Abstract
The tumor suppressor p53 is activated in response to cellular stress to prevent malignant transformation by activation of the DNA repair machinery to preserve the cell, or by induction of apoptosis to eliminate the cell should the damage prove irrevocable. The gene encoding p53 frequently undergoes inactivating mutations in many human cancers, but WT p53 is often expressed at high levels in melanoma, which, as judged from the malignant nature of the disease, fails to act as an effective tumor suppressor. Here we show that p53 directly up-regulates microRNA-149* (miR-149*) that in turn targets glycogen synthase kinase-3α, resulting in increased expression of Mcl-1 and resistance to apoptosis in melanoma cells. Although deficiency in miR-149* undermined survival of melanoma cells and inhibited melanoma growth in a mouse xenograft model, elevated expression of miR-149* was found in fresh human metastatic melanoma isolates, which was associated with decreased glycogen synthase kinase-3α and increased Mcl-1. These results reveal a p53-dependent, miR-149*–mediated pathway that contributes to survival of melanoma cells, provides a rational explanation for the ineffectiveness of p53 to suppress melanoma, and identifies the expression of miR-149* as a mechanism involved in the increased expression of Mcl-1 in melanoma cells.
Keywords: cell survival, endoplasmic reticulum stress
Melanoma cells have largely adapted to endoplasmic reticulum (ER) stress (1, 2). This not only renders melanoma cells resistant to ER stress-induced apoptosis, but also plays a role in resistance of melanoma cells to various therapeutic agents (3, 4). A major adaptive mechanism of melanoma cells to ER stress appears to be up-regulation of the antiapoptotic Bcl-2 family protein Mcl-1, which plays an essential role in antagonizing the proapoptotic BH3-only proteins PUMA and Noxa that are also up-regulated by the ER stress response, otherwise referred to as the unfolded protein response (UPR) (5). The latter refers to a range of signaling pathways in response to accumulation and aggregation of unfolded and/or misfolded proteins in the ER lumen of cells under ER stress. Importantly, the Mcl-1 expression increases with melanoma progression in vivo (6), and is correlated with the expression of GRP78, an indicator of activation of the UPR (7), and the expression of LDH5, an indicator of hypoglycemia that is one of the causes of ER stress (8). However, the mechanism by which Mcl-1 is up-regulated by the UPR remains elusive.
The tumor suppressor p53 is activated in response to cellular stress to prevent malignant transformation by activation of the DNA repair machinery to preserve the cell, or by induction of apoptosis to eliminate the cell should the damage prove irrevocable (9, 10). The gene encoding p53 frequently undergoes inactivating mutations in many human cancers (11), but mutational inactivation of p53 is uncommon, and WT p53 is often expressed at high levels in melanoma (12), which, as judged from the malignant nature of the disease, fails to act as an effective tumor suppressor. This, along with inappropriate activation of survival signaling pathways such as the RAF/MEK/REK and PI3K/Akt pathways, is important for melanoma progression and resistance to treatment (2, 5, 13,14–15). Although some p53 downstream targets have been shown to be dysregulated, the mechanisms by which the tumor suppressing function of p53 is counteracted in melanoma remain to be elucidated.
microRNAs (miRNAs) are endogenous small (19–24 nt long) noncoding RNAs that regulate gene expression in a sequence specific manner. This is primarily accomplished through binding to 3′UTR of target mRNAs, either targeting the transcripts for degradation or blocking their translation (16). Like conventional protein-coding mRNA, miRNAs are transcribed by RNA polymerase II, spliced, and polyadenylated to produce pri-miRNAs, which contain a stem-loop structure that is recognized and excised by the RNA interference machinery to generate hairpin “precursor” miRNAs (premiRNA) that are approximately 70 nt long. PremiRNAs are cleaved by the RNase III Dicer into a approximately 22-nt miRNA duplex: one strand (miRNA*) of the duplex is often degraded shortly, whereas the other strand serves as a mature miRNA. Nevertheless, there is increasing evidence showing that not all miRNA*s are short-lived. Some of them have been shown to play important roles in regulating gene expression as efficiently as its complementary mature miRNAs (17).
Although a number of miRNAs have been shown to be important components of the p53 tumor suppressor network in various cell types (18–23), the potential interaction between miRNAs and p53 in melanoma cells, in which the expression of p53 may result in distinct biological consequences, have not be established. We report here that p53 mediates a prosurvival pathway by up-regulation of miRNA-149* (miR-149*) that in turn targets glycogen synthase kinase-3α (GSK3α), leading to stabilization of Mcl-1 in melanoma cells under ER stress. We demonstrate that deficiency in miR-149* enhances sensitivity of melanoma cells to apoptosis and retards melanoma growth in a xenograft mouse model. Moreover, we show that the vast majority of metastatic melanomas express elevated levels of miR-149* that is associated with decreased GSK3α and increased Mcl-1. Thus, the p53–miR-149*–GSK3α–Mcl-1 pathway may be an important apparatus in melanoma biology.
Results and Discussion
miR-149* Is Up-Regulated in Melanoma Cells in Response to ER Stress.
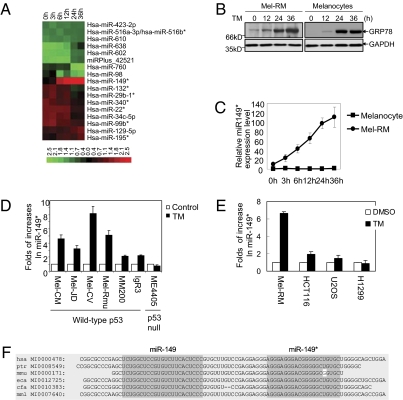
Because Mcl-1 is a major adaptive mechanism of melanoma cells to ER stress and the expression of Mcl-1 is frequently regulated by posttranscriptional mechanisms (24–26), we sought to determine if miRNAs are involved by use of expression profiling in the WT p53 melanoma cell line Mel-RM under pharmacological ER stress that is known to cause up-regulation of Mcl-1 (5). Among the changes in miRNAs triggered by tunicamycin (TM), which induces ER stress by inhibition of glycosylation, an increase in miR-149* was the most pronounced and sustained (Fig. 1 A and B and Table S1), which was subsequently confirmed by using quantitative PCR analysis (Fig. 1C).
Fig. 1.
miR-149* is up-regulated by ER stress in melanoma cells. (A) Mel-RM cells were treated with TM at 3 μM for indicated periods. The filtered miRNA array data were subjected to unsupervised hierarchical clustering analysis. The metric was set as the Euclidean distance. (B) Induction of ER stress by TM (3 μM for indicated periods) in Mel-RM cells and melanocytes as shown by up-regulation of GRP78 measured by Western blot analysis. (C) Quantitative RT-PCR (qRT-PCR) analysis of total RNA shows that TM (3 μM for indicated periods) up-regulates miR-149* in Mel-RM cells but not in melanocytes. (D and E) qRT-PCR analysis shows that TM (3 μM, 12 h) up-regulates miR-149* in a panel of WT p53 melanoma cell lines (D), but not in the p53-null melanoma cell line ME4405, colon cancer (HCT116), osteosarcoma (U2OS), and lung cancer (H1299) cells (E). (F) miR-149 and miR-149* sequences from six species of mammals were compared by using ClustalW serve (www.ch.embnet.org/software/ClustalW.html). Identical amino acid residues are shaded in dark gray. Values are presented as mean ± SD (n = 3 in C–E).
Of note, treatment of Mel-RM cells with TM also resulted in an increase in miR-149, albeit to a lesser extent (Fig. S1 and Table S1). This concurrent induction of miR-149* and miR-149 by ER stress is intriguing, as one strand of the miRNA duplex, in particular, miRNA*, resulting from cleavage of premiRNAs by Dicer, is commonly degraded (27). Although how the complementary pair of miR149/149* are both preserved in melanoma cells remains unknown, the unusual occurrence of miR-149*, particularly its increase in response to ER stress, implicates that it may have a functional role in adaptation of melanoma cells to stress conditions.
We then focused on examination of the effect of ER stress on the expression of miR-149* in melanoma cells. Up-regulation of miR-149* by TM was observed in another six WT p53 melanoma cell lines, but not in the p53-null melanoma cell line ME4405 (Fig. 1D) (28), suggesting this is a common response of melanoma cells harboring WT p53 to ER stress. The effect of ER stress on miR-149* appeared specific for melanoma cells, as it was not significant in melanocytes and cancer cell lines of nonmelanocyte origin such as colon cancer (HCT116), osteosarcoma (U2OS), and p53-deficient lung cancer (H1299) after treatment with TM (Fig. 1 D and E). Collectively, these results indicate that the expression of miR-149* can be induced by ER stress in melanoma cells.
p53 Transcriptionally Regulates miR-149* in Melanoma Cells Under ER Stress.
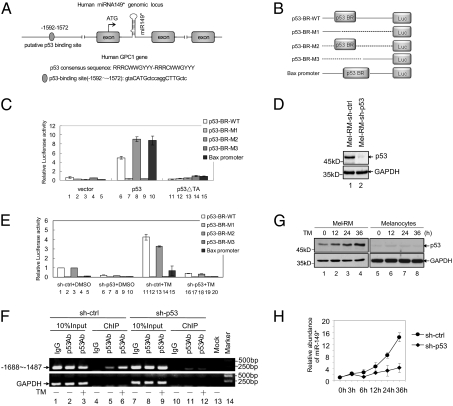
miR-149 and miR-149* are highly conserved among different mammalian species (Fig. 1F), further suggesting that they may have biological functions preserved during evolution. By in silico analysis, it was found that the pri-miR-149 is embedded within the first intron of the human glypican 1 (GPC1) gene (miRBase; http://www.mirbase.org/). Examination of the flanking genomic DNA region identified a putative p53 binding site located 1,592 to 1,572 bp upstream of the GPC1 translational start site (Fig. 2A). This p53 binding region (p53-BR) is transcriptionally responsive to ER stress, as treatment with TM increased the transcriptional activity of the luciferase reporters containing the p53-BR, but had no effect on those lacking the region (Fig. 2 B and E). When WT p53 was cotransfected, the transcriptional activity of the p53-BR was enhanced, but it was limited when a p53 mutant lacking the transactivation domain (p53ΔTA) was introduced (Fig. 2 B and C). In addition, knockdown of p53 by shRNA markedly inhibited activation of the p53-BR–containing luciferase reporters, confirming that the response of the p53-BR to ER stress is mediated by p53 (Fig. 2 D and E). This was supported by binding of p53 to this region, which was enhanced by the addition of TM (Fig. 2F). Therefore, the putative p53-BR is responsive to the cellular levels of p53 under ER stress. Consistently, treatment with TM up-regulated the endogenous GPC1 mRNA levels (Fig. S2), suggesting that the GPC1 gene may be a unique p53 transcriptional target in melanoma cells.
Fig. 2.
miR-149* is a transcriptional target of p53. (A) Schematic illustration that the premiR-149 is embedded within the first intron of the human GPC1 gene. A putative p53-BR located 1,592 to 1,572 bp upstream of the GPC1 translational start site is also illustrated. (B) A schematic illustration of pGL3-basic based reporter constructs used in dual luciferase assays to examine the transcriptional activity of the p53-BR in response to ER stress. Dotted lines indicated deleted region. (C) Mel-RM cells were cotransfected with the indicated reporter constructs and Renilla luciferase plasmids. Twenty-four hours later, transcriptional activity was determined by luciferase assays. A Bax promoter reporter was included as a control. (D) Western blot analysis showing stable knockdown of p53 by shRNA. (E) Mel-RM cells with or without p53 knocked down by shRNA were cotransfected with the indicated reporter constructs and Renilla luciferase plasmids. Twenty-four hours later, cells were treated with TM (3 μM) for another 24 h, followed by measurement of reporter activity by using luciferase assays. (F) ChIP analysis shows binding of p53 to the p53-BR. Input and immunoprecipitated DNA (by an antibody against p53) from Mel-RM cells before and after treatment with TM (3 μM) for 24 h were amplified by PCR with the primer pair complementary to −1,688 to −1,487 of the GPC1 gene. An anti-IgG antibody was used as a negative control. (G) Western blotting analysis of p53 in whole-cell extracts from Mel-RM cells and melanocytes before and after treatment with TM (3 μM) for indicated periods. (H) qRT-PCR analysis shows that shRNA knockdown of p53 inhibited up-regulation of miRNA-149* by TM (3 μM for indicated periods) in Mel-RM cells. Values are shown as mean ± SD (n = 3 in C, D, and H). Ctrl, control; sh, shRNA; p53ΔTA, mutant p53 lacking the transactivation domain.
Similar to induction of miR-149* by ER stress (Fig. 1 C–E and Fig. S1), the p53 levels were increased by TM in Mel-RM cells, but not in melanocytes, HCT116, U2OS, and H1299 cells (Fig. 2G and Fig. S3), suggesting an association between the endogenous expression of p53 and miR-149* in melanoma cells under ER stress. This was further demonstrated in Mel-RM cells with p53 knockdown by shRNA. Deficiency in p53 not only attenuated the binding of p53 to the p53-BR in ChIP assays (lanes 5 and 6 vs. 11 and 12, Fig. 2F), but also reduced the expression of miR-149* in response to TM (Fig. 2H). Moreover, knockdown of p53 similarly inhibited TM-induced up-regulation of the GPC1 mRNA in Mel-RM cells (Fig. S2). Together, these data demonstrate that binding of p53 to the p53-BR at the GPC1 gene transcriptionally up-regulates miR-149* in melanoma cells under ER stress.
Intriguingly, p53-mediated up-regulation of miR-149* appeared highly specific to melanoma cells under ER stress, as induction of p53 by the genotoxic drug doxorubicin or the pharmacological p53 activator nutlin3 did not cause any significant change in the expression of miR-149* (Fig. S4). This suggests that p53 is necessary but not sufficient to activate the p53-BR at the GPC1 gene promoter. It is conceivable that some ER stress-responsive transcription cofactors may be required for p53 to up-regulate miR-149* expression in melanoma cells. Similarly noticeable was that induction of transcriptional targets of p53 is also selective in melanoma cells under ER stress, in that, except for Noxa, other canonical p53 targets including p21, MDM2, Bax, and Bad were not increased by TM along with GPC1 and miR-149* (Fig. S2). This further indicates that the transactivation activity of p53 in melanoma cells under ER stress is regulated by additional mechanisms that are yet to be identified.
miR-149* Down-Regulates Cellular Levels of GSK3α.
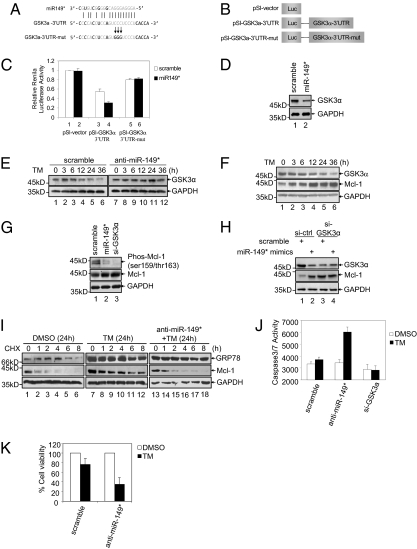
We searched the MicroCosm and miRBase databases for the potential targets of miR-149*. Among the candidates identified, the 3′UTR of GSK3α contains a putative region (nucleotides 132–152) that matched perfectly to the miR-149* “seed” region (Fig. 3A). Because GSK3α is known to target Mcl-1 for phosphorylation and subsequent proteasomal degradation (29), we examined if GSK3α is indeed down-regulated by miR-149*, which may be involved in up-regulation of Mcl-1 in melanoma cells under ER stress (5). To this end, we introduced luciferase reporter plasmids of the 3′UTR of GSK3α into Mel-RM cells (Fig. 3 B and C). The reporter activity was markedly suppressed by the presence of the 3′UTR of GSK3α, which was reversed when the 3′UTR was mutated (Fig. 3C), suggesting that the 3′UTR of GSK3α was inhibited by endogenous miR-149*. In support, introduction of anti–miR-149* into Mel-RM cell increased (Fig. S5)—whereas the addition of miR-149* mimics further reduced—the reporter activity (Fig. 3C). Therefore, miR-149* targets the 3′UTR of GSK3α in melanoma cells. Consistently, introduction of miR-149* mimics into Mel-RM cells down-regulated the GSK3α protein levels (Fig. 3D). Moreover, the expression of GSK3α in Mel-RM cells after exposure to TM decreased progressively, whereas this reduction was blocked by pretreatment with anti-miR-149* (Fig. 3E). Collectively, these results substantiate that GSK3α is a bona fide target of miR-149* that is modulated in melanoma cells subjected to ER stress.
Fig. 3.
miR-149* targets GSK3α and up-regulates Mcl-1 in melanoma cells. (A) Schematic illustration of base paring between miR-149* and the 3′UTR of GSK3α. Substitution of three consecutive cytosine bases with guanine (CCC to GGG) for the mutant reporter construct is also shown. (B) Schematic illustration of pSI-CHECK2–based luciferase reporter constructs used for examining the effect of miR-149* on the 3′UTR of GSK3α (C) Mel-RM cells were cotransfected with the indicated reporter constructs and Renilla luciferase plasmids. Twenty-four hours later, reporter activity was measured by using luciferase assays. (D) Mel-RM cells were transfected with scramble or miR-149* mimics. Thirty-six hours later, whole cell lysates were subjected to Western blot analysis. (E) Mel-RM cells were transfected with scramble and anti–miR-149* oligonucleotide. Two hours later, cells were treated with TM (3 μM) for the indicated periods. Whole-cell lysates were then subjected to Western blot analysis. (F) Whole-cell lysates from Mel-RM cells treated with TM (3 μM) for the indicated periods were subjected to Western blot analysis. (G) Mel-RM cells transfected with indicated oligonucleotides. Twenty-four hours later, cells were treated with MG132 (25 μM) for another 6 h. Whole-cell lysates were then subjected to Western blot analysis. Phosphorylation of Mcl-1 was probed by a Ser159/Thr163 phosphorylation-specific antibody. (H) Mel-RM cells were transfected with the indicated oligonucleotides. Thirty-six hours later, whole-cell lysates were subjected to Western blot analysis. (I) Mel-RM cells with or without transfection with anti–miR-149* oligonucleotides for 24 h were treated with DMSO or TM (3 μM) for another 24 h before the addition of cycloheximide (200 μg/mL) for the indicated periods. Whole-cell lysates were subjected to Western blot analysis. (J) Mel-RM cells were transfected with scramble, anti-miR-149* or si-GSK3α. Twenty-four hours later, cells were treated with TM (3 μM) for another 36 h. Whole-cell lysates were then subjected to measurement of caspase 3/7 activity. (K) Mel-RM cells transfected with scramble or anti–miR-149* oligonucleotides were treated with TM (10 μM) for 36 h. Cell death was analyzed mainly by microscopic counting. Values are presented as mean ± SD (n = 3 in C, J, and K). si, siRNA.
Among other predicted targets, E2F1 has been shown to be down-regulated by miR-149*, which played a role in induction of apoptosis in neuroblastoma and HeLa cells (30). However, although induction of ER stress by TM in melanoma cell also resulted in a decrease in E2F1 along with up-regulation of miR-149* (Fig. 1E and Fig. S6), suggesting that miR-149* may similarly target E2F1 in melanoma cells under ER stress, this did not appear to be sufficient to trigger apoptosis. Furthermore, over-expression of E2F1 did not alter the levels of Mcl-1, nor did it impinge on induction of apoptosis, as shown by the lack of effects on PARP cleavage (Fig. S6). This is in contrast to over-expression of GSK3α that decreased Mcl-1 and rendered melanoma cells sensitive to ER stress-induced apoptosis (Fig. S6). Therefore, the antiapoptotic effect of miR-149* in melanoma cells under ER stress is unlikely mediated by E2F1, even though it is targeted by miR-149* resulting from ER stress.
Up-Regulation of Mcl-1 in Melanoma Cells Under ER Stress Is Associated with Increased Expression of miR-149*.
Upon exposure of Mel-RM cells to TM, the GSK3α protein levels decreased in parallel with the increase in Mcl-1 (Fig. 3F), suggesting that up-regulation of Mcl-1 in the cells under ER stress may be associated with the decreased expression of GSK3α. In support of this, introduction of miR-149* mimics inhibited the phosphorylation of Mcl-1 and enhanced the stability of the Mcl-1 protein (lane 2, Fig. 3G), recapitulating the effect of knockdown of GSK3α (lane 3, Fig. 3G). Knockdown of GSK3α in combination with miR-149* mimics did not further increase the levels of Mcl-1 (Fig. 3H), suggesting that down-regulation of GSK3α is required for miR-149*-mediated up-regulation of Mcl-1. The role of miR-149* in regulation of the expression of Mcl-1 was further confirmed by inhibition of endogenous miR-149*. The half-life of the endogenous Mcl-1 was prolonged upon exposure to TM, which was attenuated when miR-149* was inhibited (Fig. 3I). Therefore, stabilization of the Mcl-1 protein as a corollary of miR-149*-mediated down-regulation of GSK3α contributes to up-regulation of Mcl-1 in melanoma cells under ER stress. Consistently, inhibition of miR-149* promoted activation of caspase 3/7 and enhanced cell death in Mel-RM cells after exposure to TM (Fig. 3 J and K and Fig. S7), indicating that the increased levels of miR-149* in melanoma cells upon ER stress blocks apoptotic signaling. The effect of inhibition of miR-149* on TM-induced apoptosis was recapitulated by knockdown of p53, whereas introduction of miR-149* mimics inhibited PARP cleavage induced by TM in cells with p53 knocked down (Fig. S7).
Because miR-149 was also up-regulated, albeit to a lesser extent, by ER stress in melanoma cells, we examined if miR-149 has a similar role in protection of melanoma cells from ER stress-induced apoptosis. Introduction of neither anti–miR-149 nor miR-149 mimics altered sensitivity to ER stress-induced apoptosis in melanoma cells with or without p53 knockdown, indicating that miR-149 does not impinge on survival of melanoma cells under ER stress (Fig. S7). Taken together, results from these studies identify a p53-dependent, miR-149*-mediated prosurvival signaling pathway that contributes to the increased expression of Mcl-1 and resistance to apoptosis in melanoma cells upon ER stress.
To further confirm the prosurvival role of p53 in melanoma cells under ER stress, we exposed Mel-RM and ME4405 (p53-null) cells to TM at an increased concentration (10 μM). Although TM did not induce PARP cleavage in Mel-RM cells even at this high dose, cleavage of PARP was evident in ME4405 cells and Mel-RM cells with p53 knockdown as well (Fig. S8), suggesting that melanoma cells lacking p53 are more sensitive to ER stress-induced apoptosis. Introduction of p53 into ME4405 cells inhibited cleavage of PARP, indicative of inhibition of apoptosis. This was associated with up-regulation of Mcl-1 (Fig. S8). These data substantiate the role of p53-dependent up-regulation of Mcl-1 in protection of melanoma cells from ER stress-induced apoptosis.
GSK3α is known to regulate other targets besides Mcl-1. For example, it has been reported to inhibit β-catenin expression in Xenopus embryos and human bronchial smooth muscle cells (31–33). However, ER stress did not notably trigger changes in the expression of β-catenin, even when GSK3α is overexpressed (Fig. S9), indicating that GSK3α does not regulate β-catenin in melanoma cells. In support of this, regulation of the activity of the pTOP-flash luciferase construct, an established indicator of Wnt/β-catenin activity (34), in melanoma cells under ER stress appeared to be independent of GSK3α, in that over-expression of GSK3α did not reverse the slight increase in the pTOP-flash activity in melanoma cells triggered by TM (Fig. S9). Therefore, protection of melanoma cells from ER stress-induced apoptosis by down-regulation of GSK3α is primarily mediated by Mcl-1 rather than β-catenin.
Biological Significance of p53-Dependent, miR-149*–Mediated Pathway in Melanoma.
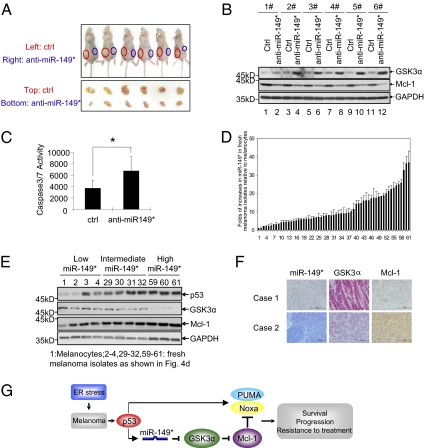
To determine the functional significance of the p53–miR-149*–GSK3α–Mcl-1 pathway on melanoma biology in vivo, tumor growth of Mel-RM cells with or without miR-149* stably inhibited by anti–miR-149* was examined after s.c. transplantation into nu/nu mice. Deficiency in miR-149* resulted in marked retardation of tumor initiation and growth in vivo (Fig. 4A and Fig. S10). Examination of tumors from postmortem animals indicated that they maintained their respective phenotypes, low GSK3α/high Mcl-1 for controls and high GSK3α/low Mcl-1 for those deficient in miR-149* (Fig. 4B). The latter also displayed comparatively high levels of caspase 3/7 activity (Fig. 4C), mirroring the reduced ability of the cells to survive stress conditions.
Fig. 4.
Biological significance of the p53–miR-149*–GSK3α–Mcl-1 pathway in melanoma. (A) Representative photographs of xenografts of Mel-RM cells stably infected with lentivirus possessing scramble or anti-–iR-149* oligonucleotides grown in nude mice for 36 d. (B) Whole-cell lysates of ex vivo tumors of Mel-RM cells stably possessing scramble or anti–miR-149* oligonucleotides were subjected to Western blotting analysis. (C) Whole-cell lysates of ex vivo tumors of Mel-RM cells stably possessing scramble or anti–miR-149* oligonucleotides were subjected to caspase 3/7 activity. Values are presented as mean ± SD (n = 6). Two-tailed Student t test: *P < 0.05, **P < 0.001. (D) qRT-PCR analysis of the expression of miR-149* in 60 fresh melanoma isolates (isolates 2–61) relative to melanocytes (1). (E) Western blot analysis of GSK3α and Mcl-1 in melanocytes and fresh melanoma isolates with relatively low, intermediate, and high levels of expression of miR-149*. (F) Representative photomicrographs of ISH (miR-149*, blue) and IHC (GSK3α, red; and Mcl-1, brown) studies on melanoma tissue sections. (Scale bar: 100 μm.) (G) A schematic illustration of the proposed model depicting a unique p53-mediated prosurvival pathway in melanoma.
To further validate the biological importance of the the p53–miR-149*–GSK3α–Mcl-1 signaling pathway in melanoma in vitro, we analyzed the expression of miR-149* in a panel of 60 WT p53 fresh metastatic melanoma isolates, and found that 59 of 60 samples expressed constitutively higher levels of miR-149* relative to pooled melanocytes of three different lines (HEMa-LP, HEMa-DP, and HEMn-LP; Fig. 4D). Examination of representative fresh melanoma isolates sampled by relatively low (n = 4), intermediate (n = 4), and high (n = 3) levels of miR-149* showed that melanomas with low miR-149* expression displayed relatively low levels of p53, high levels of GSK3α, and low levels of Mcl-1, whereas intermediate and high miR-149* expression associated with progressively more p53 and Mcl-1 and less GSK3α (Fig. 4E). In situ analysis of melanoma tissue sections (n = 18) similarly demonstrated that melanomas displaying relatively high levels of miR-149* had low levels of GSK3α and high levels of Mcl-1, and conversely, those with relatively low levels of miR-149* had high levels of GSK3α and low levels of Mcl-1 (Fig. 4F and Fig. S11). Therefore, the expression of the key elements of the p53–miR-149*–GSK3α–Mcl-1 pathway in human melanoma in vivo is in accordance with the regulatory model identified in vitro (Fig. 4G). Notably, a similar association in the expression of p53, miR-149*, GSK3α, and Mcl-1 was not observed in HCT116, U2OS, and H1299 cells even when ER stress was induced (Fig. 1E and Fig. S3), suggesting that this prosurvival signaling pathway is not common in other cancer types.
p53 has previously been shown to transcriptionally regulate a number of miRNAs such as miR-34a, miR-34b, miR-34c, and miR-107, and can also modulate miRNA biogenesis independently of transcription (18–23). These have been proposed to contribute to its tumor suppressing function in some cancers (35–37). Despite the presence of high levels of WT p53 in some melanomas (38), the disease is highly malignant and largely resistant to available therapeutic agents (3). Here we show that, instead of being a functionally active tumor suppressor, p53 mediates a prosurvival pathway through up-regulation of miR-149* in melanoma cells under ER stress. The latter is a constitutive pressure encountered by melanoma cells in vivo and a state to which melanoma cells have adapted for survival (1) This has been previously shown by increased expression of GRP78, a commonly used indicator of activation of the UPR, with melanoma progression and resistance of melanoma cells to ER stress-induced apoptosis (7). Additionally, to verify that p53—but not its variants or homologues—transactivates miR149*, p63, p73, and two p53 transcriptional variants—p53-Δ40 and p53-Δ133 (the latter serves to counteract the proapoptotic function of p53) (39)—were included for testing their transactivation abilities. As shown in Fig. S12, only WT p53 was shown to significantly enhance miR149* promoter activity, suggesting that p53 is the bona fide regulator of miR149* expression. Up-regulation of Mcl-1 is known to be induced by ER stress and is one of the major prosurvival mechanisms of melanoma cells (5, 40). Thus, an unexpected function of p53 in promoting Mcl-1 expression via miR-149* and GSK3α provides an advantage for melanoma cell survival and conceivably resistance to treatment.
Despite our clear evidence showing a prosurvival role of miR-149* in melanoma cells, miR-149* has been reported to induce apoptosis by inhibiting Akt1 and E2F1 in other cell types (30). This suggests that, like p53, miR-149* has distinct biological functions in melanoma. Although functional consequences of miR expression are commonly tissue- and cell type-specific (41), p53 regulation of its target genes also occurs in a cell type- and stimulus-dependent manner (42, 43). Although the mechanisms involved remain to be defined, our results indicate that melanoma-specific up-regulation of miR-149* by p53 in cells under ER stress is a result of selective activation of its transcription along with the host gene (the GPC1 gene). Many mechanisms are known to regulate promoter selectivity of p53 target genes, such as posttranslational modifications of p53, interactions of p53 with cofactors, and SNPs in putative p53 response elements (42–44). Further studies are required to address whether any of these is involved in specific regulation of miR-149* by p53 in melanoma cells.
Because most patients from whom fresh melanoma isolates were obtained and analyzed in this study are still alive, we are unable to conclude at present whether the levels of miR-149* is of significance in predicting disease progression and prognosis of patients. Nevertheless, the elevated expression of miR-149* at high frequency in melanoma makes it an attractive candidate for further investigation, either as a biomarker of melanoma progression or a therapeutic target. The failure of p53 in regulation of miRNAs that are known to be responsive to p53 in other cell types may also contribute to its ineffectiveness to suppress melanoma (Table S1).
Materials and Methods
ChIP.
Mel-RM cells were cross-linked with 1% formaldehyde for 10 min. ChIP assay was performed by using anti-p53 and the ChIP assay kit (Upstate/Millipore) according to the manufacturer's instructions. Anti-mouse IgG (ChIP assay kit) were used as controls. The bound DNA fragments were subjected to PCR reactions using the following primer pair: 5′>GTGAGACCACACAGAGAGAGAGCG<3′ and 5′>GGACCCAATCCAAGTGTGCATTTC<3′. PCR products were separated by gel electrophoresis on 2% agarose gel.
In Situ Hybridization.
For in situ hybridization (ISH), biotin-labeled probes were purchased from Exiqon for human miR-149* and a negative control scramble miRNA probe. Slides were hybridized in 20 nM of probe diluted in 200 μL of hybridization buffer at 21 °C below the manufacturer's supplied melting temperature. Hybridization was performed on a Hybridizer (S2450; Dako) overnight. Signal amplification was carried out with 1:50 biotinyl tyramide. Slides were then incubated in ExtrAvidin–alkaline phosphatase (Sigma), followed by incubation in detection buffer and then in NBT/BCIP. Slides were not counterstained.
Immunohistochemistry.
Staining of GSK3α and Mcl-1 on tissue sections by immunohistochemistry (IHC) was carried out by using previously described methods. GSK3α was detected with Liquid Permanent Red chromogen and Mcl-1 with 3,3′diaminobenzidine (DAB). Sections were counterstained with Harris hematoxylin.
Supplementary Material
Acknowledgments
The authors thank Mrs. Liqing Zhuang for her technique support in immunohistochemistry staining of Mcl-1 expression on tissue sections. This work was supported by National Natural Science Foundation of China Grants 31030046; Ministry of Science and Technology of China Grants 2010CB912804, 2011CB966302, and 2011CBA01103; Chinese Academy of Sciences Grant XDA01020104; and by the National Health and Medical Research Council (NHMRC), the Cancer Council of New South Wales, and Cancer Institute New South Wales, Australia. X.D.Z. is supported by an NHMRC senior research fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019312108/-/DCSupplemental.
References
- 1.Hersey P, Zhang XD. Adaptation to ER stress as a driver of malignancy and resistance to therapy in human melanoma. Pigment Cell Melanoma Res. 2008;21:358–367. doi: 10.1111/j.1755-148X.2008.00467.x. [DOI] [PubMed] [Google Scholar]
- 2.Jiang CC, et al. Inhibition of MEK sensitizes human melanoma cells to endoplasmic reticulum stress-induced apoptosis. Cancer Res. 2007;67:9750–9761. doi: 10.1158/0008-5472.CAN-07-2047. [DOI] [PubMed] [Google Scholar]
- 3.Griffith TS, Chin WA, Jackson GC, Lynch DH, Kubin MZ. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J Immunol. 1998;161:2833–2840. [PubMed] [Google Scholar]
- 4.Soengas MS, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 5.Jiang CC, et al. Up-regulation of Mcl-1 is critical for survival of human melanoma cells upon endoplasmic reticulum stress. Cancer Res. 2008;68:6708–6717. doi: 10.1158/0008-5472.CAN-08-0349. [DOI] [PubMed] [Google Scholar]
- 6.Zhuang L, et al. Mcl-1, Bcl-XL and Stat3 expression are associated with progression of melanoma whereas Bcl-2, AP-2 and MITF levels decrease during progression of melanoma. Mod Pathol. 2007;20:416–426. doi: 10.1038/modpathol.3800750. [DOI] [PubMed] [Google Scholar]
- 7.Zhuang L, et al. Expression of glucose-regulated stress protein GRP78 is related to progression of melanoma. Histopathology. 2009;54:462–470. doi: 10.1111/j.1365-2559.2009.03242.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang L, et al. Lactate dehydrogenase 5 expression in melanoma increases with disease progression and is associated with expression of Bcl-XL and Mcl-1, but not Bcl-2 proteins. Mod Pathol. 2010;23:45–53. doi: 10.1038/modpathol.2009.129. [DOI] [PubMed] [Google Scholar]
- 9.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 10.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 11.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 12.Sparrow LE, Soong R, Dawkins HJ, Iacopetta BJ, Heenan PJ. p53 gene mutation and expression in naevi and melanomas. Melanoma Res. 1995;5:93–100. doi: 10.1097/00008390-199504000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Dhawan P, Singh AB, Ellis DL, Richmond A. Constitutive activation of Akt/protein kinase B in melanoma leads to up-regulation of nuclear factor-kappaB and tumor progression. Cancer Res. 2002;62:7335–7342. [PubMed] [Google Scholar]
- 14.Jinushi M, et al. Milk fat globule EGF-8 promotes melanoma progression through coordinated Akt and twist signaling in the tumor microenvironment. Cancer Res. 2008;68:8889–8898. doi: 10.1158/0008-5472.CAN-08-2147. [DOI] [PubMed] [Google Scholar]
- 15.Liu ZJ, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 2006;66:4182–4190. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- 16.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 17.German MA, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 18.Barlev NA, Sayan BS, Candi E, Okorokov AL. The microRNA and p53 families join forces against cancer. Cell Death Differ. 2010;17:373–375. doi: 10.1038/cdd.2009.73. [DOI] [PubMed] [Google Scholar]
- 19.Braun CJ, et al. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 21.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki HI, et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 23.Yamakuchi M, et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci USA. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwickart M, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 25.Warr MR, Shore GC. Unique biology of Mcl-1: Therapeutic opportunities in cancer. Curr Mol Med. 2008;8:138–147. doi: 10.2174/156652408783769580. [DOI] [PubMed] [Google Scholar]
- 26.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Guo L, Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS ONE. 2010;5:e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avery-Kiejda KA, et al. Small molecular weight variants of p53 are expressed in human melanoma cells and are induced by the DNA-damaging agent cisplatin. Clin Cancer Res. 2008;14:1659–1668. doi: 10.1158/1078-0432.CCR-07-1422. [DOI] [PubMed] [Google Scholar]
- 29.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Lin RJ, Lin YC, Yu AL. miR-149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol Carcinog. 2010;49:719–727. doi: 10.1002/mc.20647. [DOI] [PubMed] [Google Scholar]
- 31.Thomas GM, et al. A GSK3-binding peptide from FRAT1 selectively inhibits the GSK3-catalysed phosphorylation of axin and beta-catenin. FEBS Lett. 1999;458:247–251. doi: 10.1016/s0014-5793(99)01161-8. [DOI] [PubMed] [Google Scholar]
- 32.Yost C, et al. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 33.Nunes RO, et al. GSK-3/beta-catenin signaling axis in airway smooth muscle: Role in mitogenic signaling. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1110–L1118. doi: 10.1152/ajplung.00500.2007. [DOI] [PubMed] [Google Scholar]
- 34.Xie W, Jin L, Mei Y, Wu M. E2F1 represses beta-catenin/TCF activity by direct upregulation of Siah1. J Cell Mol Med. 2008;13:1719–1727. doi: 10.1111/j.1582-4934.2008.00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 38.Box NF, Terzian T. The role of p53 in pigmentation, tanning and melanoma. Pigment Cell Melanoma Res. 2008;21:525–533. doi: 10.1111/j.1755-148X.2008.00495.x. [DOI] [PubMed] [Google Scholar]
- 39.Bourdon JC, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolter KG, et al. Therapeutic window for melanoma treatment provided by selective effects of the proteasome on Bcl-2 proteins. Cell Death Differ. 2007;14:1605–1616. doi: 10.1038/sj.cdd.4402163. [DOI] [PubMed] [Google Scholar]
- 41.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 42.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 43.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 44.Tomso DJ, et al. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc Natl Acad Sci USA. 2005;102:6431–6436. doi: 10.1073/pnas.0501721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.