Abstract
Helper T-cell activation generally requires the coreceptor CD4, which binds MHC class II molecules. A remarkable feature of the CD4–MHC class II interaction is its exceptionally low affinity, which ranges from KD = ∼200 μM to >2 mM. Investigating the biological role of the much lower affinity of this interaction than those of other cell–cell recognition molecules will require CD4 mutants with enhanced binding to MHC class II for testing in models of T-cell development. To this end, we used in vitro-directed evolution to increase the affinity of human CD4 for HLA-DR1. A mutant CD4 library was displayed on the surface of yeast and selected using HLA-DR1 tetramers or monomers, resulting in isolation of a CD4 clone containing 11 mutations. Reversion mutagenesis showed that most of the affinity increase derived from just two substitutions, Gln40Tyr and Thr45Trp. A CD4 variant bearing these mutations bound HLA-DR1 with KD = 8.8 μM, compared with >400 μM for wild-type CD4. To understand the basis for improved affinity, we determined the structure of this CD4 variant in complex with HLA-DR1 to 2.4 Å resolution. The structure provides an atomic-level description of the CD4-binding site on MHC class II and reveals how CD4 recognizes highly polymorphic HLA-DR, -DP, and -DQ molecules by targeting invariant residues in their α2 and β2 domains. In addition, the CD4 mutants reported here constitute unique tools for probing the influence of CD4 affinity on T-cell activation and development.
The analysis of leukocyte surface molecules involved in cell–cell recognition has been critical to advancing our understanding of immunological phenomena, such as T-cell activation. From the wide range of interactions examined to date, it has emerged that leukocyte surface molecules interact with remarkably low affinities, with dissociation constants (KD's) generally between 1 and 100 μM, mainly due to fast off-rates (koff >1 s−1) (1–3). For example, T-cell receptors (TCRs) bind cognate peptide–MHC (pMHC) complexes with KD's between 1 and 100 μM, compared with ∼10 nM for typical antibody–antigen interactions. The low affinities and rapid kinetics (both on- and off-rates) of TCR binding to pMHC are believed to allow T cells to scan with high speed and sensitivity the numerous self-pMHC complexes on antigen-presenting cells (APCs) to detect and respond to antigens expressed at low numbers (4).
Of all leukocyte cell–cell recognition molecules characterized so far, the T-cell coreceptor CD4 is the most enigmatic in terms of its binding properties (1). The interaction of CD4 with MHC class II molecules greatly augments cytokine production by helper T cells (5) and substantially reduces the number of antigenic peptides on APCs required for T-cell triggering (6). Surprisingly, however, CD4 binds MHC class II with exceptionally low affinity compared with other cell–cell recognition molecules (KD = 1–100 μM) (1–3), including the T-cell coreceptor CD8, which binds MHC class I molecules. For the CD4–MHC class II interaction, KD's have been variously estimated to range from ∼200 μM (for human CD4 binding to mouse MHC class II) (7) to >2 mM (for human CD4 binding to human MHC class II) (1). By comparison, the affinity of CD8 for MHC class I ranges from ∼10 μM (for mouse CD8 binding to mouse MHC class I) (8, 9) to ∼200 μM (for human CD8 binding to human MHC class I) (10).
The striking difference in the binding properties of the CD4 coreceptor compared with other cell–cell recognition molecules is likely to be of considerable importance for regulating T-cell activation and development. One hypothesis is that evolution has finely tuned the affinity of CD4 for MHC class II to enable peripheral T cells to respond effectively to the very low abundance of foreign pMHC molecules on APCs (sensitivity), yet avoid activation by the far greater number of self-pMHC molecules (discrimination), which could result in autoimmunity. In addition, evolution may have calibrated the affinity of CD4 to help ensure that developing T cells undergo appropriate thymic selection whereby weak interactions with self-pMHC promote T-cell survival (positive selection), and strong interactions induce apoptosis (negative selection) (11, 12).
A direct approach to addressing the biological role of the remarkably low affinity of CD4 for MHC class II involves generating animals transgenic for high-affinity CD4 mutants. Would expression of such mutants result in deletion of T cells in the thymus that would otherwise have been positively selected? Conversely, would the presence of T cells in the periphery expressing high-affinity CD4 increase the incidence of autoimmune disease? However, no CD4 mutants with enhanced binding to MHC have been described. Accordingly, we used in vitro-directed evolution by yeast surface display (YSD) to increase the affinity of human CD4 for the MHC class II molecule HLA-DR1 to the low micromolar range. These variants represent unique tools for investigating the influence of CD4 affinity on T-cell activation and development in animal models.
To understand the basis for improved binding, we determined the structures of two affinity-matured CD4 mutants in complex with HLA-DR1 to high resolution (<2.5 Å). The only previous structure of a CD4–MHC class II complex is for human CD4 bound to the mouse MHC class II molecule I-Ak (13). However, its low resolution (4.3 Å) precluded a detailed description of the CD4–MHC class II interface. The CD4–HLA-DR1 structures reported here permitted us to define the CD4-binding site on HLA-DR1 at the atomic level and to explain the ability of CD4 to recognize highly polymorphic HLA-DR, -DP, and -DQ molecules, which is central to CD4 function (5).
Results and Discussion
Yeast Display of CD4 and Design of Mutant Library.
The YSD system for affinity maturation relies on expression of a library of mutants on the surface of yeast, followed by selection of variants with improved affinity (14). The extracellular portion of CD4 consists of four Ig-like domains (D1–D4). To display CD4 on yeast, we fused CD4 to the C terminus of the yeast agglutinin protein Aga2p (14). We first tested the D1 domain of CD4 alone for yeast display, because D1 is the only CD4 domain directly involved in binding MHC class II (13). However, only weak staining was observed with the conformation-dependent anti-human CD4 D1 domain monoclonal antibody (mAb) RPA-T4 (Fig. S1). In marked contrast, a CD4 construct comprising domains D1 and D2 reacted strongly with this mAb, which confirmed proper folding of CD4 D1–D2 on the yeast cell surface. We therefore carried out directed evolution using the CD4 D1–D2 construct.
The structure of human CD4 D1–D2 bound to mouse I-Ak (13) served as a guide for designing CD4 mutant libraries for affinity maturation. In the complex, the C′′ strand and DE helix of CD4 D1 interact with the α2 and β2 domains of I-Ak. On the basis of this information, 12 CD4 residues in the presumed interface with HLA-DR1 were mutated by overlap PCR with degenerate primers to create a CD4 D1–D2 mutant library of 4 × 107 clones. These residues were at CD4 positions 35, 40, 42–48, 59, 60, and 63.
Affinity Selection of CD4 Mutant Library.
Because the affinity of the CD4–MHC class II interaction is exceedingly low (1, 7), we sorted the CD4 library by flow cytometry using fluorescent-labeled HLA-DR1 tetramers, rather than monomers, to augment the avidity of the selecting ligand. Following two rounds of sorting with decreasing concentrations of HLA-DR1 tetramers (1 μM, then 0.1 μM), the library was subjected to one more round of sorting with HLA-DR1 monomers (4 μM) (Fig. 1A). After three rounds of sorting, most yeast cells bound HLA-DR1, and these were plated to obtain single clones. DNA sequencing showed that a single CD4 mutant clone (A1) dominated this enrichment process and that all other mutants differed only slightly from A1. Remarkably, the A1 clone contained mutations in 11 of 12 putative HLA-DR1–contacting residues (Fig. 1B).
Fig. 1.
Affinity maturation of human CD4 by yeast surface display. (A) The CD4 mutant library was sorted by 1 μM HLA-DR1 tetramer for round 1 (Rd1), 0.1 μM HLA-DR1 tetramer for round 2 (Rd2), and 4 μM HLA-DR1 monomer for round 3 (Rd3). The positive population (boxed) is shown in the gate. (B) Amino acid sequence alignment of wild-type (WT) CD4 and the dominant A1 mutant from Rd3 around the positions targeted for mutagenesis (residues 35, 40, 42–48, 59, 60, and 63). Mutated residues are shaded; only Arg59 is wild type.
Reversion Mutagenesis of Affinity-Matured CD4.
To pinpoint which of the 11 mutations in CD4 A1 had been selected because they improved binding to HLA-DR1, and not for other reasons (e.g., because they increased surface expression of CD4), we performed reversion mutagenesis. Thus, each mutated residue in CD4 A1 was individually converted to the corresponding wild-type residue. Yeast cells displaying each reversion mutant were then tested for binding to HLA-DR1 tetramers by flow cytometry. Surprisingly, replacement of Trp45 in CD4 A1 by the wild-type threonine residue (W45T) completely abolished binding (not shown), indicating that this mutation was critical for increased affinity. The 10 other single amino acid changes had much less pronounced effects on binding to HLA-DR1 tetramers, although the mutants showed some variation.
To test the CD4 reversion mutants under more stringent conditions, we next examined binding to HLA-DR1 monomers, rather than tetramers, to eliminate avidity effects (Fig. 2A). For each mutant, we plotted the ratio of binding relative to the parental CD4 A1 clone (Fig. 2B). The mutations were divided into three categories. In the first category were P35K, Y40Q, L43F, W45T, and L48P, which reduced binding to HLA-DR1 monomers by >80%, as well as S47G, which reduced binding by 56%. Mutations at these six positions therefore contribute substantially to the improved affinity of CD4 A1. In the second category were R46K, R60S, and R63D, which decreased binding to a lesser extent (15–25%), but which are nevertheless involved in affinity maturation. In the third category were G42S, which had no effect on binding, and T44L, which actually increased binding by CD4 A1 about twofold and is by inference detrimental to affinity maturation. The rank order of the observed maturation mutations is therefore the following: T45W >> Q40Y, P48L > K35P, F43L > G47S > K46R, S60R, D63R > S42G > L44T.
Fig. 2.
Reversion mutagenesis of affinity-matured CD4. (A) Dot plots showing labeling of yeast cells displaying wild-type CD4, affinity-matured CD4 A1 mutant, or CD4 A1 reversion mutants by 1.5-μM SAPE-conjugated HLA-DR1 monomer. Each of the 11 mutated residues in CD4 A1 was individually back-mutated to wild type (A1/position/wild type). (B) Fluorescence intensity of each CD4 A1 reversion mutant was normalized by that of the parental CD4 A1 clone and plotted against the mutation.
On the basis of these results, we sought to identify the minimum number of mutations in CD4 required for reasonably tight (low micromolar) binding to MHC class II. First, mutations Q40Y and/or P48L were introduced into CD4 T45W, and HLA-DR1 binding was tested on the yeast surface. Whereas Q40Y/T45W (designated CD4-DM, for double mutant) could bind HLA-DR1 tetramers, T45W/P48L could not (Fig. 3A). Because Q40Y/T45W/P48L bound no better than CD4-DM, we next introduced six other mutations (K35P, F43L, K46R, G47S, S60R, D63R) individually into CD4-DM and tested their ability to confer binding to HLA-DR1 monomers (Fig. 3B). Surprisingly, S60R and D63R, the two mutations expected to be least important on the basis of reversion mutagenesis, increased binding the most, and, when combined, appeared to act cooperatively. Therefore, Q40Y/T45W/S60R/D63R (designated CD4-TM, for tetramutant) and CD4-DM were chosen for further characterization.
Fig. 3.
Mutagenesis of wild-type CD4. (A) Dot plots showing binding of HLA-DR1 tetramers to yeast cells displaying wild-type CD4, affinity-matured CD4 A1, or CD4 bearing the mutations Q40Y/T45W (CD4-DM), T45W/P48L, or Q40Y/T45W/P48L. Yeast cells were labeled with SAPE-conjugated HLA-DR1 tetramers (1 μM) and anti–c-myc mAb 9E10, followed by AF488-conjugated goat anti-mouse secondary antibody. The boxed populations show HLA-DR1 tetramer binding. (B) Dot plots showing binding of HLA-DR1 monomers to yeast cells displaying wild-type CD4, CD4 A1, or CD4-DM (Q40Y/T45W) with the mutations K35P, F43L, K46R, G47S, S60R, D63R, or S60R/D63R (CD4-TM). Cells were labeled with SAPE-conjugated HLA-DR1 monomers (4 μM) and anti–c-myc mAb 9E10.
We also tested affinity-matured human CD4 mutants for cross-reactivity with mouse MHC class II molecules. However, yeast cells displaying CD4 A1 and CD4-DM did not bind I-Ad or I-Ek tetramers (Fig. S2), indicating that affinity maturation was specific for the selecting ligand HLA-DR1.
Affinity of Engineered CD4 Mutants for HLA-DR1.
We used surface plasmon resonance (SPR) to measure the affinity of CD4-TM and CD4-DM for HLA-DR1. These mutants were produced as full-length ectodomains (CD4 D1–D4) by secretion from baculovirus-infected insect cells. We also attempted to produce soluble CD4 A1 by this method, but the protein was not secreted, presumably due to one (or more) of the seven mutations not present in CD4-TM. Biotinylated HLA-DR1 was directionally coupled to a streptavidin-coated biosensor surface, and different concentrations of soluble CD4-TM, CD4-DM, or wild-type CD4 were flowed over the surface. Wild-type CD4 showed no detectable binding to HLA-DR1, even at concentrations as high as 400 μM (Fig. S3A), implying a KD of >400 μM. This result is consistent with a previous SPR estimate of the affinity of human CD4 for MHC class II (KD >2 mM) (1). In striking contrast, CD4-DM and CD4-TM bound HLA-DR1 with KD's of 8.8 and 5.3 μM, respectively (Fig. S3 B and C). For CD4-TM, this represents at least a 75-fold improvement in affinity, assuming a KD of >400 μM for the wild-type interaction (or at least a 380-fold improvement, assuming a wild-type KD of >2 mM). The finding by SPR that CD4-TM effectively bound HLA-DR1 no tighter than CD4-DM was unexpected, given our results for these mutants displayed on yeast (Fig. 3B), but could reflect secondary factors such as greater stability or expression of CD4-TM on the yeast cell surface (14).
Structure of Affinity-Matured CD4 in Complex with HLA-DR1.
All our attempts to crystallize wild-type CD4 bound to HLA-DR1 yielded only crystals of the individual proteins, presumably due to the extremely weak interaction between them (Fig. S3A). However, both CD4-TM D1–D2 and CD4-DM D1–D2 crystallized readily with HLA-DR1. We determined the structures of the CD4-TM–HLA-DR1 and CD4-DM–HLA-DR1 complexes to 2.1 and 2.4 Å resolution, respectively (Table S1). Superposition of the two complexes gave a root mean squared (rms) difference of 0.5 Å for 546 α-carbon atoms. Because CD4-DM contains only two mutations relative to wild-type CD4, further analysis was based on the CD4-DM–HLA-DR1 complex, unless stated otherwise.
The overall topology of the affinity-matured CD4–HLA-DR1 complex is similar to that of wild-type human CD4 D1–D2 bound to mouse I-Ak (13): superposition of the two complexes gave an rms difference of 2.1 Å for 519 α-carbon atoms. CD4 recognizes HLA-DR1 through its membrane-distal D1 domain, which contacts the membrane-proximal α2 and β2 domains of the MHC class II molecule (Fig. 4A). No interactions were observed between CD4 D2 and HLA-DR1. The complex buries a total solvent-accessible surface of 1230 Å2, of which 34% is contributed by the HLA-DR1 α2 domain and 66% by the β2 domain. By comparison, the complex between CD4 and HIV gp120 buries a much larger surface area (2197 Å2) (15), even though HIV gp120 and HLA-DR1 bind to overlapping sites on the CD4 D1 domain (Fig. S4), as noted previously (16). This difference in buried surfaces at least partly explains the far higher affinity of CD4 for HIV gp120 (KD = 5 nM) (17) than for HLA-DR1.
Fig. 4.
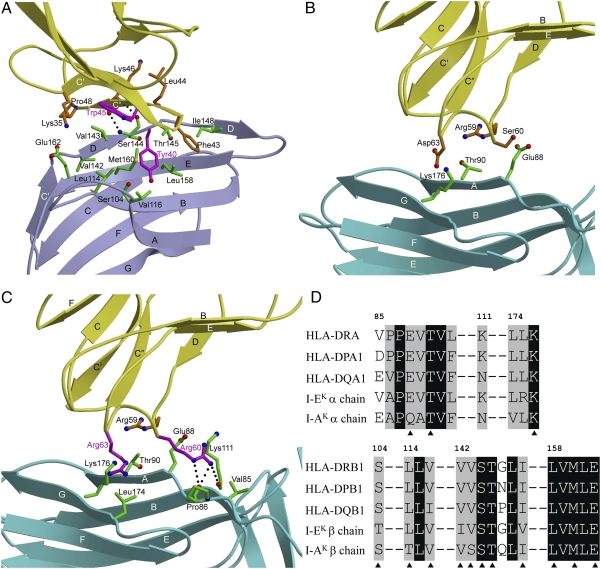
Structure of the human CD4–HLA-DR1 complex. (A) CD4 (yellow) contacts both the α2 (cyan) and β2 (blue) domains of the MHC class II molecule through its D1 domain. The HA peptide bound to HLA-DR1 is red. The strands of the β-sheets of the interacting variable Ig-like domains are labeled. (B) The CD4–HLA-DR1 binding interface. The two regions of CD4-DM (region 1: residues 35–48; region 2: residues 59–63) that contact HLA-DR1 are drawn in stick representation with carbon atoms in yellow, oxygen atoms in red, and nitrogen atoms in blue. The HLA-DR1 molecular surface that interacts with CD4-DM is shown with the α2 domain in cyan and the β2 domain in blue.
In the complex, 11 CD4 residues contact 14 HLA-DR1 residues through predominantly hydrophobic interactions, with no bound water molecules in the interface (Table 1). All these residues also interact with HIV gp120, which contacts an additional 10 residues on CD4 (Fig. S4) (15). CD4 uses two discontinuous regions to engage HLA-DR1 in a concavity formed by the α2 and β2 domains (Fig. 4B). Region 1, composed of β-strands C′ and C′′, exclusively contacts the β2 domain. Notably, the CD4-DM mutations Gln40Tyr and Thr45Trp are located on strands C′ and C′′, respectively. Region 2, comprising a short 310 helix between β-strands D and E, binds solely to the α2 domain.
Table 1.
Interactions between CD4-DM and HLA-DR1
| HLA-DR1 |
|||
| CD4-DM | Hydrogen bonds | Distance (Å) | Van der Waals contacts |
| Lys35 | Glu162β | ||
| Tyr40* | Ser104β, Leu114β, | ||
| Val116β, Met160β | |||
| Phe43 | Thr145β, Ile148β, | ||
| Leu158β | |||
| Leu44 | Ser144β | ||
| Trp45† | Val143β, Ser144β | ||
| Lys46 | Lys46 N–Ser144 O | 2.77 | Val143β, Ser144β |
| Lys46 O–Ser144 N | 2.78 | ||
| Gly47 | Val142β, Val143β | ||
| Pro48 | Val142β | ||
| Arg59 | Glu88α, Thr90β | ||
| Ser60 | Glu88α | ||
| Asp63 | Lys176α | ||
*Gln40 in wild-type CD4.
†Thr45 in wild-type CD4.
In region 1, β-strand C′′ of CD4 is in an approximate anti-parallel direction with β-strand D and the following peptide segment of DR1 β2, from Val142 to Thr145 (Fig. 5A). At the center of this region, the main-chain N and O atoms of CD4 Lys46 form hydrogen bonds to the main-chain O and N atoms, respectively, of DR1 β2 Ser144, bringing strand C′′ of CD4 into close proximity to strand D of DR1 β2. As a consequence, CD4-DM residues Leu44, Trp45 (Thr in wild type), Lys46, Gly47, and Pro48 make numerous van der Waals contacts with DR1 β2 residues Val142, Val143, Ser144, and Thr145, mainly involving main-chain atoms, that together account for more than half (27 of 42) of the total contacts between CD4 and DR1 β2 (Table 1). The remaining contacts are mediated by the side chains of CD4-DM residues Lys35, Tyr40 (Gln in wild type), and Phe43. The aromatic ring of Phe43 at the start of the C′′ strand inserts into a hydrophobic pocket formed by the DR1 β2 residues Thr145, Ile148, and Leu158, which are conserved across MHC class II molecules (see below). In region 2 (Fig. 5B), CD4-DM residues Arg59, Ser60, and Asp63 contact the DR1 β2 residues Glu88, Thr90, and Lys176.
Fig. 5.
The CD4–HLA-DR1 binding interface. (A) Close-up view of the interactions between region 1 of affinity-matured CD4-DM (yellow) and the HLA-DR1 β2 domain (blue). The side chains of interacting residues are shown in ball-and-stick representation with carbon atoms in brown (CD4) or green (HLA-DR1), oxygen atoms in red, and nitrogen atoms in blue. The mutated Tyr40 and Trp45 residues of CD4-DM are in magenta. Hydrogen bonds are drawn as dotted black lines. (B) Interactions between region 2 of CD4-DM (yellow) and the HLA-DR1 α1 domain (cyan). (C) Interactions between region 2 of affinity-matured CD4-TM and the HLA-DR1 α1 domain. The mutated Arg60 and Arg63 residues of CD4-TM are in magenta. (D) Sequence alignment of the CD4-contacting regions of the α- and β-chains of different human (DRA*0101/DRB1*0101, DPA1*0104/DPB1*01011, DQA1*01012/DQB1*0401) and mouse (I-Ak, I-Ek) MHC class II alleles. Residues that contact CD4 in the CD4-DM–HLA-DR1 structure are denoted by triangles. White characters on a black background show residues that are strictly conserved across human or mouse MHC class II molecules. The remaining residues are black.
Conformational Changes in CD4 and HLA-DR1 upon Complex Formation.
Superposition of the D1 domain of free wild-type CD4 (18) onto the D1 domain of CD4-DM in the complex with HLA-DR1 gave an rms difference in α-carbon positions of 0.7 Å, indicating close overall similarity. However, several side-chain rearrangements are observed at the binding interface (Fig. S5). Thus, the side chain of CD4 Phe43 is rotated by ∼60° around the Cα–Cβ axis to optimize interactions with HLA-DR1. In addition, the side chain of CD4-DM Tyr40 points in a different direction from that of Glu40 in unbound wild-type CD4 to avoid steric clashes with DR1 β2 Met160.
Similarly, there are no major conformational changes in HLA-DR1 due to binding CD4; superposition of the α2 and β2 domains of free HLA-DR1 (19) onto the α2 and β2 domains of HLA-DR1 in the complex with CD4 gave an rms difference in α-carbon positions of 0.8 Å. The only notable structural adjustment involves HLA-DR1 β-strand D and the following peptide segment (residues 142–148), in which the peptide segment is shifted by ∼1.5 Å away from CD4 to prevent steric clashes between DR1 β2 Val142 and CD4 Trp45 (Fig. S5).
Basis for Increased Affinity of CD4 Mutants.
Although the structure of wild-type CD4 bound to HLA-DR1 is unknown, and that of the CD4–I-Ak complex (13) is of too low resolution for detailed analysis, the effects of mutations in CD4 on the binding interface with HLA-DR1 may be understood by superposing wild-type CD4 (18) onto affinity-matured CD4 mutants in the CD4-DM–HLA-DR1 and CD4-TM–HLA-DR1 structures to construct a hypothetical wild-type complex that assumes no conformational changes (Fig. S6A).
The β2 domain of HLA-DR1 is a two-layer β-sandwich that exhibits the chain topology of I-set Ig domains, with the front and back sheets composed of β-strands C′CFG and ABED, respectively (Fig. 5A). CD4-contacting residues are distributed across all four ABED strands, which together form a broad and relatively flat binding surface. In the modeled wild-type CD4–HLA-DR1 complex (Fig. S6A), there exists a deep groove on the CD4 side of the interface that is not occupied by any HLA-DR1 residue. In the mutant CD4-DM–HLA-DR1 and CD4-TM–HLA-DR1 structures (Fig. S6B), this groove is filled by Trp45 (Thr in wild-type CD4), which contributes most to increasing affinity (see above). The bulky side chain of CD4 Trp45 makes multiple hydrophobic contacts with the side chain of DR1 β2 Val143 (Table 1). In addition, CD4 Trp45 is located at the center of the interface with HLA-DR1 (Fig. S6B), and it is well established that central residues typically contribute substantially more than peripheral ones to the energetics of protein–protein interactions (20, 21). The second most important mutation for tighter binding is CD4 Gln40Tyr. In the CD4-DM–HLA-DR1 and CD4-TM–HLA-DR1 structures, Tyr40 is surrounded by apolar DR1 β2 residues Leu114, Val116, Leu158, and Met160 (Fig. 5A), resulting in increased hydrophobic interactions at the mutation site relative to wild type. A hydrogen bond linking the side chains of CD4 Tyr40 and DR1 β2 Ser104 provides further stabilization (Table 1). Together, the Gln40Tyr and Thr45Trp mutations in CD4 act to improve the geometrical fit with HLA-DR1 on the basis of the calculations of the shape correlation statistic (Sc) for this interface (22). Thus, the Sc value for the mutant CD4-DM–HLA-DR1 complex is 0.74 (Sc = 1.0 for interfaces with perfect fits), but only 0.68 for the modeled wild-type complex.
Because CD4-DM and CD4-TM bind HLA-DR1 with virtually identical KD's (8.8 and 5.3 μM, respectively) (Fig. S3 B and C), the Ser60Arg and Asp63Arg mutations in region 2 of CD4-TM (Fig. 5C) do not affect (increase or decrease) complex stability. This accommodation is surprising, given the nonconservative nature of both substitutions. Although substitution of Asp63 in CD4-DM by Arg63 in CD4-TM does not appreciably alter contacts with HLA-DR1 (Fig. 5 B and C), replacement of Ser60 by Arg60 results in the formation of three new hydrogen bonds at the mutation site: CD4-TM Arg60 Nε–O DR1 α2 Pro86, CD4-TM Arg60 Nη2–O DR1 α2 Pro86, and CD4-TM Arg60 Nη2–O DR1 α2 Lys111 (Fig. 5C). The lack of a net contribution to complex stabilization by these hydrogen bonds may be explained by their location at the periphery, rather than at the center, of the protein–protein interface, because solvent-exposed hydrogen bonds are typically weaker than buried hydrogen bonds and are sometimes even energetically neutral (23).
Basis for CD4 Recognition of Multiple MHC Class II Alleles.
Polymorphism is a hallmark of MHC class II molecules, which, in humans, are encoded by three separate loci—HLA-DR, -DQ, and -DP. For HLA-DR, most variability derives from the β-chain, with >700 known alleles at the population level, whereas there are only three α-chain variants. In contrast, both α- and β-chains of HLA-DQ and -DP are polymorphic (24). The CD4-DM–HLA-DR1 structure readily explains the remarkable ability of CD4 to recognize highly polymorphic MHC class II molecules.
Fig. 5D shows sequence alignments of the α- and β-chains of selected HLA-DR, -DP, and -DQ alleles in the regions where HLA-DR1 interacts with CD4-DM. For the β-chains, all 11 CD4-contacting residues are absolutely conserved in these human MHC class II molecules, with the sole exception of Val116, which is Ile in HLA-DQ. However, Val116 makes only a single van der Waals contact with CD4 in the CD4-DM–HLA-DR1 complex, which involves the side-chain hydroxyl group of the mutant CD4 Tyr40 residue (Fig. 5A). In the case of wild-type CD4, the shorter side chain of Gln40 would not be expected to contact Val/Ile116 of the HLA-DR, -DP, or -DQ β-chain. For the α-chains, all three CD4-contacting residues (Glu88, Thr90, Leu176) are invariant across human MHC class II molecules. We therefore conclude that the remarkable cross-reactivity of CD4 is attributable to the exclusive targeting of nonpolymorphic residues in the concavity formed by the α2 and β2 domains of HLA-DR, -DP, and -DQ. We further conclude that CD4 engages HLA-DP and -DQ in the same manner as it does HLA-DR. Also of note is that the cross-reactivity of human CD4 extends to mouse I-A and I-E MHC class II molecules (25). For I-Ak, 11 of 14 putative CD4-contacting residues are identical to those of HLA-DR1, whereas, for I-Ek, 12 of 14 are identical (Fig. 5D). Moreover, all nonidentical residues are conservatively substituted in both molecules.
Conclusion
The ability of in vitro evolution to dramatically increase the affinity of CD4 for HLA-DR1 through just two mutations clearly demonstrates that the CD4 scaffold is capable of much tighter binding to MHC class II than is observed in nature. One interpretation of this result is that increased CD4 affinity confers no survival advantage to the host and is therefore not evolutionarily selected in vivo. Alternatively, evolution may have calibrated the affinity of CD4 for MHC class II to ensure that developing T cells undergo appropriate thymic selection, such that too high an affinity would result in deletion of T cells that would normally be positively selected, thereby restricting the size or diversity of the peripheral T-cell repertoire. It is also possible that evolution has placed an upper limit on CD4 affinity to avoid activation of peripheral T cells by self-peptides, which could result in autoimmunity. These issues may now be addressed in vivo by generating mice transgenic for the high-affinity human CD4 mutants reported here. For this purpose, mice lacking endogenous MHC class II and CD4 molecules, but expressing various HLA-DR alleles, have been described (26).
Materials and Methods
Vector Construction and Yeast Transformation.
Gene segments encoding CD4 D1 and CD4 D1-D2 were cloned into the yeast surface display vector pCTCON2-2Sfi (gift of Zeev Pancer, University of Maryland School of Medicine, Baltimore). The resulting constructs were used to transform yeast EBY100 cells (SI Materials and Methods).
Construction of Targeted CD4 Mutant Library.
CD4 residues (35, 40, 42–48, 59, 60, and 63) were mutated using degenerate primers. Yeast EBY100 cells were transformed by electroporation with the mutated CD4 D1-D2 gene (SI Materials and Methods).
Flow Cytometry of CD4 Mutant Library.
The CD4 D1-D2 mutant library was labeled with fluorescent HLA-DR1 tetramers and sorted on a BD FACSAria II sorter (SI Materials and Methods).
Protein Expression and Purification.
Soluble HLA-DR1 was prepared by in vitro folding from bacterial inclusion bodies. Soluble human CD4 D1–D4 or CD4 D1–D2 mutants were expressed in baculovirus-infected insect cells (SI Materials and Methods).
Affinity Measurements.
The binding of HLA-DR1 to CD4 (wild type or mutants) was measured by SPR using a BIAcore T100 biosensor (SI Materials and Methods).
Crystallization and Structure Determination.
Purified HLA-DR1 and CD4 D1–D2 Q40Y/T45W/S60R/D63R mutant (CD4-TM) or CD4 D1–D2 Q40Y/T45W mutant (CD4-DM) were concentrated to 10 mg/mL in 0.01 M Tris (pH 8.0) and 0.02 M NaCl. Crystals of the CD4-TM–HLA-DR1 complex grew at room temperature in 0.1 M sodium cacodylate (pH 6.5), 0.2 M ammonium sulfate, and 15% (wt/vol) polyethylene glycol (PEG) 8000. Crystals of the CD4-DM–HLA-DR1 complex grew under the same conditions, except with 12% (wt/vol) PEG 8000. For data collection, CD4-TM–HLA-DR1 and CD4-DM–HLA-DR1 crystals were cryoprotected with 25% (vol/vol) glycerol before flash cooling. X-ray diffraction data were collected to 2.1 Å resolution for CD4-TM and 2.4 Å resolution for CD4-DM at beamline X29 of the Brookhaven National Synchrotron Light Source. All data were indexed, integrated, and scaled with the program HKL2000 (27). The structures of the CD4-TM–HLA-DR1 and CD4-DM–HLA-DR1 complexes were determined by molecular replacement (SI Materials and Methods). Data collection and refinement statistics are presented in Table S1.
Supplementary Material
Acknowledgments
We thank H. Robinson (Brookhaven National Synchrotron Light Source) for X-ray data collection. We are grateful to Z. Pancer (University of Maryland School of Medicine) for advice on affinity maturation. Mouse I-Ad and I-Ek tetramers were provided by the National Institutes of Health Tetramer Core Facility at Emory University. Support for beamline X29 comes from the Office of Biological and Environmental Research and the Office of Basic Energy Sciences of the US Department of Energy and from the National Center for Research Resources of the National Institutes of Health. This work was supported by grants from the National Institutes of Health (Grants AI036900 and AI073654). X.X.W. was supported by the Irvington Institute Fellowship Program of the Cancer Research Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3S4S and 3S5L).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109438108/-/DCSupplemental.
References
- 1.Davis SJ, et al. The nature of molecular recognition by T cells. Nat Immunol. 2003;4:217–224. doi: 10.1038/ni0303-217. [DOI] [PubMed] [Google Scholar]
- 2.van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 3.Cole DK, et al. Human TCR-binding affinity is governed by MHC class restriction. J Immunol. 2007;178:5727–5734. doi: 10.4049/jimmunol.178.9.5727. [DOI] [PubMed] [Google Scholar]
- 4.Davis MM, et al. T cells as a self-referential, sensory organ. Annu Rev Immunol. 2007;25:681–695. doi: 10.1146/annurev.immunol.24.021605.090600. [DOI] [PubMed] [Google Scholar]
- 5.Janeway CA., Jr The T cell receptor as a multicomponent signalling machine: CD4/CD8 coreceptors and CD45 in T cell activation. Annu Rev Immunol. 1992;10:645–674. doi: 10.1146/annurev.iy.10.040192.003241. [DOI] [PubMed] [Google Scholar]
- 6.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 7.Xiong Y, Kern P, Chang H, Reinherz E. T cell receptor binding to a pMHCII ligand is kinetically distinct from and independent of CD4. J Biol Chem. 2001;276:5659–5667. doi: 10.1074/jbc.M009580200. [DOI] [PubMed] [Google Scholar]
- 8.Garcia KC, et al. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature. 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang R, Natarajan K, Margulies DH. Structural basis of the CD8 α β/MHC class I interaction: Focused recognition orients CD8 β to a T cell proximal position. J Immunol. 2009;183:2554–2564. doi: 10.4049/jimmunol.0901276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyer JR, et al. T cell receptor and coreceptor CD8 alphaalpha bind peptide-MHC independently and with distinct kinetics. Immunity. 1999;10:219–225. doi: 10.1016/s1074-7613(00)80022-9. [DOI] [PubMed] [Google Scholar]
- 11.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 12.Goodnow CC, Sprent J, Fazekas de St Groth B, Vinuesa CG. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature. 2005;435:590–597. doi: 10.1038/nature03724. [DOI] [PubMed] [Google Scholar]
- 13.Wang JH, et al. Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule. Proc Natl Acad Sci USA. 2001;98:10799–10804. doi: 10.1073/pnas.191124098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gai SA, Wittrup KD. Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol. 2007;17:467–473. doi: 10.1016/j.sbi.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JH. Protein recognition by cell surface receptors: Physiological receptors versus virus interactions. Trends Biochem Sci. 2002;27:122–126. doi: 10.1016/s0968-0004(01)02038-2. [DOI] [PubMed] [Google Scholar]
- 17.Myszka DG, et al. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang JH, et al. Atomic structure of a fragment of human CD4 containing two immunoglobulin-like domains. Nature. 1990;348:411–418. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, et al. Structural basis for the presentation of tumor-associated MHC class II-restricted phosphopeptides to CD4+ T cells. J Mol Biol. 2010;399:596–603. doi: 10.1016/j.jmb.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Huang Y, Swaminathan CP, Smith-Gill SJ, Mariuzza RA. Magnitude of the hydrophobic effect at central versus peripheral sites in protein-protein interfaces. Structure. 2005;13:297–307. doi: 10.1016/j.str.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 23.Dall'Acqua W, et al. A mutational analysis of binding interactions in an antigen-antibody protein-protein complex. Biochemistry. 1998;37:7981–7991. doi: 10.1021/bi980148j. [DOI] [PubMed] [Google Scholar]
- 24.Robinson J, et al. The IMGT/HLA database. Nucleic Acids Res. 2011;39(Database issue):D1171–D1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vignali DA, Moreno J, Schiller D, Hämmerling GJ. Species-specific binding of CD4 to the β 2 domain of major histocompatibility complex class II molecules. J Exp Med. 1992;175:925–932. doi: 10.1084/jem.175.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangalam AK, Rajagopalan G, Taneja V, David CS. HLA class II transgenic mice mimic human inflammatory diseases. Adv Immunol. 2008;97:65–147. doi: 10.1016/S0065-2776(08)00002-3. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.