embryoid body morphology (Fig. 2B).
Fig. 2.
Effect of TDH inhibition on ES cell growth, colony morphology, and embryoid body morphology. (A) Feederless ES cells (E14 strain) were cultured on glass chamber slides and imaged using phase contrast microscopy. When treated with vehicle (DMSO) alone, ES cell colonies rapidly grew in size. Upon exposure to the TDH inhibitor, ES cells failed to proliferate, with colony size remaining unchanged for the first 12 h. After 24 h, clusters of densely packed cells became apparent at the surface of the colonies, indicative of cell death. (B) E14 ES cells were grown in suspension without Leukemia Inhibitory Factor (LIF) for 10 d to allow differentiation into embryoid bodies. Embryoid bodies were then treated for 24 h with vehicle or Qc1 at indicated concentrations.
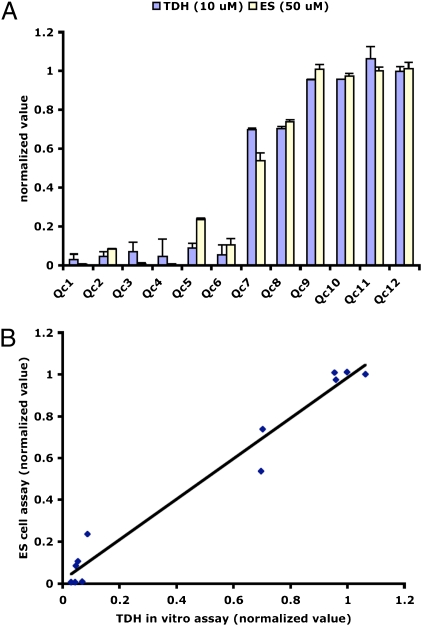
As a first attempt at probing the structure–activity relationship of the Qc compounds, we searched the UTSWMC chemical library for molecules having structures closely related to the TDH inhibitors identified in the HTS screen. Using cluster analysis, we were able to identify six compounds (Qc7–Qc12) with structures similar to the TDH inhibitors that displayed selective killing of ES cells. To test the activities of these related compounds, we carried out both TDH enzymatic assays and ES cell viability assays. Four of the related compounds (Qc9–Qc12) did not inhibit TDH activity at a concentration of 10 μM, nor did they impede ES cell growth at a concentration of 50 μM (Fig. 3A). Two of the structural analogs, Qc7 and Qc8, showed weak inhibitory activity in vitro and also inhibited ES cell proliferation when tested at a concentration of 50 μM. The TDH inhibitors identified through chemical screening inhibited enzyme activity with an IC50 of ≈0.5 μM and killed ES cells with an EC50 of ≈3 μM. Thus, a positive correlation was observed between TDH inhibition and ES cell toxicity for the structurally related group of Qc compounds (Fig. 3B). This positive correlation is consistent with the hypothesis that Qc compound-mediated ES cell death results via direct inhibition of the TDH enzyme.
Fig. 3.
Correlation between TDH inhibition and ES cell cytotoxicity for 12 Qc compounds. (A) Twelve structurally related Qc compounds were assayed for their ability to inhibit TDH in vitro (10-μM compound) and impair proliferation of ES cells (50-μM compound). The normalized values for both assays are plotted in histogram form. Qc1–Qc6 are the TDH inhibitors identified through the small molecule screen, and Qc7–Qc12 are structurally related compounds found in the UTSWMC chemical library that were not identified in the screen. Qc1–Qc6 are potent TDH inhibitors that kill ES cells with EC50 ≈3 μM. Qc7 and Qc8 are weak TDH inhibitors with EC50 >50 μM. Qc9–Qc12 did not inhibit TDH in vitro and displayed no toxicity when added to the ES cell growth medium. (B) Same as in A, with data plotted as an xy scatter plot.
Realizing that the TDH enzyme's role in ES cell metabolism was initially identified through changes in metabolite levels as a function of differentiation (16), we measured the effects of TDH inhibition on metabolite abundance. ES cells were grown free of feeder cells on gelatinized tissue culture dishes and exposed to the Qc1 inhibitor of the TDH for 0, 1, 2, 3, and 4 h. Metabolites were then extracted in 50% aqueous methanol and subjected to unbiased LC-MS/MS analysis as a means of identifying and quantitating changes in the levels of common metabolites (23). As shown in Fig. 4, only a small proportion of the 55 metabolites chosen for study changed in abundance as a function of exposure of ES cells to the Qc1 inhibitor of TDH. The two metabolites that decreased in abundance most markedly were acetyl-CoA and methyltetrahydrofolate (mTHF). By contrast, the two metabolites that increased in ES cells as a function of Qc1-mediated inhibition of the TDH enzyme were 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) and threonine. We hypothesize that acetyl-CoA and mTHF decrease in abundance in response to the Qc1 inhibitor because it prevents TDH from catabolizing threonine into acetyl-CoA and glycine. The latter catabolite of TDH-mediated degradation of threonine is known to be used to fuel the glycine cleavage system within mitochondria in support of the charging of tetrahydrofolate into mTHF. We further rationalize the increase in abundance of the AICAR intermediate in purine synthesis to reflect the fact that one carbon metabolism has been inhibited via inhibition of the TDH enzyme (as revealed by the decrease in abundance of mTHF). Finally, we hypothesize that threonine levels increase in ES cells treated with the TDH inhibitor because the enzyme is incapable of catabolizing its amino acid substrate. These findings are consistent with the conclusion that flux through the TDH pathway plays a vital role in shaping the threonine-dependent metabolic state of mouse ES cells (16).
Fig. 4.
Accumulation of threonine and AICAR and depletion of acetyl-CoA and mTHF in ES cells treated with TDH inhibitors. Feederless ES cells were treated with 10 μM of the Qc1 TDH inhibitor for 0, 1, 2, 3, and 4 h before extraction of metabolites in 50% aqueous methanol and subsequent LC-MS/MS analysis. Metabolites increasing in abundance as a function of exposure to the Qc1 inhibitor of TDH are shown in red. Metabolites decreasing in abundance are shown in green.
Inhibition of TDH Enzyme Activity in Mouse ES Cells Leads to Autophagy.
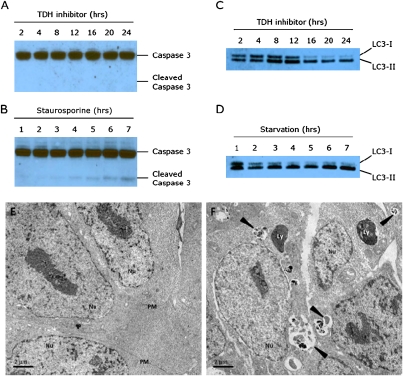
We next turned our attention to the mechanism of ES cell death resulting from TDH inhibition, hypothesizing that ES cells treated with TDH inhibitors might be dying by apoptosis. Caspase 3, a critical executioner of apoptosis, is activated by proteolytic processing of its inactive zymogen during apoptotic cell death originating from either extrinsic or intrinsic pathways (24). To assay for apoptosis in ES cells treated with TDH inhibitors, we immunoblotted cell lysates with antibodies recognizing both the uncleaved and cleaved forms of caspase 3. This analysis revealed a strong 35-kDa band representing the full-length caspase 3 zymogen present in all samples tested. However, after 24 h of TDH inhibition, when widespread cell death was clearly visible, we could not detect any activated (cleaved) caspase 3 (Fig. 5A). In control experiments, lysates from ES cells treated with staurosporine, a molecule known to induce apoptosis, contained caspase 3 cleavage products that became apparent 4 h after treatment (Fig. 5B). We conclude that TDH inhibitor-mediated cell death does not result from increased apoptosis.
Fig. 5.
TDH inhibition results in elevated autophagy but not apoptosis. (A) Protein lysates from ES cells treated with 10 μM TDH inhibitor were extracted in 0.5% Nonidet P-40, separated using SDS/PAGE, and immuoblotted using an antibody specific for caspase 3. Despite visible cell death, no cleaved caspase 3 could be detected after 24 h of TDH inhibitor treatment. (B) Control Western blot showing cleaved caspase 3 in response to treatment with 1 μM staurosporine. (C) ES cell lysates were immunoblotted with an antibody to LC3. After 16 h of TDH inhibitor treatment, most of the LC3 protein was in the LC3-II (lipidated) form, indicative of increased autophagic activity. (D) Control Western blot showing increased autophagy in response to nutrient starvation. ES cells were cultured in HBSS and harvested at the times indicated. (E and F) Electron microscopy of ES cells treated with TDH inhibitor. Mouse ES cells were cultured on plastic coverslips for 24 h with or without TDH inhibitor. Cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer and embedded in Embed-812 Resin. Thin sections (70–90 nm in thickness) were stained with 2% aqueous uranyl acetate and lead citrate and examined by transmission electron microscopy. (E) Control ES cells treated with DMSO only. (F) ES cells cultured in the presence of 10 μM TDH inhibitor. Arrowheads indicate autophagic compartments. Nu, nucleus; PM, plasma membrane; Ly, dense lysosomes.
An alternative pathway that might eventually impact viability is cellular autophagy. Autophagy (self-eating) is the process by which bulk cytoplasmic proteins and organelles reach lysosomes for degradation (25, 26). Elevated autophagy can provide a protective benefit to cells by salvaging nutrients from dispensable cytoplasmic contents so that vital processes can be maintained (27, 28). During autophagy, a protein designated light chain 3 (LC3) is conjugated to phosphatidylethanolamine and becomes associated with autophagosomes, and this lipidation can serve as a marker for autophagy (29). To assay for autophagy, we cultured ES cells between 2 and 24 h in the presence of the Qc1 TDH inhibitor and immunoblotted cell lysates with an anti-LC3 antibody (Fig. 5C). Initially, a substantial fraction of the total LC3 protein was in the LC3-I (cytoplasmic) form. After 16 h of TDH inhibition, most LC3 was converted to the LC3-II (lipid-modified) form, indicative of increased autophagic activity. This pattern was similar to that of ES cells grown under conditions of nutrient starvation (Fig. 5D).
As a complementary approach, we examined the morphology of ES cells treated with the Qc1 TDH inhibitor using transmission electron microscopy. To directly visualize autophagy, we cultured feederless mouse ES cells on gelatinized coverslips in the presence of 10 μM of Qc1. Cells were fixed in glutaraldehyde, postfixed in osmium tetroxide, dehydrated with ethanol, and embedded in resin polymerized by heat. Control ES cells treated with DMSO appeared as small, densely packed cells with prominent nuclei. A few mitochondria were visible in these cells, but otherwise the cytoplasm was relatively clear (Fig. 5E). By contrast, multiple distinct autophagic structures were visible in ES cells treated with the Qc1 TDH inhibitor (Fig. 5F). Less mature autophagosomes present in these cells appeared to contain whole organelles, whereas more mature autophagosomes contained partially degraded (electron dense) cytoplasmic contents. Thus, two independent experimental methods gave evidence that specific inhibition of the TDH enzyme in ES cells results in cellular autophagy. Given that this process precedes loss of viability, we speculate that the induced autophagic state is insufficient to prevent ES cell death.
Discussion
In this study we used a pharmacological approach to demonstrate that metabolic flux through the TDH pathway is required for the viability and self-renewal of mouse ES cells. This result was predicated on our previous discovery that mouse ES cells are uniquely dependent upon the presence of threonine in the culture medium (16). This finding, coupled with the observation that the TDH enzyme is robustly and specifically expressed in ES cells (16), led us to hypothesize that mouse ES cells exist in a threonine-dependent metabolic state. We hereby show that selective chemical perturbation of that state causes cellular autophagy and death.
The classic inducer of autophagy is nutrient starvation (25–27). Our results indicate that insufficient production of mitochondrial acetyl-CoA and glycine in ES cells results in a cellular state resembling starvation. We have previously shown that mouse ES cells make use of the high-flux backbone (HFB) of metabolism to drive rapid rates of proliferation (16). In HFB metabolism, as described for rapidly growing bacteria (30), glycine generated via threonine catabolism is used to fuel one-carbon metabolism for purine biosynthesis. The other catabolite derived from TDH-mediated breakdown of threonine, acetyl-CoA, enters the TCA cycle for ATP production during HFB metabolism (30). Thus, TDH inhibition might be predicted to result in a shortage of nucleotides for DNA replication as well as a decline in energy production. It seems that ES cells deprived of their ability to consume threonine attempt to degrade nonessential cellular components to overcome these deficiencies. Under conditions of prolonged TDH inhibition, ES cells are unable to maintain their high proliferative rate, display autophagic features typically associated with nutrient starvation, and eventually die.
This study provides evidence that knowledge of a cell's specialized metabolic state can be used to preferentially target that cell for growth inhibition or death. The success of this approach requires detailed understanding of how metabolic flux in a given cell differs from that commonly used in nature. Once a unique metabolic state has been characterized sufficiently, a targeted drug screen can be performed to identify chemotherapeutics that inhibit key enzymes responsible for generating and maintaining that state. It is hoped that the approach used in this study might be used to selectively kill other cell types having growth properties dependent upon specialized metabolic pathways, perhaps including cancer cells. Finally, it is possible that the growth of pathogens that rely on TDH-mediated catabolism of threonine might be impeded by derivatives of the Qc class of chemicals discovered and described in this study.
Methods
Materials.
Chromatographic reagents for protein purification were obtained from GE Healthcare. All other chemicals and reagents were obtained from Sigma-Aldrich except where specified.
Protein Expression and Purification.
Mouse TDH was expressed as a GST fusion protein in Escherichia coli strain Rosetta. The cells were cultured at 37 °C until the A600 nm reached 0.6 and were then induced with 0.2 mM isopropyl β-d-thiogalactoside (Promega) for 16 h at 20 °C. The cells were suspended in 50 mM Tris⋅Hcl (pH 8.0) containing 50 mM NaCl, 1 mM DTT (Promega), and 1 mg/mL lysozyme, incubated on ice for 30 min, and sonicated (Fisher Scientific Sonic Dismembrator Model 500). After spinning in an ultracentrifuge at 80 × g (Beckman rotor Ti75) for 30 min at 4 °C, the supernatant was incubated with glutathione Sepharose resin for 2 h and, after washing, eluted with 10 mM reduced glutathione. TDH was further purified using Superdex 200 and MonoQ chromatography (Amersham).
Enzymatic Activity Measurements.
TDH activity was determined by measuring the rate of formation of NADH at 25 °C. The standard assay mixture contained 100 nM purified TDH, 50 mM Tris⋅HCl (pH 8.0), 2 mM NAD+, 2 mM l-threonine, 50 mM NaCl, and 1 mM DTT (Promega) in a final volume of 50 μL. The reaction was initiated by the addition of a mixture containing both substrates, and the absorbance of the reaction mixture at 340 nm was recorded continuously on a Bio-Tek Synergy HT microplate reader.
Similar absorbance assays were used to measure activity of other dehydrogenase enzymes. Hydroxysteroid dehydrogenase activity was assayed using 30 μg/mL purified enzyme, 0.3 mM NAD+, and 0.00005% testosterone in 50 mM Tris⋅HCl buffer (pH 8.0). The alcohol dehydrogenase reaction contained 30 μg/mL purified enzyme, 8 mM NAD+, and 300 mM ethanol in 50 mM Tris⋅HCl (pH 8.0). The lactate dehydrogenase assay used 30 μg/mL enzyme, 200 μM NADH, and 3 mM sodium pyruvate in 0.2 M Tris⋅HCl (pH 7.3). Glucose-6-phosphate dehydrogenase activity was determined using a reaction mixture containing 30 μg/mL enzyme, 0.2 mM NADP+, and 3 mM glucose-6-phosphate in 50 mM Tris⋅HCl (pH 7.8) with 3 mM MgCl2.
High-Throughput Screening.
Approximately 200,000 drug-like synthetic chemicals were screened using the NADH absorbance assay in 384-well UV transparent plates (Corning). The primary assay was performed as described above, with 5 μM of individual library test compounds in pure DMSO (1% DMSO final concentration) added to the reaction mixture. The reaction was initiated with the addition of 5 μL per well of a mixture containing 10 mM NAD+ and 10 mM threonine. Total reaction volume was 50 μL per well. The positive control was in column 1 of each plate, and DMSO controls were in columns 2 and 23. The reaction was allowed to proceed for 30 min at room temperature, at which point NADH production was measured by reading the absorbance at 340 nm.
For screen validation, compounds determined to inhibit TDH were cherry picked from 5-mM compound stock library plates. The cherry picking of compounds was done with the use of a 384-pin array Biomek FX (Beckman Instruments) high-precision robot with a Span 8 pod. Resupply of confirmed hits was obtained from ChemDiv and ChemBridge. Purity of compounds was analyzed by LC/MS, and all inhibitors were found to be >95% pure. For cell viability assays, cells were plated in gelatinized Costar 384-well plates using an automated dispenser at 1,000 cells per well in 50 μL of media per well. Six hours after plating, 0.5 μL of compounds was added. The plates were then incubated for 24 (ES) or 48 (NIH 3T3 and HeLa cells) hours before measuring cell viability using CellTiter-Glo reagent (Promega). All solutions were dispersed robotically using a Biomek FX robot.
LC-MS/MS Analysis of ES Cell Metabolites.
Feederless ES cells were grown on 60-cm2 gelatinized cell culture dishes in the presence of 10 μM TDH inhibitor for 2 h. Dishes were washed in cold PBS, and metabolites were extracted in 1 mL of 50% aqueous methanol that was maintained at −20 °C. Cells were dounced in a glass homogenizer and centrifuged at 18,000 × g for 20 min to pellet cell debris, and 0.9 mL of the supernatant was transferred to a new tube and stored at −80 °C until analysis. Samples were dried on a Savant Speed Vac Plus SC210A for 2 h. Metabolites were reconstituted in 100 μL of 5 mM ammonium acetate and filtered using 0.2 μm PVDF microspin tubes (W. R. Grace). Samples were infused into an Applied Biosystems 3200 QTRAP triple quadrupole-linear ion trap mass spectrometer, and data were processed using Analyst software (23).
Western Blotting.
ES cell proteins were extracted using a 30-min incubation of ES cells in cold 50 mM Hepes (pH 7.4), 50 mM NaCl, 0.5% Nonidet P-40, 1 mM DTT, 100 μM phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture. Protein concentrations were determined by Bio-Rad protein assay, and 30 μg total protein was loaded in each lane of a 12% SDS/PAGE gel. Caspase 3 and LC3 antibodies were purchased from Cell Signaling. Antibodies were diluted 1:1,000 in 5% wt/vol nonfat dry milk (caspase 3) or 5% wt/vol BSA (LC3) in tris-buffered saline with 0.05% Tween 20 (TBST) and incubated with nitrocellulose filters at 4 °C with gentle shaking overnight. ES cells were nutrient-starved using Hank's Buffered Salt Solution (Invitrogen).
Transmission Electron Microscopy.
ES cells were cultured on plastic coverslips for 24 h with 10 μM TDH inhibitor or vehicle-only control (DMSO). Specimens were fixed for 60 min in 2.5% glutaraldehyde in 0.1 M cacodylate buffer and postfixed with 1% osmium tetroxide for 15 min. Cells were dehydrated in ethanol and embedded in Embed-812 resin (Electron Microscopy Sciences) overnight at 70 °C. Thin sections (70–90 nm in thickness) were cut on a Leica Ultracut E ultramicrotome and placed on 200-mesh copper grids. Sections were stained with 2% aqueous uranyl acetate and lead citrate and examined in an FEI Tecnai G2 Spirit Biotwin transmission electron microscope, operated at 120 kV. Digital images were captured with a Gatan 2Kx2K multiport readout post-column CCD.
Acknowledgments
We thank Shuguang Wei, Chun Hui Bu, and Bruce Posner for extensive help with high-throughput screening; Tom Januszewski and Chris Gilpin for assistance with electron microscopy; Ben Tu for help with mass spectrometry; and Bill Kaelin, Eileen White, and Jim Wells for critical review of the manuscript. This work was supported by unrestricted funds supplied to S.L.M. by an anonymous donor.
Footnotes
The authors declare no conflict of interest.
References
- 1.Smyth HF, Carpenter CP. Water-soluble derivatives of p-aminobenzene-sulfonamide (sulfanilamide) Science. 1938;87:350–351. doi: 10.1126/science.87.2259.350-a. [DOI] [PubMed] [Google Scholar]
- 2.Roland S, Ferone R, Harvey RJ, Styles VL, Morrison RW. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J Biol Chem. 1979;254:10337–10345. [PubMed] [Google Scholar]
- 3.Tally FP, Sutter VL, Finegold SM. Metronidazole versus anaerobes. In vitro data and initial clinical observations. Calif Med. 1972;117:22–26. [PMC free article] [PubMed] [Google Scholar]
- 4.Carson LE, Campbell CC. The inhibitory effect of three antihistamine compounds on the growth of fungi pathogenic for man. Science. 1950;111:689–691. doi: 10.1126/science.111.2895.689. [DOI] [PubMed] [Google Scholar]
- 5.Nyfeler R, Keller-Schierlein W. [Metabolites of microorganisms. 143. Echinocandin B, a novel polypeptide-antibiotic from Aspergillus nidulans var. echinulatus: Isolation and structural components] Helv Chim Acta. 1974;57:2459–2477. doi: 10.1002/hlca.19740570818. [DOI] [PubMed] [Google Scholar]
- 6.Petranyi G, Ryder NS, Stütz A. Allylamine derivatives: New class of synthetic antifungal agents inhibiting fungal squalene epoxidase. Science. 1984;224:1239–1241. doi: 10.1126/science.6547247. [DOI] [PubMed] [Google Scholar]
- 7.Prichard RK. Mode of action of the anthelminthic thiabendazole in Haemonchus contortus. Nature. 1970;228:684–685. doi: 10.1038/228684a0. [DOI] [PubMed] [Google Scholar]
- 8.Maizels RM, Denham DA. Diethylcarbamazine (DEC): Immunopharmacological interactions of an anti-filarial drug. Parasitology. 1992;105(Suppl):S49–S60. doi: 10.1017/s0031182000075351. [DOI] [PubMed] [Google Scholar]
- 9.Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 10.Li MC, Hertz R, Bergenstal DM. Therapy of choriocarcinoma and related trophoblastic tumors with folic acid and purine antagonists. N Engl J Med. 1958;259:66–74. doi: 10.1056/NEJM195807102590204. [DOI] [PubMed] [Google Scholar]
- 11.Brockman RW, Shaddix S, Laster WR, Jr, Schabel FM., Jr Inhibition of ribonucleotide reductase, DNA synthesis, and L1210 leukemia by guanazole. Cancer Res. 1970;30:2358–2368. [PubMed] [Google Scholar]
- 12.Old LJ, Boyse EA, Campbell HA, Daria GM. Leukaemia-inhibiting properties and L-asparaginase activity of sera from certain South American rodents. Nature. 1963;198:801. doi: 10.1038/198801a0. [DOI] [PubMed] [Google Scholar]
- 13.Horowitz B, et al. Asparagine synthetase activity of mouse leukemias. Science. 1968;160:533–535. doi: 10.1126/science.160.3827.533. [DOI] [PubMed] [Google Scholar]
- 14.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 15.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dale RA. Catabolism of threonine in mammals by coupling of L-threonine 3-dehydrogenase with 2-amino-3-oxobutyrate-CoA ligase. Biochim Biophys Acta. 1978;544:496–503. doi: 10.1016/0304-4165(78)90324-0. [DOI] [PubMed] [Google Scholar]
- 18.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 19.Linstead DJ, Klein RA, Cross GAM. Threonine catabolism in Trypanosoma brucei. J Gen Microbiol. 1977;101:243–251. doi: 10.1099/00221287-101-2-243. [DOI] [PubMed] [Google Scholar]
- 20.Aoyama Y, Motokawa Y. L-Threonine dehydrogenase of chicken liver. Purification, characterization, and physiological significance. J Biol Chem. 1981;256:12367–12373. [PubMed] [Google Scholar]
- 21.Epperly BR, Dekker EE. L-threonine dehydrogenase from Escherichia coli. Identification of an active site cysteine residue and metal ion studies. J Biol Chem. 1991;266:6086–6092. [PubMed] [Google Scholar]
- 22.Ishikawa K, Higashi N, Nakamura T, Matsuura T, Nakagawa A. The first crystal structure of L-threonine dehydrogenase. J Mol Biol. 2007;366:857–867. doi: 10.1016/j.jmb.2006.11.060. [DOI] [PubMed] [Google Scholar]
- 23.Tu BP, et al. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci USA. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson DW, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 25.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B, Yuan J. Autophagy in cell death: An innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almaas E, Kovács B, Vicsek T, Oltvai ZN, Barabási AL. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]