Abstract
Bacterial cells possess multiple cytoskeletal proteins involved in a wide range of cellular processes. These cytoskeletal proteins are dynamic, but the driving forces and cellular functions of these dynamics remain poorly understood. Eukaryotic cytoskeletal dynamics are often driven by motor proteins, but in bacteria no motors that drive cytoskeletal motion have been identified to date. Here, we quantitatively study the dynamics of the Escherichia coli actin homolog MreB, which is essential for the maintenance of rod-like cell shape in bacteria. We find that MreB rotates around the long axis of the cell in a persistent manner. Whereas previous studies have suggested that MreB dynamics are driven by its own polymerization, we show that MreB rotation does not depend on its own polymerization but rather requires the assembly of the peptidoglycan cell wall. The cell-wall synthesis machinery thus either constitutes a novel type of extracellular motor that exerts force on cytoplasmic MreB, or is indirectly required for an as-yet-unidentified motor. Biophysical simulations suggest that one function of MreB rotation is to ensure a uniform distribution of new peptidoglycan insertion sites, a necessary condition to maintain rod shape during growth. These findings both broaden the view of cytoskeletal motors and deepen our understanding of the physical basis of bacterial morphogenesis.
Keywords: bacterial cytoskeletal dynamics, cell-wall organization, peptidoglycan synthesis, cell growth
Cytoskeletal proteins play an important role in bacterial morphogenesis (1). Of the bacterial cytoskeletal proteins, the widely conserved actin homolog MreB is particularly important for bacterial cells to elongate and maintain a rod-like shape. MreB forms polymers that are associated with the cell membrane and distributed along the length of the cell in many rod-shaped bacteria (2). These polymeric MreB structures are essential for the maintenance of rod-like cell shape, as their disruption leads to cell rounding. Although MreB is essential for proper morphogenesis, bacterial cell shape is ultimately determined by the shape of the peptidoglycan cell-wall sacculus, which in turn is controlled by the cell-wall synthesis machinery. The cell wall, which is composed of stiff glycan strands cross-linked by flexible peptide linkers, forms a load-bearing structure that can counteract the intracellular turgor pressure. Cell-wall assembly requires peptidoglycan subunits to be synthesized, polymerized into glycan strands by transglycosylase enzymes, and cross-linked into the existing cell-wall network by transpeptidase enzymes. MreB directly or indirectly associates with a number of proteins that have been implicated in cell-wall assembly, such that MreB is believed to act upstream of the cell-wall assembly machinery to direct the synthesis enzymes to the sites of cell-wall insertion.
Previous studies have demonstrated that MreB structures are dynamic in Bacillus subtilis and Caulobacter crescentus (3–7). To date, no motor proteins have been shown to either move along or transport MreB, such that these dynamics were interpreted as resulting from MreB polymerization. Here, we demonstrate that Escherichia coli MreB is also dynamic and that it moves persistently in a nearly circumferential direction. Interestingly, this MreB rotation is not driven by its own polymerization, but rather requires cell-wall synthesis. These findings indicate that a motor whose activity depends on cell-wall assembly rotates MreB. Furthermore, the coupling of MreB rotation to cell-wall synthesis suggests that MreB may not merely act upstream of cell-wall assembly. Indeed, computational simulations suggest that coupling MreB rotation to cell-wall synthesis can help cells maintain rod-like morphology.
Results and Discussion
MreB Structures Persistently Rotate Around the Long Axis of the Cell.
To gain insight into both the dynamics and function of MreB, we carefully imaged the movement of MreB structures in E. coli. These MreB structures have previously been interpreted as forming a continuous helix that extends from pole to pole (8). We imaged a natively expressed, functionally complementing MreB–red fluorescent protein (Rfp) sandwich fusion (MreB–Rfpsw) (9), which is the only copy of MreB in these cells. If imaged in a plane close to the cell center, MreB–Rfpsw often displays spots at the cell boundaries and sometimes displays diagonal bands crossing the cells, which are compatible with previous experiments (Fig. S1) (9). However, if imaged closer to the bottom plane of the cell (Inset of Fig. 1C), the MreB–Rfpsw fusion also displays heterogeneous spots (Fig. 1). These spots are still much brighter than single fluorescent proteins, indicating that each spot constitutes multiple MreB molecules. When cells are treated with the small molecule A22, which binds MreB monomers and thereby uniformly decreases the polymerization of MreB structures (10), the number of bright spots is reduced and the background fluorescence increases, indicating that the spots consist of polymerized MreB (Fig. S2). The heterogeneous spotty localization is consistent with the recent report that no long helical filaments are observed in E. coli by electron cryotomography (11) (Fig. 1 and Movies S1 and S2).
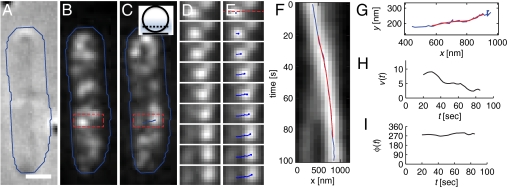
Fig. 1.
MreB persistently moves perpendicularly to the cell’s long axis in a representative cell. (A–C) DIC (A) and smoothed fluorescence images of the initial (B) and final (C) time points of a 100-s-long time lapse of two adjacent E. coli cells expressing MreB–Rfpsw. The blue line indicates a rough cell outline and the scale bar is 1 μm. (Inset of C) Sketch of the position of the focal plane at approximately one-fourth of the cell diameter (the point spread function is indicated in blue). (D and E) Intermediate snapshots, taken every 12.5 s, of the rectangular part of the lower cell highlighted in B and C. The resulting spot trajectory is highlighted in blue in E. (F) Kymograph of the interpolated fluorescence intensity along the dashed line in the first panel of E. Images were taken every 2.5 s. The horizontal positions of the raw (blue) and smoothed (red) trajectories are displayed. (G) The raw (blue) and smoothed (red) trajectory in the xy plane. (H and I) MreB trajectory velocity v (H) and orientation relative to the long cell axis ϕ (I) as a function of time t.
To quantitatively characterize MreB dynamics, we took advantage of the MreB–Rfpsw spots as fiducial markers that enabled us to measure the velocity and orientation of MreB dynamics with high spatial and temporal resolution (see SI Methods for details). We first computationally identified the trajectories of individual MreB spots with subpixel resolution using a tracking algorithm that identifies spots in each time frame and then connects proximal spots in subsequent frames (detailed in SI Methods and illustrated in Fig. 1). The resulting raw trajectories were then smoothed (Fig. 1G) to extract the instantaneous in-plane spot velocity, v (Fig. 1H), and orientation with respect to the long cell axis, φ (Fig. 1I). In the time course shown in Fig. 1 D and E, the bright spot initially on the left side moves persistently across the cell, reaching a maximum velocity of v ≈ 10 nm/s at the cell center (Fig. 1H and Movies S1 and S2). Throughout the time course, the direction of motion φ points roughly perpendicular to the long cell axis (Fig. 1I). Given the cell’s cylindrical symmetry, this perpendicular motion corresponds to a rotation of MreB about the cell’s long axis. Consistent with such circumferential motion, the in-plane velocity decreases close to the cell edges (Fig. 1H).
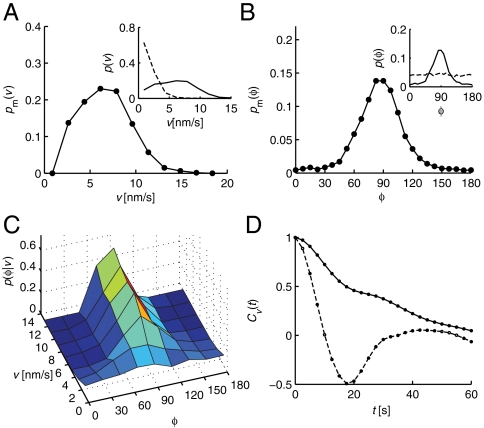
To statistically characterize MreB motion and assess the variability of spot dynamics, we computationally identified approximately 2,000 trajectories in time courses of 537 cells, of which we analyzed 119 long, cell-diameter spanning trajectories. These trajectories were analyzed in two different ways. In the first approach, we calculated the probability distribution of motion velocity and angle from individual time-point data (see Methods and SI Methods for details). We characterized the typical dynamics of the moving MreB spots in the central regions of the cells by first measuring the probability distributions of all instantaneous spot velocities p(v) and velocity orientations p(φ) (see Insets of Fig. 2 A and B). In order to restrict our analysis to the moving MreB structures, we segregated the MreB spots into mobile (m) and immobile (i) fractions using a Bayesian analysis based on p(v) and the velocity distribution of immobile spots obtained from chemically fixed cells pi(v) (82% of all spots are part of the mobile fraction; see SI Methods). Based on the same analysis, we calculated the probability distributions for the fraction of mobile MreB spots in live cells, pm(v) and pm(φ) (Fig. 2 A and B). The velocity distribution of mobile spots shows a broad peak with a mean of  . The corresponding distribution of angles pm(φ) displays a pronounced peak with a mean of
. The corresponding distribution of angles pm(φ) displays a pronounced peak with a mean of  (95% confidence), where all orientations 0° < φ < 360° were mapped onto the interval [0°…180°]. Note that the slight right-handed helical bias is also detected when the data were fit to a Gaussian distribution resulting in a peak position of 87.0°. Furthermore, the angular probability distributions for fixed velocity v, p(φ|v) (normalized independently for every v) also peak near φ = 90°, and these peaks sharpen as velocities increase (Fig. 2C). Thus, MreB spots move in a nearly perpendicular direction with respect to the cell’s long axis, and have a slight bias toward right-handed helical motion.
(95% confidence), where all orientations 0° < φ < 360° were mapped onto the interval [0°…180°]. Note that the slight right-handed helical bias is also detected when the data were fit to a Gaussian distribution resulting in a peak position of 87.0°. Furthermore, the angular probability distributions for fixed velocity v, p(φ|v) (normalized independently for every v) also peak near φ = 90°, and these peaks sharpen as velocities increase (Fig. 2C). Thus, MreB spots move in a nearly perpendicular direction with respect to the cell’s long axis, and have a slight bias toward right-handed helical motion.
Fig. 2.
Computational analysis reveals that MreB spots persistently rotate about the cell’s long axis. (A) Probability distribution pm(v) of the instantaneous velocities v of the mobile MreB spots. (Inset) Raw probability distributions of the velocities of all MreB spots in live (solid line) and chemically fixed (dashed line) cells. (B) Probability distribution pm(φ) of the orientations with respect to the long cell axis φ of the mobile MreB spots. (Inset) Raw distributions analogous to A. (C) Two-dimensional conditional probability distribution p(φ|v), independently normalized for each v, of the orientation of motion for different velocities v. (D) Velocity autocorrelation function Cv(t) of MreB spots from live (solid line) and chemically fixed (dashed line) cells.
In the second approach, we used the velocity autocorrelation function Cv(t) of all but very short trajectories to determine the extent to which the motion was persistent over time (see Fig. 2D and SI Methods). The autocorrelation function obtained from live cells decays much more slowly than that obtained from chemically fixed cells (Fig. 2D) with a persistence time of 30.4 s. The corresponding 204 ± 82 nm of persistent motion (see SI Methods for details) are similar to previously reported run lengths of single MreB proteins (7). Note that this length is shorter than the true circumferential persistence or run length because of the three-dimensional nature of MreB motion. Even when we consider all trajectories irrespective of their length, the autocorrelation function from live cells decays significantly slower than that from fixed cells (Fig. S3). Together, these results suggest that the MreB structures rotate circumferentially around the long axis of the cell in a persistent manner.
Previous studies have suggested that MreB forms helical structures and that MreB motion is driven by its own polymerization dynamics (3–7). If polymerization drives MreB motion, the velocity of the mobile MreB spots should decrease upon treatment with the polymerization inhibitor A22, which uniformly decreases the rate of MreB polymerization (10). We found that in cells treated with increasing amounts of A22, the velocity distribution of mobile MreB spots pm(v) does not notably change (Fig. S2F). Meanwhile, A22 treatment reduces the number of mobile spots per cell area (Fig. S2G) and increases the background fluorescence, confirming that MreB polymerization is reduced in these cells (Fig. S2 A–D and Movie S3). Note that cells were imaged before A22 treatment had a visible effect on cell shape. The A22-independence of the MreB spot velocity indicates that the observed MreB dynamics is not caused by MreB polymerization. Thus, MreB is actively moved by an as-yet-uncharacterized enzyme or a complex of enzymes, which we refer to as a “motor.”
MreB Rotation Requires Cell-Wall Synthesis.
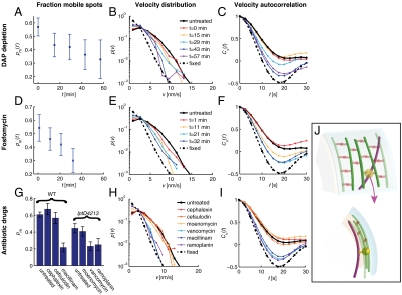
Because MreB moves circumferentially, its driving motor is expected to also move circumferentially. Recent experiments and theory indicate that cell-wall synthesis complexes are linked, at least transiently, to MreB, and that these complexes processively insert long glycan strands into the roughly circumferentially organized peptidoglycan network (12–14). MreB motion might therefore depend on peptidoglycan assembly. To test this hypothesis, we used two independent approaches to reduce the availability of peptidoglycan subunits and thereby inhibit cell-wall synthesis. First, we used an asd-1 mutant that cannot synthesize the essential peptidoglycan component diaminopimelic acid (DAP) (15). We grew asd-1 MreB–Rfpsw cells in the presence of exogenously supplied DAP and then imaged MreB motion at different time points after removing DAP. As a second independent method, we treated cells for varying durations with the antibiotic fosfomycin, which targets the essential peptidoglycan-subunit synthesis enzyme MurA (16). The DAP depletion and fosfomycin treatments produced the same qualitative results (Movie S4): The fraction of mobile spots decreased with time (Fig. 3 A and D), the velocity distributions of all MreB spots gradually approached that of fixed cells (Fig. 3 B and E), and the persistence time of the velocity autocorrelation functions gradually decreased (Fig. 3 C and F). The reduction in MreB motion by both DAP depletion and fosfomycin treatment is observed well before any effect on cell morphology, indicating that MreB motion depends on the availability of peptidoglycan subunits.
Fig. 3.
MreB rotation requires peptidoglycan substrate availability and the activity of transglycosylases and transpeptidases. For each dataset, we determined the fraction of mobile MreB spots pm(t) (A, D, and G), the distribution of the velocities of all MreB spots p(v) (B, E, and H), and the velocity autocorrelation function Cv(t) (C, F, and I). Nonmutant cells (solid black line) and chemically fixed cells (dashed line) serve as references in the latter two analyses. For A–C, asd-1 mutant cells were depleted of DAP at time zero and analyzed at the subsequent indicated time points. For D–F, cells were treated with the MurA inhibitor fosfomycin at time zero and analyzed at subsequent indicated time points. For G–I, nonmutant (WT) and lptD4213 mutant cells were analyzed between 10–20 min after treatment with the indicated antibiotic drug. (J) Top (Upper) and side (Lower) views of a model for cell-wall-synthase-driven MreB rotation. Here, we propose that the transmembrane cell-wall synthesis complex (yellow) acts in the periplasm (between the cyan outer membrane and peach inner membrane) to polymerize glycan strands (green) and form peptide cross-links (orange) to incorporate the strands into the existing peptidoglycan network, thereby causing the associated MreB filaments (magenta) to rotate in the cytoplasm.
MreB Rotation Depends upon Both Peptidoglycan Transglycosylation and Transpeptidation.
Cell-wall synthesis comprises the formation of glycan strands (transglycosylation) and peptide cross-links between them (transpeptidation). We thus treated cells with the previously characterized transglycosylation substrate inhibitor ramoplanin (17) and the transpeptidation substrate inhibitor vancomycin (18) [for effective drug treatment we used a lptD4213 mutant (19, 20) with increased outer-membrane-permeability]. Both of these antibiotics rapidly blocked MreB motion (Fig. 3 G–I, Fig. S4, and Movie S5). Specifically, the fraction of mobile spots decreased (Fig. 3G) and the velocity of the moving spots was reduced (Fig. S4 C and F). A dose-response analysis of ramoplanin and vancomycin indicated that their inhibition of MreB rotation increases as a function of drug concentration (Fig. S4 A–F) and sets in close to their minimum inhibitory concentrations (MICs) (Table S1). The effect can be observed as soon as the cells can be imaged (Fig. S4 J and K). There is no reduction in total MreB levels (Fig. S5A) indicating that the effect is not secondary to MreB levels. To make sure that the reduction in MreB rotation is not due to transcriptional downregulation, we also treated cells with the RNA polymerase inhibitor rifampicin (21, 22), which had no detectable effect on MreB rotation (Fig. S5 B–D). We thus conclude that both peptidoglycan transglycosylation and transpeptidation are required for MreB rotation.
To identify the specific transpeptidase required for MreB motion, we treated cells with the enzyme-specific antibiotics mecillinam (23, 24) (targeting PBP2), cefsulodin (24) (targeting PBP1a in a pbp1b mutant), and cephalexin (24) (targeting PBP3) (Movie S6). Of these drugs, only mecillinam blocked MreB motion and the dose-dependent mecillinam effect (Fig. S4 G–I) was rapid (Fig. S4 J and K) and set in close to its MIC (Fig. S4G and Table S1). Whereas cefsulodin had no significant effect on MreB dynamics, cephalexin treatment actually led to a slight increase in the fraction of mobile MreB spots, which could reflect PBP3 inhibition leading to increased substrate availability for PBP2. The role of PBP2 in MreB rotation is consistent with the reduced MreB rotational velocity in a pbp2ts mutant (Fig. S6 and Movies S7 and S8). Therefore, PBP2 is required for MreB rotation, whereas the transpeptidase activities of PBP1a, PBP1b, and PBP3 are not.
In contrast to the multiple specific inhibitors available for transpeptidases, the only specific transglycosylase inhibitor available is moenomycin, which targets the transglycosylase activities of both PBP1a and PBP1b. Moenomycin treatment had no detectable effect on MreB rotation (Movie S5). We also analyzed deletion mutants of two other transglycosylase proteins, PBP1C and MtgA, and found no effect of these deletions on MreB rotation (Fig. S7). Because inhibition of all transglycosylases inhibits MreB rotation, it either requires the activity of an as-yet-unknown transglycosylase or the known transglycosylases redundantly mediate MreB rotation.
MreB Rotation Could Result from Coupling MreB to the Cell-Wall Synthesis Machinery.
Because MreB rotation requires cell-wall synthesis, we sought to compare the speed of MreB rotation to the speed of cell-wall synthases, the complex of enzymes that constitute the cell-wall synthesis enzymes (25). To predict the speed of cell-wall synthases, we developed a geometric model of cell-wall growth that derives the rotation speed of a synthase based on the cellular elongation rate, the width of a glycan strand, and the number of active synthases in the cell (see SI Methods for details). Approximating the number of active synthases by the total number of PBP2 molecules in the cell, the rotation period of a synthase is predicted to be 2.8 ± 1.8 min at a temperature of 35 °C (25). To compare this calculated rotation period to the observed rotation period of MreB, we took movies of MreB–Rfpsw cells at a similar temperature of 36.5 °C (all measurements of MreB rotation reported above were performed at 21.5 °C). At this temperature, we observed a higher mean rotation speed of 18.6 ± 8.1 nm/s (Fig. S8 A and B). This speed corresponds to a rotation period of 2.5 ± 1.4 min, within error of our estimate of the period of synthase rotation.
Our model makes the testable prediction that the MreB rotation speed depends linearly on bacterial growth rate if the number of active synthases remains constant. We thus determined the MreB rotation speed at three different measurement temperatures (shortly after taking cells out of the 37 °C culture, which presumably leaves the number of active synthases unchanged) and found the expected linear relationship (Fig. S8 A and B and Table S2). Growth rate can also be modulated by growth media. Unlike upon acute temperature changes, the number of PBP2 molecules, estimated as equal to the number of active synthases, scales roughly proportionally with growth rate in different growth media (26). Our model thus predicts that MreB rotation speed should not change in these conditions. As predicted, MreB velocities are very similar in three different media (Fig. S8C and Table S3), despite the fact that these conditions span a fourfold difference in growth rates. The agreement between our model and experiments thus suggests that MreB rotation is spatially coupled to the rotation of cell-wall synthases.
A thermodynamic calculation indicates that peptidoglycan synthesis alone could provide the energy required to generate the necessary force to drag MreB around the cell (Fig. 3J; see SI Methods for details). For example, processive glycan-strand synthesis could result from a synthesis complex that moves along with the tip of the growing strand, and a natural way to keep the synthesis complex and the tip end together is if the growing strands drives the movement of the complex. However, whether the energy for MreB rotation is provided by the cell-wall synthesis enzymes themselves or a mechanism that indirectly requires cell-wall synthesis remains unknown.
MreB Rotation May Facilitate the Maintenance of Rod-Like Cell Shape.
Whereas previous studies have suggested that MreB associates with cell-wall synthases to pattern cell-wall insertion (23, 25), our results demonstrate that cell-wall synthases are also essential for MreB motion. This mutual feedback between MreB and cell-wall assembly may play an important role in elongation. Specifically, MreB rotation could uniformly distribute peptidoglycan insertion sites over the entire cell surface, which is a requirement for maintaining rod shape (27). To test the viability of this hypothesis, we performed computational simulations of cell elongation in the presence or absence of MreB rotation (Fig. 4). The simulations were based on a biophysical model that has been supported by experiments (13, 27). Here, MreB is modeled as either a single long helix or multiple short helical fragments extending the length of the cell (Fig. 4A), and MreB constitutes potential initiation sites for new peptidoglycan strand insertion into the existing cell wall. We find that MreB rotation enables the cell to maintain its rod shape over several doubling times irrespective of the specific pitch and the number of independently moving MreB filaments (Fig. 4 B–D). In stark contrast, cells that lack MreB rotation display shape distortions (Fig. 4 B–D) whose severity increases as a function of decreasing MreB helix angle (the angle between the helix and the cell axis) (Fig. 4C). Whereas these distortions are reduced with increasing MreB fragmentation (Fig. 4D), they are still further reduced in the presence of rotating MreB structures (Fig. 4D). Moreover, even though cells with nonrotating helices of large helix angle (small pitch) appear to retain their rod shape, by comparison to cells with a rotating helix, this shape is more sensitive to environmental stresses such as osmotic shock (Fig. 4 E and F). These simulations thus suggest that MreB rotation contributes to the robust maintenance of rod shape.
Fig. 4.
Computational simulations suggest that MreB rotation improves rod-like cell shape maintenance. (A) Schematic of cell-wall growth simulations in which the synthesis of new glycan strands (blue) initiates from a single continuous (Upper) or a fragmented (Lower) helical MreB structure (yellow). The glycan strands and peptides are shown in green and red, respectively. The helix angle in A and B is 60°. (B) Threefold elongation of the cell wall on the left by inserting strands from a nonrotating (Upper) or rotating (Lower) single MreB helix. (C) Cell width variability as a function of decreasing MreB helix angle, with (magenta) and without (cyan) MreB rotation. The circles correspond to the examples illustrated in B. (D) Cell diameter variability as a function of an increasing number of MreB fragments for a fixed helix angle of 60° (the helical angle of each fragment). (E) Cell walls were grown from a single nonrotating (Upper) and rotating (Lower) MreB helix with a helix angle of 72° until length doubled, and then were osmotically shocked from 1 atm (Left) to 2 atm (Right). (F) Cell diameter variability as a function of osmotic shock (1 atm corresponds to no osmotic shock).
Conclusion
In summary, we have demonstrated that MreB structures rotate around the long cell axis, that this rotation requires cell-wall synthesis, and that MreB rotation may help maintain proper cell morphology. Though challenging, reconstitution experiments should help determine if any of the cell-wall synthesis proteins are themselves motors or if their activities are only indirectly required for an as-yet-unknown motor. The process of cell-wall assembly is highly conserved across bacterial species, such that the functional and physical interdependence of MreB rotation and cell-wall synthesis may be a general mechanism for bacterial morphogenesis.
The biophysical approach taken here will also help elucidate the physical principles that govern other cell biological processes. For example, MreB rotation could be harnessed to transport other cytoplasmic or inner membrane proteins. MreB rotation could also potentially contribute to motility (28). Because cell-wall assembly has proven difficult to directly image, our results indicate that MreB rotation can provide a useful real-time assay for cell-wall morphology and synthesis. In particular, if MreB and cell-wall synthesis are physically coupled, as suggested by our model, the slight right-handed helical orientation of the MreB rotation predicts that newly inserted cell-wall material has the same right-handed helical twist. Our results also raise the possibility that other organisms with cell walls, such as plants and fungi, harness the extracellular energy of cell-wall assembly to power intracellular protein motion.
Methods
Strain Construction.
All strains used except the temperature-sensitive pbp2ts strain were derived by phage transduction and traditional selection methods from the MreB–Rfpsw strain, which is also known as FB83 in Bendezu et al. (9). The pbp2ts mutant used is the strain LMC582 harboring the chromosomal pbpa137ts mutant (29). Into this strain we transformed the MreB–Rfpsw–encoding plasmid pFB262 (9) (detailed in SI Methods).
Sample Preparation and Microscopy.
Cells were grown to exponential phase in liquid culture [M63 minimal medium (30)] at 37 °C and imaged on agarose pads using a modified Nikon TE2000 inverted microscope with both differential interference contrast (DIC) and epifluorescence modules. Cells were imaged at 21 °C every 2.5 s for a duration of 3.3 min. Laser-based feedback was used to overcome drift of the sample in 3D by monitoring the forward scattered light pattern of the detection laser sent through a coverslip-bound polystyrene bead and moving the sample stage (see SI Methods for details).
Image Analysis.
Fluorescent images were analyzed using a custom Matlab code. First, we segmented the images using a semiautomated approach based on standard image processing tools to obtain a coordinate system for each cell. MreB spots in each time frame were identified with subpixel resolution as the local maxima of the denoised (low-pass filtered) and spline-interpolated images. All intensity maxima exceeding a minimum intensity threshold were then connected into raw trajectories based on their mutual interframe distance. To obtain instantaneous velocity vectors we applied a third-order Savitzky–Golay derivative filter to the x and y components of the noisy, raw trajectories. To restrict the analysis to MreB motion in the focal plane, we excluded velocity vectors that originated near the cell borders or were part of small, potentially noisy trajectories (see SI Methods and Fig. S9 for details).
Computational Model of Cell-Wall Growth.
Cell-wall growth was simulated computationally using a biophysical model that incorporates peptidoglycan mechanics and turgor pressure (27). New material was inserted in between already existing strands by successively assembling glycan subunits into long glycan strands and cross-linking them to the surrounding strands. Sites of strand initiation were chosen to lie in close spatial proximity to a helical path representing MreB. We compared intracellular variability in width between cell walls for which the MreB was nonrotating or rotating at an angular velocity of 5 revolutions per doubling of the original cell length (see SI Methods for details).
Supplementary Material
Acknowledgments.
We thank Felipe Bendezu, Piet de Boer, Suzanne Walker, Daniel Kahne, Tanneke den Blaauwen, Natacha Ruiz, and Tom Silhavy for providing strains and helpful suggestions. We also thank Angela Zippilli and the rest of the members of the Gitai lab for technical assistance and advice. Z.G., J.W.S., and N.S.W. are collaboratively supported by National Institute of General Medical Sciences Center for Quantitative Biology/National Institutes of Health Grant 2P50 GM-071508. In addition, support has come from a Human Frontier Science Program Postdoctoral Fellowship (to S.v.T.), National Institutes of Health Director’s New Innovator Awards DP2OD004389 (to Z.G.) and DP2OD006466 (to K.C.H.), a Human Frontier Science Program Young Investigator Award (to Z.G.), the Pew Charitable Trusts and National Science Foundation Award PHY-0844466 (to J.W.S.), and National Institutes of Health Grant GM075000 (to K.C.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108999108/-/DCSupplemental.
References
- 1.Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- 2.Shaevitz JW, Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harb Perspect Biol. 2010;2:a000364. doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard JF, Rutenberg AD. Steady-state helices of the actin homolog MreB inside bacteria: Dynamics without motors. Phys Rev E. 2007;76:031916. doi: 10.1103/PhysRevE.76.031916. [DOI] [PubMed] [Google Scholar]
- 4.Biteen JS, et al. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carballido-Lopez R, Errington J. The bacterial cytoskeleton: In vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell. 2003;4:19–28. doi: 10.1016/s1534-5807(02)00403-3. [DOI] [PubMed] [Google Scholar]
- 6.Defeu Soufo HJ, Graumann PL. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 2004;5:789–794. doi: 10.1038/sj.embor.7400209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bean GJ, et al. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swulius MT, et al. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem Biophys Res Commun. 2011;407:650–655. doi: 10.1016/j.bbrc.2011.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci USA. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci USA. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- 15.Hatfield D, Hofnung M, Schwartz M. Genetic analysis of the maltose A region in Escherichia coli. J Bacteriol. 1969;98:559–567. doi: 10.1128/jb.98.2.559-567.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCoy AJ, Sandlin RC, Maurelli AT. In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J Bacteriol. 2003;185:1218–1228. doi: 10.1128/JB.185.4.1218-1228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang X, et al. The mechanism of action of ramoplanin and enduracidin. Mol Biosyst. 2006;2:69–76. doi: 10.1039/b515328j. [DOI] [PubMed] [Google Scholar]
- 18.Barna JC, Williams DH. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann G, Honikel KO, Knusel F, Nuesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 22.Riva S, Silvestri LG. Rifamycins: A general view. Annu Rev Microbiol. 1972;26:199–224. doi: 10.1146/annurev.mi.26.100172.001215. [DOI] [PubMed] [Google Scholar]
- 23.Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 24.Wientjes FB, Nanninga N. On the role of the high molecular weight penicillin-binding proteins in the cell cycle of Escherichia coli. Res Microbiol. 1991;142:333–344. doi: 10.1016/0923-2508(91)90049-g. [DOI] [PubMed] [Google Scholar]
- 25.Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: New insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dougherty TJ, Kennedy K, Kessler RE, Pucci MJ. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J Bacteriol. 1996;178:6110–6115. doi: 10.1128/jb.178.21.6110-6115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furchtgott L, Wingreen NS, Huang KC. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Mol Microbiol. 2011;81:340–353. doi: 10.1111/j.1365-2958.2011.07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nan B, et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woldringh CL, et al. Autoradiographic analysis of peptidoglycan synthesis in shape and cell division mutants of Escherichia coli LMC500. In: Actor P, Daneo-Moore L, Higgins ML, Salton MRJ, Shockman GD, editors. Antibiotic Inhibition of Bacterial Cell Surface Assembly and Function. Washington, DC: American Society for Microbiology; 1988. pp. 66–78. [Google Scholar]
- 30.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor; 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.