Abstract
Nitrification is a fundamental component of the global nitrogen cycle and leads to significant fertilizer loss and atmospheric and groundwater pollution. Nitrification rates in acidic soils (pH < 5.5), which comprise 30% of the world's soils, equal or exceed those of neutral soils. Paradoxically, autotrophic ammonia oxidizing bacteria and archaea, which perform the first stage in nitrification, demonstrate little or no growth in suspended liquid culture below pH 6.5, at which ammonia availability is reduced by ionization. Here we report the discovery and cultivation of a chemolithotrophic, obligately acidophilic thaumarchaeal ammonia oxidizer, “Candidatus Nitrosotalea devanaterra,” from an acidic agricultural soil. Phylogenetic analysis places the organism within a previously uncultivated thaumarchaeal lineage that has been observed in acidic soils. Growth of the organism is optimal in the pH range 4 to 5 and is restricted to the pH range 4 to 5.5, unlike all previously cultivated ammonia oxidizers. Growth of this organism and associated ammonia oxidation and autotrophy also occur during nitrification in soil at pH 4.5. The discovery of Nitrosotalea devanaterra provides a previously unsuspected explanation for high rates of nitrification in acidic soils, and confirms the vital role that thaumarchaea play in terrestrial nitrogen cycling. Growth at extremely low ammonia concentration (0.18 nM) also challenges accepted views on ammonia uptake and metabolism and indicates novel mechanisms for ammonia oxidation at low pH.
Keywords: acidophile, nitrification
Approximately 30% of the world's soils are acidic (pH < 5.5), including more than 50% of potential arable land (1). Although low pH reduces the rates of many soil ecosystem processes, a metastudy of gross nitrification in almost 300 soils showed weak negative correlation with soil pH and some of the highest rates in acid soils (2). This is paradoxical given the physiology of cultivated soil nitrifiers. Nitrification involves the oxidation of ammonia to nitrite and, subsequently, to nitrate. Ammonia oxidation in soil was traditionally considered to be dominated by autotrophic β-proteobacteria, but most cultivated bacterial ammonia oxidizers do not grow in suspended batch culture below pH 6.5 (3, 4). This is believed to be through reduced availability of NH3 (pKa for NH3:NH4+, 9.25), the substrate for ammonia monooxygenase, which catalyzes conversion of ammonia to hydroxylamine (5, 6). In addition, in the absence of nitrite oxidation, ammonia oxidizers may be inhibited by nitric and nitrous acids, whose production from nitrite increases under acid conditions, and by generation of nitric oxide and nitrogen dioxide from nitrous acid, which is unstable and reactive at low pH.
Several mechanisms have been proposed to explain this paradox, including pH-neutral microenvironments, urease activity (7, 8), heterotrophic nitrification (9), biofilm and aggregate formation (10, 11), and close interactions between ammonifiers and ammonia oxidizers in soil aggregates (12). These mechanisms are difficult to demonstrate in soil and none fully explains high rates of nitrification in soils with pH less than 5.5. An additional explanation is the existence of acidophilic ammonia oxidizers, but all attempts to isolate such organisms have failed, although an enrichment was obtained at low pH (10) in which aggregate formation was required for ammonia oxidation. Attempts to cultivate acidophilic ammonia oxidizers preceded the discovery of ammonia-oxidizing archaea, which fall within the thaumarchaeal lineage. These organisms appear to contribute significantly to ammonia oxidation in many soils in which thaumarchaeal amoA (ammonia monooxygenase subunit A) genes and transcripts outnumber equivalent bacterial genes (13–17). Their activity, relative to that of bacterial ammonia oxidizers, also appears to increase in soil pH transects with decreasing soil pH (13).
The aim of this study was to determine whether ammonia oxidation in acid soils resulted from the existence of acidophilic thaumarchaea by attempting their enrichment in low-pH, mineral salts medium containing ammonia, following inoculation with acid soil in which thaumarchaea are believed to drive nitrification (13, 14).
Results
Enrichment of an Obligate Acidophilic Thaumarchaeal Ammonia Oxidizer.
To determine whether ammonia oxidation in acid soils was caused by acidophilic autotrophic ammonia oxidizers, we attempted enrichment of acidophilic archaeal ammonia oxidizers by inoculation of several acidic soils into mineral salts medium containing inorganic ammonium and adjusted to pH 4.5. An ammonia oxidizer enrichment culture was successfully obtained from an agricultural soil that had been maintained at pH 4.5 since 1961 and in which ammonia oxidation appears to be dominated by thaumarchaea (14). The enrichment contained a large number and diversity of heterotrophic “contaminants,” and attempts were made to obtain a pure culture by successive subculturing in mineral salts medium, inclusion of streptomycin, and filtration. This led to a highly enriched culture of a thaumarchaeal ammonia oxidizer that is stable, in terms of its physiology and the coenriched bacterial community composition. A pure archaeal culture was also obtained by filtration, but failed to grow in the absence of the cocultured bacteria, and addition of pyruvate did not enable the growth of the organism in pure culture, as reported recently for the cultivation of a neutrophilic ammonia oxidizer from soil (18). The cultivated archaeal strain grew exponentially in liquid batch culture, as demonstrated by coordinate, exponential increases in nitrite concentration and thaumarchaeal amoA gene abundance, assessed by quantitative PCR (qPCR), and by a decrease in ammonia concentration (Fig. 1). Cell activity and yield were estimated as 11 fmol·cell−1·h−1 and 4 × 105 cells·μM ammonia−1 (assuming that all nitrite was derived from ammonia oxidation), compared with respective values of 0.53 fmol·cell−1·h−1 and 4 × 105 cells·μM ammonia−1 reported for Nitrosopumilus maritimus (10).
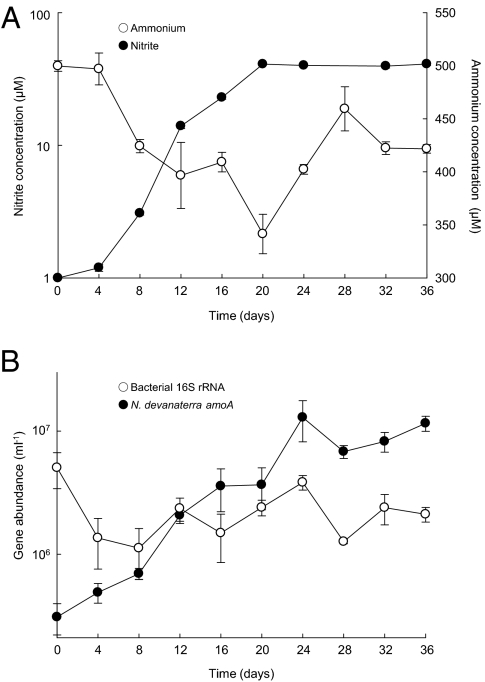
Fig. 1.
Growth of, and ammonia oxidation by, an acidophilic thaumarchaeon in inorganic liquid medium at pH 4.5 containing 500 μM ammonium. (A) Exponential increase in nitrite concentration and accompanying decrease in ammonia concentration. (B) Exponential increase in thaumarchaeal amoA gene abundance, and growth of coenriched bacteria. Data are presented as mean and SE of triplicate cultures.
Maximum amoA gene abundance was 1.3 ± 0.47 × 107, and nitrite concentration did not increase to more than 41 μM, despite supply of 500 μM ammonium. Nitrite production and increases in thaumarchaeal amoA abundance were completely inhibited in the presence of 0.01% (10 Pa) acetylene (Fig. S1), which is a suicide substrate for ammonia monooxygenase (19) that is commonly used to inhibit autotrophic ammonia oxidation. qPCR provided no evidence of the presence of bacterial ammonia oxidizers (i.e., no amplification of 16S rRNA or amoA genes) or the presence of known nitrite oxidizers.
Ammonia concentration decreased during growth of the enriched culture, but conversion to nitrite was not stoichiometric. This is presumably because of the activity of coenriched bacteria, which could have used ammonia for growth and/or generated ammonium by mineralization of organic nitrogen carried over with the inoculum. The relative abundance of thaumarchaeal and bacterial 16S rRNA genes varied during growth, with bacterial abundance decreasing before thaumarchaeal growth began and remaining constant thereafter (Fig. 1B); bacteria presumably used organic carbon from the inoculum. The bacterial community contained organisms closely related to cultivated strains of Burkholderia, Cupriavidus, Bradyrhizobium, Rhizobium, Mesorhizobium, and Elusimicrobium, most of which have been isolated from root nodules and/or have the potential for nitrogen fixation (Table S1).
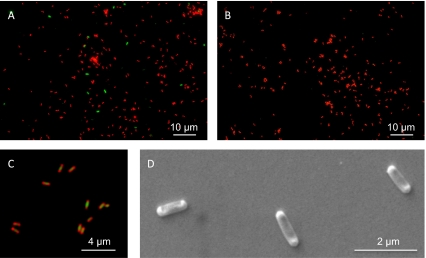
Assuming one amoA gene copy per thaumarchaeal genome (20, 21) and an estimated average of at least three 16S rRNA genes per bacterial genome (Table S1), qPCR analysis indicates that the thaumarchaeon represented approximately 90% of the cell abundance present during stationary phase. SEM of independent stationary-phase cultures indicated even greater dominance by small rod-shaped cells (0.33 ± 0.01 μm wide and 0.89 ± 0.05 μm long) with a central indentation and electron-dense poles (Fig. 2D). These cells are morphologically very similar to the thaumarchaeon N. maritimus (22) and are clearly distinguishable from larger bacterial cells that were observed in SEM images of early growth phase cultures, but very rare in SEM images from stationary phase. To confirm the identity of the small rod-shaped cells, FISH analyses were performed by using archaeal and bacterial probes ARCH915 and EUB338, respectively (Fig. 2 A and B). The thaumarchaeon dominated cultures and represented more than 99% of all cells entering stationary phase. As observed previously for rod-shaped thaumarchaeal cells (22), ribosomes were largely concentrated at the poles, with SYBR Green 1 counterstaining revealing localization of the genome at the center and to one side of the cell (Fig. 2C).
Fig. 2.
Morphology and dominance of N. devanaterra in an acidic ammonia oxidizing culture. FISH images with doubly labeled oligonucleotide probes at (A) midexponential and (B) onset of stationary phase growth of N. devanaterra. Cells were cohybridized with Cy3-labeled ARCH915 (red) and FLUOS-labeled EUB338 (green) probes. (C) Cells probed with ARCH915 (red) and counterstained with DNA stain SYBR green 1. (D) SEM image of N. devanaterra cells.
Phylogenetic Analysis and Physiological Characteristics of Enriched Soil Thaumarchaeon.
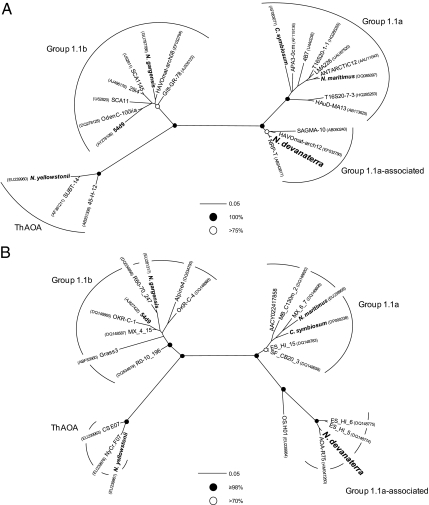
Phylogenetic analysis of 16S rRNA and amoA genes amplified from the enriched ammonia oxidizer places it within a group 1 lineage (Fig. 3) distinct from other cultivated thaumarchaea, with 89.8% and 77.4% identity to the 16S rRNA and amoA genes of N. maritimus, respectively. It does, however, have a relatively deep-branching association with group 1.1a organisms, forming a well supported monophyletic lineage distinct from group 1.1b and the Nitrosocaldus yellowstonii lineage, and is therefore described here as group 1.1a-associated (Fig. 3). The amoBCxA gene order was identical to that in N. maritimus, and a gene encoding 4-hydroxybutyryl-CoA dehydratase (hcd), a key enzyme and diagnostic of archaeal autotrophy (23), was also present.
Fig. 3.
Phylogenetic analysis of N. devanaterra gene sequences with other lineages known to contain ammonia-oxidizing archaea (including group 1.1a, group 1.1b, and the thermophilic AOA lineage). (A) Maximum-likelihood analysis of 16S rRNA gene sequences using estimated variable sites (599 unambiguously aligned positions). Circles at major nodes represent the most conservative value of bootstrap support (maximum likelihood, parsimony, and distance methods; 1,000 replicates each) or posterior probabilities (Bayesian analysis; 200,000 generations). The scale bar represents 0.05 changes per nucleotide position. (B) Maximum-likelihood tree of inferred AmoA sequences (162 unambiguously aligned positions). Circles at major nodes represent the most conservative value of bootstrap support (maximum likelihood, parsimony, and distance methods; 1,000 replicates each) or posterior probability (Bayesian analysis; 100,000 generations). (Scale bar: 0.05 changes per amino acid position.)
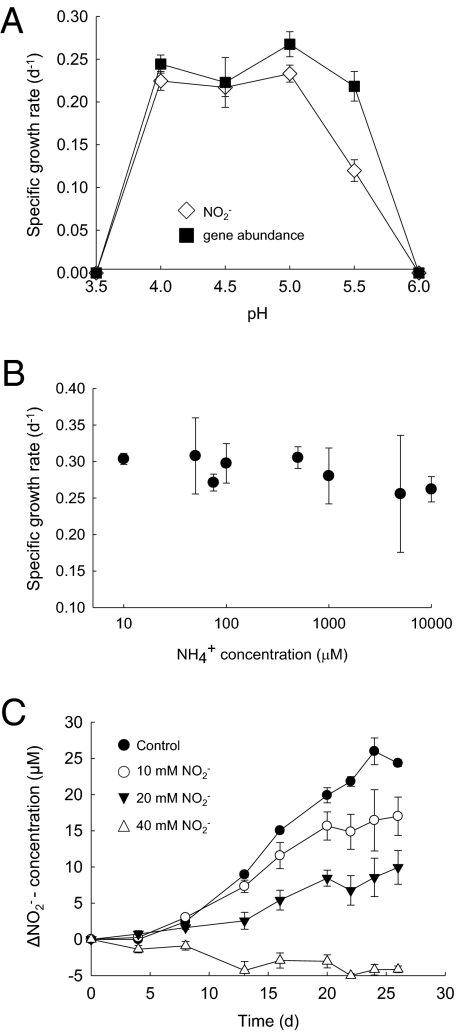
The enrichment grew within the temperature range 20 to 30 °C, with highest maximum specific growth rate (0.37 d−1) at 25 °C (Fig. S2). No or little nitrite was detected after incubation for 28 d at 35 °C or at 15 °C, despite enrichment from a temperate soil. The influence of pH was investigated by batch growth of the enriched thaumarchaeal ammonia oxidizer in liquid batch culture with initial pH adjusted to 3.5 to 7. Growth was restricted to the pH range 4.0 to 5.5 (Fig. 4A) when supplied with 500 μM ammonium, with no detectable changes in nitrite concentration or amoA abundance at pH 3.5 or at pH values greater than 5.5, and the organism may therefore be considered an obligate acidophile. Growth was optimal in the pH range of 4.0 to 5.0, with a maximum specific growth rate of 0.27 ± 0.012 d−1 or 0.23 ± 0.01 d−1, calculated from exponential increases in 16S rRNA gene abundance and nitrite concentration, respectively. These values are approximately 40% of the reported specific growth rate of 0.65 d−1 reported for N. maritimus (10). Maximum thaumarchaeal 16S rRNA gene abundance and nitrite concentration were observed at pH 5.0 and the pH response curve was shallow, with specific growth rates greater than 80% of the maximum. Reduced pH is believed to limit growth by reducing ammonia concentration and increasing nitrous/nitric oxide/NOx toxicity (10, 12). To assess the former, the thaumarchaeal ammonia oxidizer was grown with initial concentrations in the range of 10 μM to 10 mM total ammonium (equivalent to 0.18 nM to 1.8 μM ammonia) at pH 4.5. Surprisingly, specific growth rate measured by nitrite accumulation decreased with increasing ammonium concentration (correlation analysis, P = 0.046) (Fig. 4B), and no growth was detected at 50 and 100 mM ammonium. Standard batch culture growth was investigated in medium containing 500 μM ammonium, but oxidation of ammonium was never complete. Growth of the enrichment always ceased when nitrite concentration reached approximately 40 μM, and there was evidence of reduced specific growth rate as this concentration was approached (Fig. 1). To determine whether incomplete growth was caused by nitrite toxicity, the thaumarchaeal enrichment was grown in medium with initial nitrite concentrations of 0 to 40 μM. Growth was reduced at 10 and 20 μM nitrite, and was completely inhibited at 40 μM nitrite (Fig. 4C).
Fig. 4.
The influence of pH and initial ammonium and nitrite concentrations on thaumarchaeal ammonia oxidizer growth. (A) Specific growth rate [from log-linear plots of nitrite concentration (diamonds) or thaumarchaeal 16S rRNA gene abundance (squares)] during exponential growth in medium containing 500 μM ammonium, initial pH 3.5 to 7.5. (B) Specific growth rate (from log-linear plots of nitrite concentration) during growth in medium containing initial ammonium concentration in the range of 10 μM to 10 mM, pH 4.5. (C) Changes in nitrite concentration during growth in medium containing 500 μM ammonium and initial nitrite concentration in the range of 10 to 40 μM, pH 4.5. Data are presented as mean and SE of triplicate cultures for all growth experiments.
Autotrophic Growth and Ammonia Oxidation in Acid Soil.
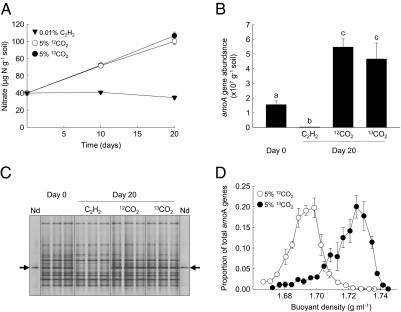
To determine the ecological significance of the thaumarchaeal ammonia oxidizer, a stable isotope microcosm experiment was performed in microcosms containing acidic soil (pH 4.5) from which the enrichment was obtained. Microcosms were incubated for 20 d in the presence of 5% (vol/vol) 12C- or 13C-CO2 or 0.01% (vol/vol) acetylene (Fig. 5). Nitrification occurred in microcosms without acetylene, with increases in nitrate concentration (Fig. 5A) and thaumarchaeal amoA abundance (Fig. 5B). Acetylene completely inhibited ammonia oxidation, leading to accumulation of ammonium through mineralization of organic nitrogen, and no change in nitrate concentration (Fig. 5A). Nitrification and thaumarchaeal growth were accompanied by an increase in the relative intensity of a band in denaturing gradient gel electrophoresis (DGGE) profiles representing phylotype(s) identical to N. devanaterra (Fig. 5C). This change in DGGE profiles was not observed in microcosms in which nitrification was inhibited by acetylene, in which N. devanaterra-like phylotypes decreased in relative intensity. This was confirmed by quantitative PCR using a group 1.1a-associated specific assay, with N. devanaterra-like amoA genes decreasing to 1.6% of their original abundance after 20 d in the presence of acetylene. This may be a result of reduction in cell integrity in the absence of ammonia oxidation-derived energy, but could also result from predation or phage infection that is not apparent in uninhibited microcosms because of a net increase in cell numbers. This contrasts with growth in liquid culture in which acetylene inhibited growth but cell numbers remained relatively constant. Stable isotope probing provided further evidence for autotrophic growth of this phylotype. Comparison of the distribution in CsCl gradients of genomic DNA extracted from microcosms incubated with 98% 12CO2 or 99% 13CO2 demonstrated an increase in the buoyant density of N. devanaterra-like genomic DNA from approximately 1.69 g·mL−1 to 1.73 g·mL−1 (Fig. 5D). Although this approach lacks precision, a difference of 0.04 g·mL−1 would be expected between unlabeled 12C- and fully labeled 13C-DNA (24), thus indicating that N. devanaterra grows in soil using mainly inorganic carbon.
Fig. 5.
Autotrophic growth and ammonia oxidation by N. devanaterra phylotypes in an acidic soil. Triplicate microcosms were incubated with 5% 12C- or 13C-CO2 or 0.01% acetylene for 0, 10, or 20 d. (A) Changes in nitrate concentration. (B) Abundance of group 1.1a-associated amoA genes at 0 and 20 d. Different letters indicate statistically significant differences (P < 0.05). (C) DGGE analysis of thaumarchaeal amoA PCR products from soil and from N. devanaterra (Nd). Arrows indicate the migration of N. devanaterra phylotypes. Each lane represents an individual microcosm. (D) Distribution of group 1.1a-associated thaumarchaeal genomic DNA after separation and fractionation in CsCl gradients. Vertical and horizontal bars represent SEs of proportional abundance and buoyant density of fractions, respectively, derived from triplicate CsCl gradients representing triplicate microcosms.
Discussion
Previous explanations for the common observation of relatively high rates of nitrification in soil, despite only neutrophilic growth of ammonia oxidizer laboratory isolates in standard laboratory culture, have been difficult to demonstrate in soil. For example, many bacterial ammonia oxidizers have ureolytic activity (25), which enables growth on urea at low pH (7, 8), but urea will derive mainly from urine patches in grazed land and is usually converted rapidly to ammonia by extracellular soil enzymes and ureolytic heterotrophs. Ureolytic activity may therefore enable ammonia oxidation only in acid soils under restricted conditions. Other explanations involve growth in biofilms or in multicellular aggregates, and acidophilic growth in such microenvironments is difficult to demonstrate unequivocally. In addition, these explanations are based on physiological characteristics of bacterial ammonia oxidizers, which are often undetectable in acid soils (15, 26, 27), and predate demonstration of the capacity for ammonia oxidation by thaumarchaea in soil (28). Here we report the discovery of an obligately acidophilic ammonia oxidizer, Candidatus Nitrosotalea devanaterra, that does not grow at neutral pH, but whose growth and activity at low pH are similar to those of soil bacterial ammonia oxidizer enrichments and isolates at neutral pH. Phylogenetic analysis of 16S rRNA and amoA genes places the organism within the group 1.1a-associated thaumarchaea and we propose the following candidate status:
“Nitrosotalea devanaterra” gen. et sp. nov.
Etymology.
Nitrosus (Latin masculine adjective), nitrous; talea (Latin feminine noun), slender rod; devana (Latin), Aberdeen; terra (Latin feminine noun), earth, soil. The name reflects its ability to oxidize ammonia to nitrite, its morphology and its site of origin.
Locality.
Acidic agricultural soil, Craibstone, Aberdeen, Scotland, United Kingdom.
Diagnosis.
A chemolithoautotrophic ammonia oxidizer of the kingdom Thaumarchaeota, appearing as straight rods with diameter 0.33 ± 0.01 μm and length 0.89 ± 0.05 μm.
The discovery of Nitrosotalea devanaterra does not exclude a role for bacterial ammonia oxidizers in nitrification in acid soils, nor involvement of urease activity or biofilm and aggregate formation in enabling acidophilic nitrification. It does, however, provide the most parsimonious explanation for ammonia oxidation in acid soils, which is reinforced by the common occurrence of closely related phylotypes in acid soils. This is further reinforced by demonstration of autotrophic ammonia oxidation by closely related phylotypes in acid soil microcosms.
The current inability to eliminate heterotrophs from the N. devanaterra enrichment culture limits the scope of physiological studies, but, in common with other ammonia oxidizers, activity is inhibited by acetylene in liquid and in soil. It is intriguing that many of the coenriched heterotrophs have the potential for nitrogen fixation or are associated with legume root nodules. This suggests a possible commensal or symbiotic association, particularly as their removal prevented growth. However, the enrichment cultivation conditions are unlikely to favor nitrogen fixation, their presence may be fortuitous, and additional enrichments are required to assess the significance of this finding.
Acidophilic ammonia oxidation suggests novel physiological mechanisms in N. devanaterra. Its growth is consistent with the half-saturation constant (Km) of 133 nM total ammonium reported for N. maritimus (29), but not if measured in terms of ammonia, which is generally assumed to be the substrate for ammonia monooxygenase. Growth occurred at pH 4.5 and 0.18 nM ammonia, and specific growth rate decreased at higher ammonia concentrations, suggesting that this organism has developed potentially novel system(s) for ammonia oxidation, in addition to protective mechanisms enabling acidophilic growth. Further characterization of mechanisms for acidophilic growth and ammonia oxidation are of considerable interest in terms of the physiology and evolution of this lineage, and its discovery may even require reassessment of the paradigm of ammonia, rather than ammonium, being the substrate for the membrane-bound ammonia monooxygenase. Of particular interest will be the location of the active site, i.e., whether it faces outwards into a periplasmic-like space, where it may be significantly influenced by the pH of the external environment.
Growth of N. devanaterra was inhibited by 40 μM nitrite, which, at pH 4.5, is equivalent to 2.53 μM free nitrous acid concentration (HNO2), which is believed to be responsible for inhibition of ammonia oxidation. This concentration is lower than reported Ki values, which lie within the range of 12 to 200 μM HNO2 (30, 31), and may indicate greater sensitivity to nitrous acid. Regardless of the mechanism of inhibition, continued nitrification would require close interactions with acidophilic nitrite oxidizers to relieve nitrous acid-associated inhibition in soil. Coculture with an acidophilic nitrite oxidizer, or other means of removing nitrite, might also enable more extended growth of N. devanaterra (10).
In conclusion, enrichment of an autotrophic, obligately acidophilic ammonia oxidizing thaumarchaeon, commonly found in acid soils, and demonstration of its autotrophic growth in acid soil provide a new and convincing solution to the paradox of nitrification in acid soils. It also suggests novel mechanisms for activity and growth of ammonia oxidizers and, in particular, challenges current paradigms surrounding substrate uptake mechanisms.
Materials and Methods
Cultivation and Growth Experiments.
Ammonia oxidizer enrichment cultures were established in 100-mL Duran bottles containing 50 mL of an unbuffered mineral salts medium based on that of Tourna et al. (18) consisting of NaCl (1 g·L−1), MgCl2 (0.4 g·L−1), CaCl2 (0.1 g·L−1), KH2PO4 (0.2 g·L−1), KCl (0.5 g·L−1), 1 mL modified nonchelated trace element solution (32), 1 mL 7.5 mM NaFeEDTA, 2 mM NaHCO3, 500 μM NH4Cl and 50 mg·L−1 streptomycin. The pH of the medium was adjusted to 4.5 with HCl, before filter-sterilization through a 0.22 μm pore size, bottle-top filter. Medium was inoculated (1%, wt/vol) with an agricultural sandy loam soil (Scottish Agricultural College; grid reference NJ872104) (13). Cultures were incubated at 28 °C in the dark and ammonia oxidizer activity and growth were assessed by analysis of inorganic nitrogen and thaumarchaeal 16S rRNA and amoA gene abundance, respectively. All physiology experiments were conducted in triplicate cultures. The effect of ammonia concentration was investigated in medium containing 10, 50, 100, and 500 μM and 1, 5, 10, 50, and 100 mM NH4Cl. Inhibition of growth by nitrite was studied in medium containing initial nitrite (i.e., NaNO2) concentrations of 0, 10, 20, and 40 μM. The effects of pH and temperature were investigated by incubating cultures at pH values in the range 3.5 to 7.0 (0.5 pH intervals) and temperatures in the range of 15 to 40 °C (5 °C intervals), respectively. Incubations with acetylene were performed in 144-mL serum vial bottles containing 50 mL of culture. Bottles were crimp-sealed, and 0.01% acetylene (vol/vol) headspace concentration was established. Bottles were opened at 4-d intervals for sampling and reestablishment of acetylene concentration. Specific growth rate was calculated from log-linear plots of gene abundance or nitrite concentration for individual cultures under a variety of treatments.
Process Measurements.
Nitrite and ammonia concentrations were determined colorimetrically (33, 34). Standards consisted of NaNO2 (0.781–50 μM) or NH4Cl (15.6–500 μM) prepared in mineral salts medium. All reactions and absorbance measurements were performed in clear polystyrene 96-well microplates (Greiner Bio-One) by using a LT-4000 Microplate Reader (Labtech).
qPCR.
Thaumarchaeal growth was assessed by changes in abundance of 16S rRNA and amoA genes. DNA was extracted by using a bead-beating protocol with SDS-based buffer, phenol, chloroform, and isoamyl alcohol (35). DNA precipitation was performed as described previously (35) but with the inclusion of linear acrylamide. qPCR amplification was performed by using a MyIQ real-time PCR detection system (BioRad) with QuantiFast SYBR Green 1 PCR Master Mix (Qiagen). qPCR was performed as previously described (36) by using thaumarchaeal-specific 16S rRNA gene primers 771F and 957R (37) and group 1.1a-associated/N. devanaterra primers 167F (5′-TATCACAATCAYTGATGCTCGCAGT-3′) and 409R (5′-TCATGTTGAACAWCGACAT-3′) designed in this study by using group 1.1a-associated sequences, and discriminating against other group 1 lineages. qPCR standards for thaumarchaeal 16S rRNA gene and group 1.1a-associated amoA gene assays were a cloned 1.3-kb thaumarchaeal gene sequence and N. devanaterra amoBCxA amplicons, respectively. Bacterial qPCR was performed as described previously (36). qPCR efficiencies for the thaumarchaeal 16S rRNA gene, group 1.1a-associated amoA gene, and bacterial 16S rRNA gene assays were 97.0 to 105.1%, 91.1 to 101.2%, and 98.2 to 101.0%, respectively, with r2 values of at least 0.99 for all assays.
Sequence Analyses.
Thaumarchaeal 16S rRNA genes were amplified from enrichment culture DNA by using primers A109F (38) and 1492R (13) generating a product approximately 1.3 kb in length. The 3-kb amo gene cluster (containing amoB, amoC, a conserved amo-associated hypothetical ORF, and amoA) was amplified by using primers 616R and amo2.2R (13, 22), and entire 16S rRNA and amoA gene amplicons were sequenced along both strands. The hcd gene was amplified as described previously (23). Phylogenetic analyses were performed on both DNA and inferred amino acid sequences using distance (39), parsimony (39), maximum-likelihood (40), and Bayesian methods (41) with GTR (DNA), JTT, or mixed model (amino acid) correction with ML-estimated (40) γ-distributed variable sites. To identify cocultured bacteria, 16S rRNA genes were amplified by using primers 27F and 1492R (42) from DNA extracted from three independent cultures before cloning and transforming into competent Escherichia coli by using standard techniques. Clones were randomly selected from each library and sequenced in one direction, and phylogenetic affiliation inferred through BLASTN searches. DGGE analysis was performed on total thaumarchaeal amoA gene amplicons as described previously (13).
SEM.
Glutaraldehyde-fixed cell suspensions from a stationary culture were applied to glass coverslips coated with poly-l-lysine, allowed to adhere for 5 min, rinsed with water, and postfixed with 1% OsO4. Attached cells were then rinsed with distilled H2O, dehydrated through a graded ethanol series to absolute ethanol, and critically point-dried with liquid CO2. Coverslips were sputtered with Au before examination by using an Evo MA 10 scanning electron microscope (Carl Zeiss).
DOPE-FISH.
Cultures were fixed with paraformaldehyde, and FISH was performed by using doubly labeled oligonucleotide probes (43) using Cy3 doubly labeled probe Arch915 and the FLUOS doubly labeled probe EUB338, respectively. Counterstaining with the nucleic acid-binding dye SYBR Green I revealed a detection rate greater than 99%. Hybridizations were analyzed using a Leica DM500 microscope equipped with a DFC345 FX Peltier-cooled CCD camera (Wetzlar).
Soil Incubation and Stable Isotope Probing.
Sandy loam soil (pH 4.5) was sampled from 0 to 10 cm of an agricultural plot subject to yearly crop rotation (most recent crop was potatoes). Soil was sieved and 10 g (30% water; wt/wt) was added to 120-mL serum vial bottles before establishing 5% 12C- or 13C-CO2 headspace concentrations (vol/vol) or 0.01% acetylene (vol/vol) as described previously (28). Microcosms were established for destructive sampling in triplicate per time point and incubated at 30 °C for as long as 20 d. Extracted DNA was subjected to CsCl isopycnic density gradient centrifugation (35), fractionation, and quantification as described previously (28), but by using the group 1.1a-associated lineage amoA assay. Ammonia and nitrate concentrations were determined in 1 M KCl extracts (13).
Statistical Analysis.
The effect of treatments on specific growth rate was analyzed by one-way ANOVA and correlation analysis using SigmaPlot, version 11 (Systat Software).
Supplementary Material
Acknowledgments
We thank Maria Tourna and Christa Schleper (University of Vienna) for discussions on their unpublished data. L.E.L.-M. is funded by a Natural Environment Research Council (NERC) Postgraduate Studentship. G.W.N. is funded by NERC Advanced Fellowship NE/D010195/1. K.S. and A.V. were supported by the German Federal State of Hessen as part of the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz program.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN227488 and JN227489).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107196108/-/DCSupplemental.
References
- 1.von Uexküll HR, Mutert E. In: Plant–Soil Interactions at Low pH, Principles and Management. Date RA, Grundon NJ, Raymet GE, Probert ME, editors. New York: Kluwer; 1995. pp. 5–19. [Google Scholar]
- 2.Booth MS, Stark JM, Rastetter E. Controls on nitrogen cycling in terrestrial ecosystems: A synthetic analysis of literature data. Ecol Monogr. 2005;75:139–157. [Google Scholar]
- 3.Allison SM, Prosser JI. Urease activity in neutrophilic autotrophic ammonia-oxidizing bacteria isolated from acid soils. Soil Biol Biochem. 1991;23:45–51. [Google Scholar]
- 4.Jiang QQ, Bakken LR. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol Ecol. 1999;30:171–186. doi: 10.1111/j.1574-6941.1999.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki I, Dular U, Kwok SC. Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J Bacteriol. 1974;120:556–558. doi: 10.1128/jb.120.1.556-558.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frijlink MJ, Abee T, Laanbroek HJ, de Boer W, Konings WN. The bioenergetics of ammonia and hydroxylamine oxidation in Nitrosomonas europaea at acid and alkaline pH. Arch Microbiol. 1992;157:194–199. [Google Scholar]
- 7.de Boer W, Duyts H, Laanbroek HJ. Urea stimulated autotrophic nitrification in suspensions of fertilized, acid heath soil. Soil Biol Biochem. 1989;21:349–354. [Google Scholar]
- 8.Burton SAQ, Prosser JI. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol. 2001;67:2952–2957. doi: 10.1128/AEM.67.7.2952-2957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Killham K. Heterotrophic nitrification. In: Prosser JI, editor. Nitrification. Oxford: IRL Press; 1986. pp. 117–126. [Google Scholar]
- 10.De Boer W, Gunnewiek PJ, Veenhuis M, Bock E, Laanbroek HJ. Nitrification at low pH by aggregated chemolithotrophic bacteria. Appl Environ Microbiol. 1991;57:3600–3604. doi: 10.1128/aem.57.12.3600-3604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison SM, Prosser JI. Ammonia oxidation at low pH by attached populations of nitrifying bacteria. Soil Biol Biochem. 1993;25:935–941. [Google Scholar]
- 12.de Boer W, Kowalchuk GA. Nitrification in acid soils: Micro-organisms and mechanisms. Soil Biol Biochem. 2001;33:853–866. [Google Scholar]
- 13.Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 14.Gubry-Rangin C, Nicol GW, Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010;74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- 15.Stopnišek N, et al. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol. 2010;76:7626–7634. doi: 10.1128/AEM.00595-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leininger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 17.He JZ, et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol. 2007;9:2364–2374. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 18.Tourna M, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyman MR, Wood PM. Suicidal inactivation and labelling of ammonia mono-oxygenase by acetylene. Biochem J. 1985;227:719–725. doi: 10.1042/bj2270719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker CB, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallam SJ, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 23.Offre PO, Nicol GW, Prosser JI. Autotrophic community profiling and quantification of putative autotrophic thaumarchaeal communities in environmental samples. Environ Microbiol Rep. 2010;3:245–253. doi: 10.1111/j.1758-2229.2010.00217.x. [DOI] [PubMed] [Google Scholar]
- 24.Buckley DH, Huangyutitham V, Hsu S-F, Nelson TA. Stable isotope probing with 15N achieved by disentangling the effects of genome G+C content and isotope enrichment on DNA density. Appl Environ Microbiol. 2007;73:3189–3195. doi: 10.1128/AEM.02609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koops H-P, Böttcher B, Möller UC, Pommerening-Röser A, Stehr G. Classification of eight new species of ammonia-oxidising bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J Gen Microbiol. 1991;137:1689–1699. [Google Scholar]
- 26.Klemedtsson L, Jiang Q, Klemedtsson AK, Bakken L. Autotrophic ammonium-oxidising bacteria in Swedish mor humus. Soil Biol Biochem. 1999;31:839–847. [Google Scholar]
- 27.Schmidt CS, et al. PCR profiling of ammonia-oxidizer communities in acidic soils subjected to nitrogen and sulphur deposition. FEMS Microbiol Ecol. 2007;61:305–316. doi: 10.1111/j.1574-6941.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L-M, et al. Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci USA. 2010;107:17240–17245. doi: 10.1073/pnas.1004947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 30.Park S, Bae W. Modeling kinetics of ammonium oxidation and nitrite oxidation under simultaneous inhibition by free ammonia and free nitrous acid. Process Biochem. 2009;44:631–640. [Google Scholar]
- 31.Anthonisen AC, Loehr RC, Prakasam TBS, Srinath EG. Inhibition of nitrification by ammonia and nitrous acid. J Water Pollut Control Fed. 1976;48:835–852. [PubMed] [Google Scholar]
- 32.Widdel F, Bak F. The Prokaryotes. In: Ballows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H, editors. A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Application. 2nd Ed. New York: Springer; 1992. pp. 3352–3378. [Google Scholar]
- 33.Shinn MB. Colorimetric method for determination of nitrite. Ind Eng Chem Anal Ed. 1941;13:33–35. [Google Scholar]
- 34.Crooke WM, Simpson WE. Determination of ammonium in Kjeldahl digests of crops by an automated procedure. J Sci Food Agric. 1971;22:9–10. [Google Scholar]
- 35.Tourna M, Freitag TE, Nicol GW, Prosser JI. Stable isotope probing analysis of interactions between ammonia oxidizers. Appl Environ Microbiol. 2010;76:2468–2477. doi: 10.1128/AEM.01964-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehtovirta LE, Prosser JI, Nicol GW. Soil pH regulates the abundance and diversity of Group 1.1c Crenarchaeota. FEMS Microbiol Ecol. 2009;70:367–376. doi: 10.1111/j.1574-6941.2009.00748.x. [DOI] [PubMed] [Google Scholar]
- 37.Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol. 2003;5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- 38.Grosskopf R, Janssen PH, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 40.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 41.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 42.Lane DJ. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 43.Stoecker K, Dorninger C, Daims H, Wagner M. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl Environ Microbiol. 2010;76:922–926. doi: 10.1128/AEM.02456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.