Abstract
The P transposable element recently invaded wild Drosophila melanogaster strains worldwide. A single introduced copy can multiply and spread throughout the fly genome in just a few generations, even though its cut-and-paste transposition mechanism does not inherently increase copy number. P element insertions preferentially target the promoters of a subset of genes, but why these sites are hotspots remains unknown. We show that P elements selectively target sites that in tissue-culture cells bind origin recognition complex proteins and function as replication origins. The association of origin recognition complex-binding sites with selected promoters and their absence near clustered differentiation genes may dictate P element site specificity. Inserting at unfired replication origins during S phase may allow P elements to be both repaired and reduplicated, thereby increasing element copy number. The advantage transposons gain by moving from replicated to unreplicated genomic regions may contribute to the association of heterochromatin with late-replicating genomic regions.
Keywords: genome evolution, cell cycle, DNA replication, pre-replication complex
Transposable elements comprise or have molded a significant fraction of most metazoan genomes (1). The potency of DNA transposons makes them particularly attractive as tools for understanding genome evolution and as agents of genetic manipulation (2, 3). The Drosophila P element, a 3-kb DNA transposon encoding a transposase, ranks among the best-understood eukaryotic transposons (Fig. 1A; reviewed in ref. 4) and has been used widely in Drosophila molecular genetic research. P elements entered Drosophila melanogaster by horizontal transfer from a distantly related Drosophila group about 80 y ago and spread rapidly throughout all natural populations (5), many of which now contain 30–50 intact or internally deleted element copies. Despite transposing by a cut-and-paste mechanism that does not inherently increase copy number (6, 7), a single P element introduced within a small inbreeding laboratory population can spread rapidly throughout all its members, giving rise in each individual to a collection of genomic elements resembling those found in wild strains (8, 9). Stability is achieved eventually by the production of truncated transposase proteins (10, 11) and by piwi-interacting RNA (piRNA)-mediated mechanisms, some of which are maternally inherited (12).
Fig. 1.
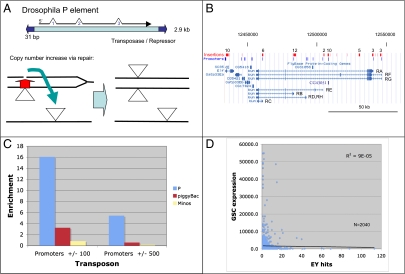
P element insertions frequently coincide with promoters. (A) P element structure and proposed mechanism of copy number increase via homologous repair from sister strands (ref. 6). Lines represent two chromosomes, one replicating. After one copy of a replicated P element (triangle) transposes (arrow) to a new site (Left), the double-strand break is repaired (red) from the sister, regenerating the lost element, and increasing copy number (Right). (B) Drosophila genomic region surrounding the bunched (bun) locus. Transcripts (blue) are labeled (RA–RH), and promoters are shown above as blue boxes. P element insertions are indicated by red vertical bars or boxes with the number of insertions stated above. Most clustered insertions overlap promoters. (C) The enrichment level of P element (blue), piggyBac (red), and Minos (yellow) insertions in promoter regions. (D) Lack of correlation between mRNA levels in early germ cells (2,040 expressed genes) and P targeting.
The Gene Disruption Project (GDP) has accumulated extensive data on the site specificity of several transposons, including P elements, as a byproduct of generating gene disruption strains for public use in about two-thirds of all Drosophila genes (13, 14). By analyzing more than 100,000 individual transposition events from more than 10 independent screens based on a variety of engineered P elements, the highly nonrandom selectivity of P element insertion has been thoroughly documented. Regardless of the starting site or the nature of added internal sequences, 30–40% of all P element insertions are found in essentially the same 200–400 “hotspot” loci. More than 70% are near gene promoters, but clustered tissue-specific genes rarely are mutated. Although P element integration sites are GC rich, their primary sequences do not explain this insertional bias (15). Other well-studied DNA transposons such as piggyBac share a general preference for genes and promoters, but piggyBac hotspots overlap P element hotspots to only a limited extent (14, 16). P elements may target genes that contain open chromatin and/or are actively transcribed in early germ cells where transposition occurs (4, 17). However, no model has been decisively supported, and the basis of P element site preferences remains largely unsolved.
Host replication presents a prime opportunity for transposons to multiply, and several prokaryotic and eukaryotic transposons coordinate movement with DNA replication (18–20). Transposons that move after replication can increase in copy number if the excised element is restored fully by homologous repair of a double-strand break using the sister strand, which still retains a donor copy (Fig. 1A). Such restoration is observed at high frequency following P element transposition (6), suggesting that P transposition occurs after DNA replication. How such coordination is achieved and whether sister strand-mediated repair is sufficient to explain rapid P element expansion remain uncertain.
Except in rare instances, metazoan DNA is replicated exactly once per cell cycle by assembling prereplication complexes (preRCs) that license sequence-nonspecific replication origins in G1 phase (reviewed in ref. 21). The six-protein origin recognition complex (ORC), other factors, and an appropriate chromatin environment facilitate binding of the minichromosome maintenance (MCM) replicative helicase (22). Conditions permissive for licensing are terminated before origin activation in S phase. Proteins at preRCs either initiate bidirectional replication or are displaced from the DNA by a passing replication fork that initiated elsewhere. In addition to recognizing origins, metazoan ORC proteins also function in epigenetic inheritance, heterochromatin formation, chromosome cohesion, and mitotic division (reviewed in ref. 23).
Genomic methods recently have begun to provide detailed insight into metazoan chromosome replication (24). As part of the modENCODE project (25), thousands of candidate replication origins active in tissue-culture cells were mapped at high resolution throughout the Drosophila genome as sites of ORC binding and were verified by comparison with early replication patterns (26, 27). Replication origins share multiple features with transcriptional promoters, such as open chromatin and high levels of histone acetylation (24, 26–28). Perhaps because of these common requirements, many ORC-binding sites in Drosophila are located at gene promoters (26, 27).
Here we show that P elements preferentially integrate into replication origins defined by ORC protein-binding sites in rapidly cycling cells. We propose that P elements coordinate transposition within the cell cycle by interacting with preRCs. These mechanisms might accelerate copy number growth not only by maximizing opportunities for sister strand-mediated repair but also by facilitating movement from replicated to unreplicated genome regions.
Results
P Elements Insert at a Subset of Promoters.
We analyzed 18,213 independent transposition events of the EY P element, which were shown previously to be representative of P transposition generally (13, 14). Comparable datasets for two other DNA transposons, the mariner-like Minos element and the piggyBac element, were used for comparison (14, 16). As expected from previous studies, P element insertions show a strong correlation with promoters. For example, in a genomic region containing the bunched hotspot gene, most sites with multiple nearby insertions are near promoters (Fig. 1B). Genomewide, however, relatively few promoters are P-element insertion targets. Defined as the annotated start site ± 100 bp, the 18,021 annotated promoters (correcting for overlaps) span 2.8% of the genome (3.34/117 Mb). The 18,213 P insertions in the collection hit only 3,254 of these promoters (18%) but they accounted for 8,219 total insertions (45%). Thus, promoters as a whole are enriched for P insertion by a factor of 16-fold (45/2.8) (Fig. 1C). For comparison, piggyBac insertions are only 3.5-fold enriched for promoter insertion, and Minos insertions show no enrichment for promoter sites (0.83-fold). Broadening the definition of promoter to encompass ± 500 bp around the start site increases the fraction of promoters hit to 24% and the fraction of insertions associated with promoters to 71% (14) but lowers P enrichment at these promoters to 5.7-fold (Fig. 1C).
These values show that P elements do not randomly target promoters per se. If they did, the Poisson distribution (with λ = 18,213/18,021 = 1.0) predicts that the 18,213 P insertions would hit 63% of the 18,021 promoters at least once, rather than 18–24%. Furthermore, individual promoters are hit at very different rates, because the top 300 hotspot genes (corresponding to less than 2% of promoters) account for more than 40% of all P insertions in the collection, consistent with previous studies (13). Within many hotspots, even individual nucleotide positions we term “hotpoints” are targeted selectively by multiple, independent insertions in both orientations (Table S1). A weak consensus sequence derived from hotpoints closely matches the preferred sequence reported previously from a diverse sample of P insertion sites (16).
Previous attempts to explain P element insertional selectivity based on developmental factors have been largely unsuccessful (13). We compared the P insertion frequency to gene expression levels measured previously by microarray (29) in Drosophila germline stem cells and found no correlation (Fig. 1D). Thus, P elements may insert preferentially in a small set of particular gene promoters as a secondary consequence of some other correlated aspect of genome organization. Table S2 lists the 100 strongest P element hotspots to assist in searching for the basis of P element site specificity.
P Elements Target ORC-Binding Sites.
Replication origins frequently colocalize with promoters. To examine whether the association of P element insertion sites with promoters might reflect a connection to replication, we compared the distribution of P insertions and the ORC-binding sites reported for three Drosophila tissue-culture cell lines (26, 27). A striking correlation was observed, even though the ORC-binding regions represent only 1.8% of the genome. For example, in the hotspot gene Hr39 (Fig. 2A), most P elements integrate within two small regions, both of which correspond to ORC-binding sites. One of the hotspots overlaps the promoter, but the other lies within intron 2. Likewise, most of the promoter-associated P insertion clusters described at bunched are associated with ORC sites that map to these same regions in tissue-culture cells (Fig. 2B).
Fig. 2.
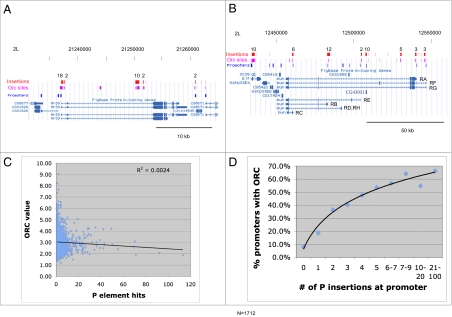
P elements target ORC-binding sites. (A) P insertions (red) in the Hr39 region align with the location of ORC-binding sites in tissue-culture cells (ref. 27,magenta boxes). A cluster of 18 insertions falls within an ORC site that overlaps the Hr39 promoter; a cluster of 10 insertions spans two adjacent ORC sites within intron 2. (B) The bun genome region as in Fig. 1B, showing ORC sites (magenta boxes). Most promoter-associated hotspots overlap ORC-binding sites. (C) The strength of individual ORC-binding sites does not correlate with their frequency of targeting by P elements. (D) Genes hit more frequently by P element insertion are more likely to contain an ORC site at their promoter.
We carried out several additional comparisons to elucidate further the relationship between promoter and ORC-site targeting. No correlation was observed between the strength of an ORC-binding site and its frequency of P insertion (Fig. 2C). In contrast, a striking relationship exists between promoter targeting and the presence of an overlapping ORC site (Fig. 2D). Genes not hit by P elements contain a promoter with an overlapping ORC site less than 10% of the time. As the number of P hits increases, genes more and more frequently contain promoters with associated ORC sites. Promoters with more than six insertions in the data set contain a coincident ORC site about 60% of the time.
The correspondence between P element insertion and ORC sites is illustrated further by the hotspot genes (Table S2). These loci mostly are conventional protein-coding genes but also include two RNA genes, mir-282 and Hsromega, and three sites outside any annotated gene. At least one ORC site that contains P insertions, usually overlapping a promoter, is present in 94% of the top 100 P hotspot genes, and in 78% such an ORC site is common (i.e., overlapping) to all three cell lines. The enrichment for insertions within a single ORC site selected from each hotspot is as high as 3,790-fold, with an average enrichment of more than 600-fold (Table S2). Strikingly, there is little or no enrichment for piggyBac or Minos insertion at these same ORC sites (average enrichment 2.7-fold and 1.1-fold, respectively).
The strength of the association between P element insertion and ORC sites suggested that a connection with replication origins, rather than promoters, is of primary significance (Fig. 3A). The 7,329 nonoverlapping ORC-binding regions within the three cell lines are 29-fold enriched for P element insertion, compared with 16-fold enrichment at promoters. Moreover, 30% of the clustered ORC sites contain at least one P element insertion, compared with 18% of (clustered) promoters, and more insertions are in ORC sites than promoters (51% vs. 45%). Among ORC sites common to all three cell lines the enrichment is 42-fold. At ORC sites that overlapped a promoter, enrichment is even higher, 54-fold, whereas nonpromoter ORC sites are still 15-fold enriched. In contrast, the enrichment of P insertions at promoters without an ORC site is only 6.5-fold, only twofold higher than the enrichment of piggyBac insertions (Fig. 3A).
Fig. 3.
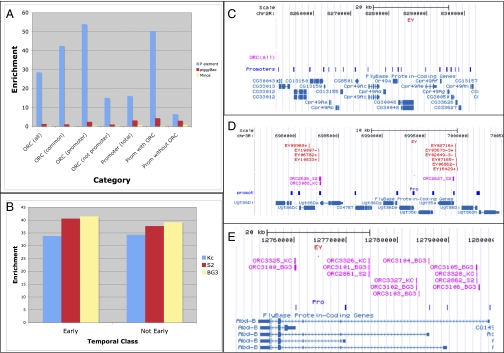
P element enrichment at ORC sites compared with promoters. (A) Enrichment of P elements (blue), piggyBac elements (red), or Minos elements (yellow) at classes of ORC sites or promoters (prom). (B) Early origins are targeted by P elements at a frequency similar to the targeting of nonearly origins in Kc (blue), S2 (red), and BG3 (yellow) cells. (C) The cuticle protein gene complex in region 49A (Cpr49A–Cpr49Ah) lacks ORC sites (purple) and P insertions (red). (D) The nine-gene Ugt86D cluster. The only P element insertions (red) are found at the two Ugt86D genes whose promoters (blue boxes) are associated with ORC sites (magenta boxes). (E) In the Abd-B region, ORC sites (purple) are not sites of P insertion (red).
We used data in which early-replicating regions of the same three cell lines were mapped by BrdU incorporation in hydroxyurea-arrested cells to investigate whether replication timing influenced P element insertion (26, 27). Mapping early-replicating regions using this approach is imprecise, and ORC-binding sites are overrepresented in early-replicating regions (27). However, when we compared the level of P enrichment in these “early” ORC sites with the level of P enrichment in nonearly sites, no differences were seen (Fig. 3B).
ORC Location Affects P Element Selectivity.
P element insertions rarely have been recovered in clustered tissue-specific genes associated with terminal differentiation (13) (e.g., clustered genes such as chorion genes, cuticle protein genes, and salivary gland secretory genes, among others). We observed a dearth of ORC sites in such gene regions, suggesting that a lack of replication origins in germ cells may be responsible for the absence of insertions. For example, in the nine-gene cuticle protein gene cluster in region 49D, not a single ORC site or P insertion was present (Fig. 3C). In the Ugt86D cluster, seven of nine genes lacked an ORC site, and none of these seven had P insertions (Fig. 3D). However, the other two genes, which appear no different in structure, had ORC sites at their promoters, and both contained multiple P insertions.
P element insertions are absent in many “repressed” genomic regions regulated by Polycomb Group (PcG) proteins (14). Consequently, we determined if such regions also were deficient in ORC-binding sites and found that they were not. For example, the Abd-B gene region of the bithorax complex contains multiple ORC sites (Fig. 3E), as do other major PcG-regulated regions, despite their lack of insertions. The repressive chromatin at PcG sites may preclude P element transpositional machinery from associating normally with local preRCs at these sites, disrupting integration in these regions.
Discussion
P Elements Integrate Preferentially at Replication Origins Defined by ORC Binding.
Our results indicate that P transposons insert selectively within ORC protein-binding sites defined in cultured cell lines, sites that frequently correspond to replication origins. This striking preference is not confined to a particular P element derivative: Essentially the same genes and hotspots are targeted by structurally diverse P element constructs (13, 14). We verified that P insertions recovered in all the major screens analyzed by the GDP preferentially target ORC-binding sites. In contrast, neither piggyBac elements nor the Minos transposon target these hotspots (14) or show significant enrichment at ORC-binding sites. Consequently, targeting replication origins appears to be a specific property of the P element transposon.
The strong replication origin preference documented here represents only a minimum estimate of the actual level with which origins are targeted within immature germ cells where P elements transpose. Insertional hotspots at some promoters lacked overlapping tissue culture cell ORC sites (Table S2), but insertions at these sites may have targeted germ-cell replication origins that are inactive or could not be mapped in cultured cells. Other hotspots were located very close to ORC sites, for example in the mbl gene, but were not counted because they fell just outside their reported boundaries. We suspect such insertions actually were associated with the ORC sites in question. The fact that germ-cell insertion sites corresponded so strongly to tissue-culture cell ORC-binding sites despite these limitations testifies to the exceptional strength of this association, shows that the ORC mapping data are accurate (27), and reveals that many of the same replication origins are used in germ cells and tissue-culture cells.
Transposition to Replication Origins May Explain P Element Selectivity.
Our results argue that a substantial fraction, and possibly all, of the P element's previously reported promoter tropism is secondary to its selectivity for replication origins. There is a strong correspondence between ORC-binding sites, regions of open chromatin, and promoters (26). This correspondence probably underlies changes in nucleosome positioning and chromatin modification that are used to activate both transcription and replication. It is possible that origins coincident with promoters differ functionally in some way that particularly attracts P element insertion. Nonetheless, promoter-unlinked ORC sites were still 15-fold enriched for insertion, suggesting that promoter association is not essential. It will be necessary to map ORC-binding sites in early germ cells to resolve whether promoters ever are targeted for insertion without the presence of a replication origin.
The complex chromatin factors that specify replication origins probably also explain P element hotspot and coldspot genes. Hotspot genes correspond to genes with one or more strong ORC-binding sites, usually near a particular gene promoter. Despite the presence of such ORC sites, not all these genes are highly transcribed, at least based on steady-state RNA accumulation (Fig. 1D). Coldspots, such as those at clustered, tissue-specific genes, now can be explained by the lack of replication origins in the vicinity of such loci.
Origin-Linked P Elements May Transpose Only Following Replication but Frequently Insert Locally.
Integrating at replication origins might help a cut-and-paste transposon increase its genomic copy number (Fig. 4). An element located at an origin could use the presence of a preRC to coordinate transposition with replication. By delaying transposition while ORC proteins remain, or by sensing some other structural change indicative of origin activation, each departing element would ensure that a copy remains on the sister strand to act as a template for repair of the resulting double-strand break. Our results predict that P elements located at genomic sites that do not function as replication origins in a particular cell would exhibit distinctive properties, including altered timing of transposition during the cell cycle.
Fig. 4.
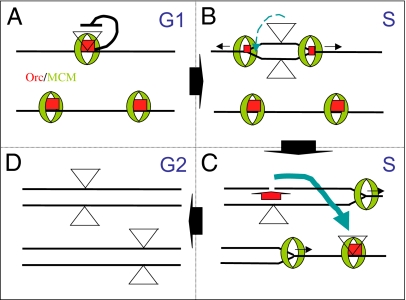
Coordination of transposition with replication may expand P element copy number and cause local transposition. (A) Timing transposition. Two chromosomes in a G1 cell containing three replication origins bound by ORC complexes (red box) and MCM complexes (green doughnut) to form preRCs are shown. The preRC is hypothesized to repress transposition of an integrated transposon (triangle), preventing excision prior to duplication when the preRC is disassembled. (B) Local transposition. Just after origin activation a derepressed P element may transpose locally (dashed blue arrow) before full preRC disassembly (small red boxes). Black arrows indicate replication fork movement. (C) More frequently, one duplicated element excises and integrates at an unfired origin on another chromosome (blue arrow), guaranteeing that it will reside in an unreplicated region. One new transposon copy is generated by homologous repair (red arrow) of the double-strand break using the remaining element on the sister strand. (D) A second new transposon copy is generated later in S phase, when the newly integrated copy replicates, doubling transposon copy number by G2.
The discovery that P elements preferentially insert at replication origins suggests an explanation for a perplexing aspect of their behavior. P elements, line maize Ac elements, and many other transposons integrate at elevated frequency near their starting site (30), a phenomenon known as “local transposition.” Replication factors and DNA structures characteristic of preRCs are likely to be attractive to insertion. For a brief period following preRC activation these factors may remain at the newly diverging forks, leading a fraction of excised elements to reintegrate rapidly near their starting site (Fig. 4B). If P elements orient to one of the two replicating strands (see below) and transpose across the origin center, this model would provide a simple explanation of the head-to-head orientation and other orientational preferences of elements during local transposition (30).
Other transposons have been shown previously to interact with DNA replication. The Ac element generally transposes after replicating, and the excised copy inserts in both unreplicated and replicated genomic regions (19). The prokaryotic element Tn10 transposes rapidly following the onset of replication, a feature shared by Ac. Rather than detecting changes in preRC proteins as proposed for the P element, Tn10 is activated when its termini become transiently hemimethylated following replication (18). One of Tn7's distinct transposition pathways detects an altered DNA structure as well as factors associated with lagging-strand DNA synthesis, which leads to a strong orientational bias in insertion (20). Recently, the prokaryotic analog of Proliferating Cell Nuclear Antigen (PCNA), the β-clamp, was shown to be such a factor and to interact with Tn7 at lagging strands (31). Interactions with replication, including integration at origins, may be a feature shared by many transposons. Features associated with P element selectivity may be diagnostic of such a connection. However, other elements likely use different methods, as shown by the disparate behavior of piggyBac and Minos elements.
Targeting Unfired Replication Origins Might Increase P Element Copy Number Further.
Our experiments suggest an additional mechanism that P elements and other transposons may use to increase their copy number beyond that which can be accomplished by efficient repair from sister strands (Fig. 4C). The ability to sense preRCs at unfired origins might allow an element to move preferentially during S phase from replicated to unreplicated genomic regions, allowing it to replicate twice in a single cell cycle. Adopting such a “replication timing” strategy could double the rate at which copy number increases compared with relying on sister-strand repair alone. Replication timing might be accomplished if, after excision, the transposition complex binds strongly to preRCs but not to elongating forks before integrating. Unlike sister-strand repair, which specifically benefits elements that excise, all types of transposons, in principle, could use replication timing to increase their copy number.
As a strategy, replication timing poses potential problems. If origin activation occurs at the same time in S phase during each cell cycle (32), elements activated by replication that target unfired preRCs might insert on average into regions replicating later than the starting site. Soon, transposons might become trapped or insertionally inactivated in very late-replicating regions of the genome. However, the problem of a shrinking target zone could be overcome by using a second transposition mechanism, such as local transposition, to allow some elements to escape replication control and reseed early origins in subsequent cell cycles. Although their origin preference suggests that P elements have the capability to practice replication timing, it remains unclear if they actually do so. We did not observe any tendency of P elements to transpose into later-firing classes of replication origins mapped in tissue-culture cells (Fig. 3B). Studies of replication and transposition in early germ cells will be required to resolve this issue.
Transposons That Time Replication and the Origin of Heterochromatin.
Does a selfish drive to increase copy number by replication timing influence the evolution of genome organization? Such a connection might rationalize the nearly universal observation that heterochromatic regions replicate late in S phase and are enriched in transposons. Any late-replicating region might experience elevated transposition, leading over time to its acquisition of heterochromatic properties. This tendency also may be used to regulate transposition. piRNA clusters are located in heterochromatin (12) where presumably they replicate very late in S phase. Such locations might increase the chance that transposons will integrate within these large transcription units and generate repressive piRNAs. In sum, by gaining insight into why a specific transposon moves nonrandomly, we may have begun to glimpse mechanisms that have profoundly influenced the evolution of metazoan genomes.
Materials and Methods
Insertion Site Mapping.
Generation and mapping of P element, piggyBac, and Minos insertions have been described (13, 14), and site data are publicly available from the GDP Website (http://flypush.imgen.bcm.tmc.edu/pscreen/index.php). The starting site for the P screen was within the CG2201 hotspot.
ORC-Binding Sites and Early Replication Regions.
The number, position, and strength of ORC sites, as well as the number and location of early origin regions, were taken from data deposited at GEO under accessions GSE17285–GSE17287 and GSE17279–GSE17281 (27). Individual sites in each file were numbered consecutively along the chromosome arms, and a suffix was added to indicate from which cell line the data originate. Copies of these files are available on the GDP Website.
Promoter Data.
We used data on 22,427 transcripts described in sequence release 5 and FlyBase gene annotation 5.22. The 18,021 unique 5′ start sites located within the 117-Mb core genome where insertions can be mapped uniquely (14) that were >25 bp from any other start sites on the same DNA strand were considered to represent distinct promoters. The conclusions reported are unaffected by recent updates in annotation.
Analysis of Enrichment.
To determine enrichment (i.e., the relative probability of transposition into a subset of the genome such as gene promoters compared with the genome as a whole), we determined the fraction of all insertions that were located within the genomic subset in question and divided by the fraction of the 117-Mb genome it comprised. To avoid counting insertions twice in the case of genomic subsets with overlapping elements, we first amalgamated all elements of the subset into nonoverlapping clusters and then calculated the enrichment for the clustered dataset.
Supplementary Material
Acknowledgments
We thank Dr. Robert Levis and all the members of the Gene Disruption Project who have contributed to generating the databases of transposon insertion sites. We also thank Robert Levis for helpful comments on the manuscript. We acknowledge support from National Institutes of Health Grant GM067858.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112960108/-/DCSupplemental.
References
- 1.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 2.Cooley L, Kelley R, Spradling AC. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- 3.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engels W. In: P Elements in Drosophila melanogaster. Mobile DNA, Berg EE, Howe MM, editors. Washington: ASM Publications; 1989. pp. 185–210. [Google Scholar]
- 5.Clark JB, Kidwell MG. A phylogenetic perspective on P transposable element evolution in Drosophila. Proc Natl Acad Sci USA. 1997;94:11428–11433. doi: 10.1073/pnas.94.21.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engels WR, Johnson-Schlitz DM, Eggleston WB, Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 7.Kaufman PD, Rio DC. P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell. 1992;69:27–39. doi: 10.1016/0092-8674(92)90116-t. [DOI] [PubMed] [Google Scholar]
- 8.Preston CR, Engels WR. Spread of P transposable elements in inbred lines of Drosophila melanogaster. Prog Nucleic Acid Res Mol Biol. 1989;36:71–85. doi: 10.1016/s0079-6603(08)60162-2. [DOI] [PubMed] [Google Scholar]
- 9.Good AG, Meister GA, Brock HW, Grigliatti TA, Hickey DA. Rapid spread of transposable P elements in experimental populations of Drosophila melanogaster. Genetics. 1989;122:387–396. doi: 10.1093/genetics/122.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rio DC, Laski FA, Rubin GM. Identification and immunochemical analysis of biologically active Drosophila P element transposase. Cell. 1986;44:21–32. doi: 10.1016/0092-8674(86)90481-2. [DOI] [PubMed] [Google Scholar]
- 11.Black DM, Jackson MS, Kidwell MG, Dover GA. KP elements repress P-induced hybrid dysgenesis in Drosophila melanogaster. EMBO J. 1987;6:4125–4135. doi: 10.1002/j.1460-2075.1987.tb02758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellen HJ, et al. The BDGP gene disruption project: Single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellen HJ, et al. The Drosophila gene disruption project: Progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thibault ST, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 17.Bownes M. Preferential insertion of P elements into genes expressed in the germ-line of Drosophila melanogaster. Mol Gen Genet. 1990;222:457–460. doi: 10.1007/BF00633856. [DOI] [PubMed] [Google Scholar]
- 18.Roberts D, Hoopes BC, McClure WR, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Greenblatt IM, Dellaporta SL. Molecular analysis of Ac transposition and DNA replication. Genetics. 1992;130:665–676. doi: 10.1093/genetics/130.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters JE, Craig NL. Tn7: Smarter than we thought. Nat Rev Mol Cell Biol. 2001;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- 21.Arias EE, Walter JC. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 22.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 23.Chesnokov IN. Multiple functions of the origin recognition complex. Int Rev Cytol. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert DM. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet. 2010;11:673–684. doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy S, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eaton ML, et al. Chromatin signatures of the Drosophila replication program. Genome Res. 2011;21:164–174. doi: 10.1101/gr.116038.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kai T, Williams D, Spradling AC. The expression profile of purified Drosophila germline stem cells. Dev Biol. 2005;283:486–502. doi: 10.1016/j.ydbio.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 30.Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks AR, et al. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell. 2009;138:685–695. doi: 10.1016/j.cell.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiratani I, Gilbert DM. Replication timing as an epigenetic mark. Epigenetics. 2009;4:93–97. doi: 10.4161/epi.4.2.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.