Abstract
A number of acute leukemias arise from fusion of the mixed lineage leukemia 1 protein (MLL) N terminus to a variety of fusion partners that have been reported to reside in one or more poorly defined complexes linked to transcription elongation through interactions with the histone H3-K79 methyltransferase DOT1 and positive transcription elongation factor b (P-TEFb). Here we first identify natural complexes (purified through fusion partners AF9, AF4, and ELL) with overlapping components, different elongation activities, and different cofactor associations that suggest dynamic interactions. Then, through reconstitution of defined, functionally active minimal complexes, we identify stable subcomplexes that, through newly defined protein-protein interactions, form distinct higher order complexes. These definitive analyses show, for example, that (i) through direct interactions with AF9 and cyclinT1, family members AF4 and AFF4 independently mediate association of P-TEFb with AF9, (ii) P-TEFb, through direct interactions, provides the link for association of ELL and ELL-associated factors 1 and 2 (EAF1 and EAF2) with AF4, and (iii) in the absence of other factors, DOT1 forms a stable complex with AF9 and does not interact with AF9•AF4•P-TEFb complexes. Finally, we show the importance of defined higher order complex formation in MLL–AF9-mediated transcriptional up-regulation and cell immortalization potential in vivo. Thus, our study provides direct mechanistic insight into the role of fusion partners in MLL fusion-mediated leukemogenesis.
The mixed lineage leukemia 1 protein (MLL) regulates HOX gene expression through the H3 lysine 4 (K4) methyltransferase activity of its C-terminal catalytic domain (1, 2). Rearrangements of the MLL gene are associated with aggressive acute leukemias (2), the most common of which are balanced chromosomal translocations that fuse N-terminal sequences in frame to over 50 different fusion partner proteins (3). MLL fusion-transformed hematopoietic progenitor cells exhibit a persistent expression of MLL target genes that is correlated with persistent H3K4 trimethylation (mediated by MLL expressed from the nonrearranged allele) and an increase in histone H3-K79 methylation by DOT1 (4, 5).
MLL fusion partners include AF9 and family member ENL, AF4 and family member AFF4, AF10, and ELL, which together account for the majority of MLL fusion protein leukemias (3). Although MLL and MLL fusion proteins are thought to be recruited to target genes through interactions of N-terminal sequences with other proteins and with DNA (1, 6), the mechanisms by which they jointly lead to abnormal gene expression and leukemogenesis are not well understood. A possible effect of MLL fusion proteins on transcription elongation was indicated by identification of ELL as a bona fide elongation factor (7) and by the demonstration of a direct association of AFF4 with positive transcription elongation factor b (P-TEFb) (8). P-TEFb is composed of cyclin-dependent kinase Cdk9 and either cyclinT1 or cyclinT2 and acts by phosphorylation of the inhibitory 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) complexes (9) and the RNA polymerase II C-terminal domain (CTD), which serves as a recruitment signal for various factors during the elongation phase of transcription. Further, AF10 was shown to interact with DOT1 and to increase H3K79 methylation at target genes in vivo (10); and recent studies have confirmed a role for DOT1 in MLL-related leukemia (ref. 11, and references therein).
Both earlier (12, 13) and more recent (14–17) studies have reported large complexes containing overlapping subunits that include DOT1 and/or P-TEFb in variable associations with MLL fusion partners AF4/AFF4, ENL/AF9, ELL and other proteins. These studies emphasized that MLL fusion partners and fusion proteins may regulate transcription elongation through recruitment of P-TEFb, as originally suggested (8), and/or through ELL. However, the presumed elongation activities of these various, potentially heterogeneous, complexes were not demonstrated. Moreover, P-TEFb and interacting MLL fusion partners are also recruited to target genes in cells transformed by a fusion protein (MLL–AF6) that fails to interact directly with any of these protein complexes (17). Hence, the overall effect of the MLL fusion proteins appears to involve more than a simple P-TEFb recruitment mechanism. In addition, and importantly, the specific activities of individual components and the exact interplay between them remain to be determined and will be crucial for understanding why different MLL fusion proteins produce different disease outcomes in vivo (18).
To further investigate these questions, we have purified and characterized, structurally and functionally, both natural MLL fusion partner-containing complexes from mammalian cells and, most importantly, reconstituted and biochemically defined subcomplexes of MLL fusion/partner proteins and transcription elongation factors. These analyses provide unique mechanistic insights into the function of MLL fusion proteins and fusion partner proteins.
Results
Biochemical Purification and Functional Analyses of Protein Complexes Associated with AF9, AF4, and ELL.
To investigate the mechanism of action of MLL fusion proteins, we first sought to identify proteins associated with representative partner proteins (AF4, AF9, and ELL). After establishing cell lines that stably express corresponding FLAG-HA-tagged proteins, corresponding complexes were purified by a rigorous two-step affinity purification method. Associated proteins were identified by mass spectrometry and confirmed by immunoblot, with the following results: First, the FLAG-HA-AF9 preparation contained some of the factors, including AF4, AFF4, P-TEFb, and DOT1, that were earlier reported to be associated with AF9 and ENL (13, 15–17) (Fig. S1A and Fig. S2A). However, previously undescribed interactions with transcription factor IID (TFIID) subunits [TATA box-binding protein (TBP) and TBP-associated factors (TAFs)] and AF10 were also observed. Second, relative to the FLAG-HA-AF9 isolate, the FLAG-HA-AF4 isolate contained overlapping factors that included AFF4, AF9, and P-TEFb, as well as unique factors that included ELL1, ELL3, ELL-associated factor 1 (EAF1), and components of the Mediator complex (Fig. S1B and Fig. S2C). Recent studies have also described the presence of ELL and EAF family members in various AF4–AF9 complexes (14–16). Third, and consistent with the FLAG-HA-AF4 data, the FLAG-HA-ELL isolate contained EAF1, ELL-associated factor 2 (EAF2), AF4, AFF4, and P-TEFb (Fig. S1C and Fig. S2E), but not Mediator. Notably, and in agreement with recent studies (15, 17) and potentially mutually exclusive interactions of some factors with AF9/ENL, the FLAG-HA-AF4 and FLAG-HA-ELL isolates did not contain DOT1. However, and in contrast to two recent studies (15, 16), this analysis failed to identify an association of AF9 or ENL with the ELL-isolated complexes. These differences, and our unique identification of TFIID and Mediator associations with the specific fusion protein complexes, may be attributed to differences in purification methodologies and/or levels of expression of ectopic proteins that favor formation or stabilization of specific complexes. Given the presence of both overlapping factors and some unique factors in any given purified complex, it is evident that the MLL fusion partner proteins may dynamically associate with other distinct/overlapping factors to form distinct (perhaps meta-stable) complexes to regulate transcription.
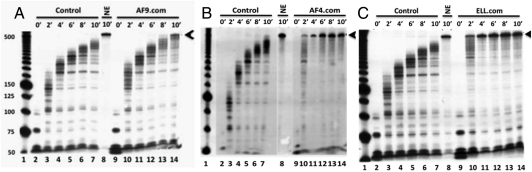
It has been assumed that MLL fusion partners play a role in transcription elongation on the basis of their association with P-TEFb. However, with the exception of studies of purified ELL, EAF1, and EAF2 (7, 19), there has been no direct evidence for this function. To this end, we employed an elongation assay (20) that involves formation of pulse-labeled early elongation complexes followed by analysis of the rate of runoff transcript formation during a chase reaction. Whereas the FLAG-HA-AF9 complex showed only a weak stimulation (Fig. 1A, lanes 10–14 vs. 2–7) when compared to a control reaction, the FLAG-HA-AF4 (Fig. 1B, lane 14 vs. lane 8) and FLAG-HA-ELL (Fig. 1C, lane 14 vs. 8) complexes robustly stimulated the rate of intermediate and runoff transcript formation. Consistent with their association with P-TEFb subunits, all three complexes showed CTD serine 2 phosphorylation activity (Fig. S2 B, D, and F). However, the absence in the elongation assays of factors (DSIF and NELF) that normally elicit a P-TEFb requirement (9) makes it likely that the observed elongation activities of the AF4 and ELL complexes are not due to P-TEFb but rather to the presence of ELL/EAF.
Fig. 1.
Functional elongation analyses of AF9-, AF4-, and ELL-associated protein complexes. (A, B, and C) In vitro transcription elongation kinetic assay on a DNA template using the purified AF9 (A), AF4 (B), and ELL (C) complexes (Fig. S1 A, B, and C, respectively). The initial pulse-labeled RNA transcripts (0′ time point) were chased for the indicated time periods to produce runoff transcripts (arrows).
Thus, our combined analyses show that whereas these various purified complexes share a common functional activity (P-TEFb-mediated CTD phosphorylation) associated with transcription elongation, their differential complements of associated factors clearly mediate differential effects on transcription elongation per se. As might be predicted by the differential disease biology of individual fusion proteins (18), these results imply functional differences for the associated factors and, more importantly, argue against the earlier proposed existence and simultaneous function of all these factors in a single large static macromolecular complex (15, 16).
Biochemical Reconstitution of Functionally Active Complexes Containing AF9, AF4 Family Member Proteins, P-TEFb, and DOT1.
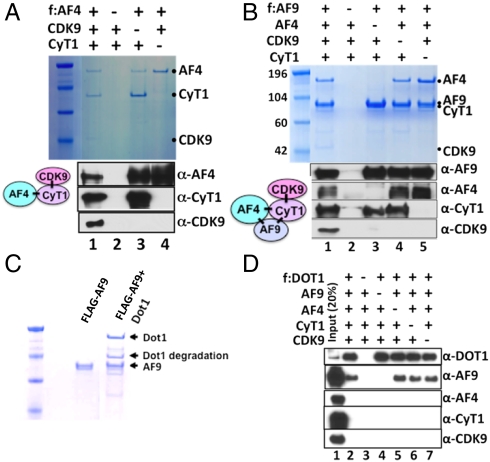
The natural affinity purified complexes described by us and others (14–17) provide important clues for functional studies but, being heterogeneous in nature, do not provide clear mechanistic insight into the specific interactions and functions of individual factors within specific (sub)complexes. For this purpose, we employed baculovirus-based expression and affinity purification methods to reconstitute specific well-defined (sub)complexes. Based on our initial observation that the AF4 family member AFF4 interacts with P-TEFb (8), we coinfected Sf9 cells with baculoviruses expressing FLAG-AF4, cyclinT1, and Cdk9 and subsequently purified a functionally active AF4•P-TEFb complex through the AF4 epitope tag (Fig. 2A and Fig. S3A). The use of various combinations of baculoviruses in this reconstitution analysis revealed that a direct interaction between AF4 and cyclinT1 mediates formation of the trimeric AF4•P-TEFb complex (Fig. 2A, lane 1 vs. lanes 3 and 4). Comparable results were observed with AFF4 (Fig. S3 B and C), confirming both our earlier (8) and more recent (15, 17) observations of an AFF4 association with P-TEFb.
Fig. 2.
Baculovirus reconstitution of higher-order protein complexes involving AF4, P-TEFb, AF9, and DOT1. (A) SDS/PAGE and Coomassie blue staining (Upper) and immunoblot (Lower) analyses of the reconstituted AF4•P-TEFb complex and corresponding subunit interactions. Sf9 cells were coinfected, as indicated, with baculoviruses that express FLAG-AF4 (f:AF4), His-cyclinT1 (CyT1), and His-Cdk9 and complexes were purified on M2 agarose. (B) SDS/PAGE and Coomassie blue staining (Upper) and immunoblot (Lower) analyses of the reconstituted AF9•AF4•P-TEFb complex and corresponding subunit interactions. (C) SDS/PAGE and Coomassie blue staining of the reconstituted AF9•DOT1 complex. (D) Immunoblot analysis indicating that DOT1 and the AF4•P-TEFb complex do not interact directly or assemble into a common complex containing AF9.
Similar reconstitution analyses have also shown that AF4 and AFF4 both interact, individually, with AF9 and P-TEFb to form corresponding higher order AF9•AF4•P-TEFb and AF9•AFF4•P-TEFb complexes (Fig. 2B and Fig. S3D). Importantly, direct subunit interaction analyses show that: (i) unlike AF4 family proteins, AF9 does not directly form a complex with P-TEFb even though it can directly interact with the isolated cyclinT1 subunit (Fig. 2B and Fig. S3D, lane 3), (ii) the stable association of P-TEFb with AF9 is absolutely dependent on the presence of an AF4 family protein (Fig. 2B and Fig. S3D, lane 3 vs. lane 1), thereby showing a direct role of AF4 family proteins in assembly of complexes containing both AF9 and P-TEFb, and (iii) direct interactions of AF9 with AF4 and AFF4 (21) are not enhanced by P-TEFb (Fig. 2B, lanes 1 and 5, and Fig. S3D, lanes 1 and 4). Importantly, further reconstitution analyses using FLAG-tagged AFF4 have shown a mutually exclusive existence of AFF4 and AF4 in the defined higher order complex containing AF9 and P-TEFb (Fig. S3E).
Up-regulation of DOT1-mediated methylation of histone H3K79 is a hallmark of several MLL fusion protein-mediated leukemias (4, 5, 10). Based on the presence of DOT1 in our purified AF9 complex (Fig. S1A), we coexpressed AF9 and DOT1 in Sf9 cells and, as expected (22), were able to purify a stoichiometric AF9•DOT1 complex (Fig. 2C). However, further reconstitution analysis showed that although DOT1 directly associates with AF9, it fails to associate with any component of the AF4•P-TEFb complex in the presence or absence of AF9 (Fig. 2D, lanes 2 and 4–7). This observation, in a well-defined system, is consistent with the finding of DOT1 in mammalian cell-derived complexes purified through AF9 or ENL, but not in complexes purified through AF4, as reported here (Fig. S1 A and B) and recently by others (15, 17). These results reinforce the conclusion that DOT1 and AF4 interactions with AF9 are direct and mutually exclusive. Thus, our biochemically defined reconstitution analyses further show, mechanistically, that AF9 also forms a distinct stoichiometric complex with DOT1 in the absence of other factors.
Biochemical Reconstitution of Functionally Active ELL-Containing Higher Order Complexes and Analysis of Their Direct Interactions with Other Complexes.
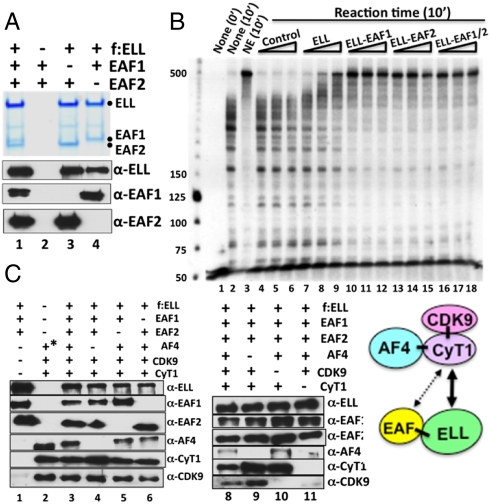
To define the basis for the observed association of ELL with AF4 and P-TEFb (Fig. S1 B and C), we first reconstituted ELL•EAF1/2 complexes (Fig. 3A, lanes 3 and 4) and showed that a direct association with either EAF1 or EAF2 strongly stimulates the transcriptional elongation activity of the recombinant ELL (Fig. 3B, compare lanes 9–11, 12–14, and 13–15 with lanes 6–8), consistent with results of an earlier study (19). Further reconstitution analyses showed that the ELL•EAF1/2 complex directly associates with the AF4•P-TEFb complex, through P-TEFb, to form a multimeric ELL•EAF1/2•AF4•P-TEFb complex (Fig. 3C). Thus, ELL•EAF1/2 interacts directly with P-TEFb in the absence of AF4, but fails to associate with AF4 and Cdk9 in the absence of cyclinT1 (Fig. 3C, lane 11). This result indicates that a direct interaction between cyclinT1 and a component(s) of ELL•EAF1/2 mediates the association of ELL•EAF1/2 with AF4•P-TEFb. In this regard, further analyses showed a direct and specific interaction of cyclinT1, but not AF4 or Cdk9, with both components of the ELL•EAF1/2 complex (Fig. S4A).
Fig. 3.
Baculovirus reconstitution of a functional ELL•EAF1/2 complex and its interaction with the AF4•P-TEFb complex. (A) SDS/PAGE, Coomassie blue staining (Upper) and immunoblot (Lower) analyses of the reconstituted ELL•EAF1/2 complex and corresponding subunit interactions. (B) In vitro transcription elongation assay (as described in Fig. 1) with purified baculovirus-expressed ELL and with purified reconstituted ELL•EAF1/2 complexes (Fig. 3A). (C) Immunoblot analysis of the reconstituted ELL•EAF1/2•AF4•P-TEFb complex and corresponding subunit interactions. The asterisk indicates protein complex purification through FLAG-AF4 in this lane.
In contrast to indications of associations of ELL•EAF1/2 with some natural AF9- and AF4-containing complexes (14–16) (Fig. S1B), our reconstitution analyses failed to show direct association of ELL and EAFs with AF9 either alone or in the presence of AF4 family members (Fig. S4 B and C, compare lane 2 to lane 4). Along with our failure to see ELL in our FLAG-HA-AF9-purified complex (Fig. S1A), these results imply that other factors that are absent in the FLAG-HA-AF9-purified complex and in the direct interaction analyses of Fig. S4 B and C, account for ELL association with the AF9 complexes described by Lin et al. (15) and in Fig. S1B.
Importantly, our observations clearly indicate that contrary to the proposed existence of the common MLL fusion partners in a single large static complex, these factors may form several distinct subcomplexes that interact dynamically with other factors/complexes through different subunits to regulate various stages of transcription (Discussion). Differential regulation of these subcomplexes could also provide a more ready explanation for the fact that distinct disease phenotypes are observed with different fusion proteins (18).
Preferential Recruitment of Common MLL Fusion Partners to an MLL Fusion Target Gene in Vivo.
Toward an understanding of the relevance of in vitro defined biochemical interactions to the transcriptional regulation of target genes by MLL fusion proteins in vivo, we used RS4;11 cells that express MLL–AF4. Comprehensive chromatin immunoprecipitation (ChIP) analyses (Fig. S5) for several fusion partners showed that (i) a preferential recruitment of MLL, AF4, AFF4, AF9, and Cdk9 to the MLL–AF4 target gene HOXA9 (probes C-E) relative to the MYC gene (probes A and B), (ii) these factors are highly enriched in the coding region of the target gene, implying a role in the regulation of transcriptional elongation, (iii) although no significant differences are observed for active histone methylation marks such as H3K4Me3 and H3K79Me2 on MYC vs. HOXA9 genes, the significant absence of all the fusion partners on the active MYC gene suggests that the MLL fusion partners are preferentially recruited to specific MLL fusion target genes during transcriptional activation. Although no direct interaction between AF4 and DOT1 was observed, an increase in H3K79 methylation in the HOXA9 coding region in an MLL–AF4 expressing cell line also suggests a DOT1 recruitment mechanism that is independent of direct interactions with AF4 or MLL–AF4 during transcriptional activation but still dependent upon the functions of MLL–AF4.
An AF9 Domain Involved in Interactions with Other Factors: Implications for MLL–AF9-Mediated Leukemia.
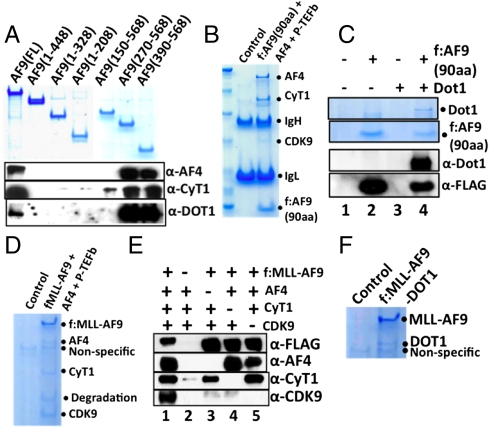
To document the relevance of AF9-dependent formation of defined higher order complexes to MLL fusion protein-mediated transcriptional up-regulation and cell transformation, we first monitored the ability of AF9 proteins with N- and C-terminal deletions (Fig. 4A, Top) to form complexes with coexpressed factors in Sf9 cells. These analyses indicated that a 120-amino acid deletion of the AF9 C terminus completely abolishes independent AF9 interactions with AF4, cyclinT1, and DOT1 (Fig. 4A, compare lanes 2 and 1) and, reciprocally, that a C-terminal 178-amino acid fragment fully retains these AF9 interactions (Fig. 4A, lane 7). Moreover, a C-terminal 90-amino acid fragment of AF9 that is sufficient for MLL–AF9-mediated leukemia (23, 24), formed defined higher order complexes both with AF4•P-TEFb and with DOT1 (Fig. 4 B and C and Fig. S6A). Importantly, in a similar analysis, a natural MLL–AF9 fusion (containing the same C-terminal 90 amino acids) formed MLL–AF9•AF4•P-TEFb and MLL–AF9•DOT1 complexes that were active, respectively, in CTD kinase and H3K79 methylation assays (Fig. 4 D and F, Fig. S6 B–F). These results provide direct evidence for gain of function of the MLL–AF9 fusion protein, through recruitment of the AF4•P-TEFb complex and DOT1, in up-regulating the HOX gene clusters during leukemogenesis. Subsequent subunit interaction analyses showed that AF4 is necessary and sufficient for assembly of a higher order complex containing both MLL–AF9 and P-TEFb (Fig. 4E, compare lanes 1 and 3). This intriguing result establishes a direct role for AF4 family proteins in MLL–AF9-mediated higher order complex assembly that likely underlies one of the mechanisms of transcriptional up-regulation by MLL–AF9 (5).
Fig. 4.
Mapping of an AF9 domain that interacts directly with other factors: Implication for MLL–AF9-mediated higher order complex assembly. (A) SDS/PAGE and Coomassie blue staining showing expression of FLAG-AF9 fragments (Upper) and immunoblot showing interactions of these fragments with coexpressed AF4, cyclinT1, and DOT1 (Lower). (B and C) SDS/PAGE and Coomassie blue staining of complexes reconstituted with AF9 (90-amino acid fragment from 479–568) and either AF4•P-TEFb (B) or DOT1 (C). (D) SDS/PAGE and Coomassie blue staining of the reconstituted MLL–AF9•AF4•P-TEFb complex. (E) Immunoblot analysis of the corresponding subunit interactions in the MLL–AF9•AF4•P-TEFb complex. (F) SDS/PAGE Coomassie blue staining of the reconstituted MLL–AF9•DOT1 complex.
Assembly of a Higher Order Complex Involving MLL–AF9 Interactions with the AF4•P-TEFb Complex is Required for MLL–AF9-Mediated Cell Transformation.
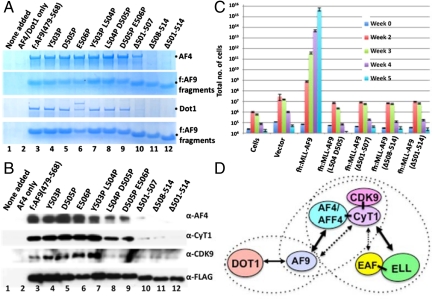
Because an interaction between a minimal AF9 fragment (residues 479–568) and AF4 is essential for MLL–AF9-mediated assembly of a complex containing P-TEFb, we identified mutants within the minimal AF9 fragment that would disrupt the AF4 interaction and, thus, higher order complex formation. This refined deletion analysis showed that C-terminal sequences starting from AF9 residue 500 are important for its interaction with AF4 (Fig. S7A). Two independent analyses of several mutations within this region showed that deletion of residues 508–514 completely abolished, and deletion of residues 501–507 significantly reduced, independent interactions of the minimal AF9 fragment with AF4 (Fig. 5A, Top and Second Rows) and with DOT1 (Fig. 5A, Third and Bottom Rows). These results thus identify a very small AF9 region that is required for both DOT1 and AF4 interactions, and provide an explanation of the failure to observe a DOT1 association with the natural AF4 protein complex by us (Fig. S1B) and others (15, 17) or with the reconstituted and better defined AF4•P-TEFb complex (Fig. 2D). A further reconstitution analysis using these minimal AF9 mutant fragments in conjunction with coexpressed AF4 and P-TEFb showed that these deletion fragments also abolished higher order complex assembly (Fig. 5B, lanes 10–12). Importantly, two of the double point mutants (L504P,D505P and D505P,E506P) retained strong DOT1 interactions in the direct interaction assay (Fig. 5A, lanes 8 and 9), but showed significantly reduced AF4 and cyclinT1 interactions and elimination of Cdk9 association in the higher order complex assembly assay (Fig. 5B).
Fig. 5.
Mutational analysis of AF9: A role for higher order complex assembly in MLL–AF9-mediated transformation. (A) Analysis of direct AF4 and DOT1 interactions with the small AF9 fragment (90 amino acids) and derived mutants. In two independent experiments, Sf9 cells were infected with baculoviruses expressing FLAG-AF9 fragments and either AF4 (Top and Second Rows) or DOT1 (Third and Bottom Rows) and corresponding complexes were purified on M2 agarose and analyzed by SDS/PAGE with Coomassie blue staining. (B) Analysis of higher order complex assembly between the small AF9 fragment or mutant derivatives and the AF4•P-TEFb complex. (C) Hematopoietic transformation assays with retroviruses carrying MLL–AF9 or the indicated mutants. (D) Diagram showing possible functionally active higher order complex formations involving AF9, AF4, AFF4, and ELL along with interacting partners DOT1, P-TEFb, EAF1, and EAF2. The dashed ovals represent the stable complexes observed in the reconstitution studies with defined factors. Note that the AF9•DOT1 and AF9•AF4•P-TEFb complexes are mutually exclusive.
To test the functional importance of these higher order interactions in MLL–AF9-mediated transformation potential in vivo, selective mutant AF9 fragments were fused to the N terminus of MLL (amino acids 1–1420). Preliminary control experiments in 293 cells showed equivalent expression and recruitment of these MLL–AF9 proteins to target HOXA9 genes in vivo (Fig. S7 B and C). Transduction of lineage-negative murine progenitor bone marrow cells with retroviruses expressing these fusion proteins showed an up-regulation of HOXA9 expression only by the wild-type MLL–AF9 protein (Fig. S7D) that contains an AF9 fragment capable of forming complexes both with the AF4•P-TEFb complex and with DOT1. Importantly, as shown by the L504P,D505P double mutant that shows a reduced AF4•P-TEFb interaction but a normal DOT1 interaction, the DOT1 interaction alone is not sufficient for HOXA9 up-regulation in the transduced bone marrow cells. Consistent with this observation, transformation analyses, which measure growth potential of the transduced cells in culture, show that the wild-type MLL–AF9 efficiently transforms the lineage-negative bone marrow precursor cells (Fig. 5C), whereas the mutants that fail to form higher order complexes also fail to transform the precursor cells. These results indicate that AF4-mediated higher order complex assembly involving MLL–AF9 and P-TEFb is critical for MLL–AF9-mediated transformation, but leave open the question of whether the DOT1–AF9 interaction is important in this process.
Discussion
As in other studies (Introduction), our affinity-purification of ectopically expressed AF9, AF4, and ELL complexes from mammalian cells have identified variable associations not only with recently described effector proteins (including P-TEFb and DOT1) but also with other protein complexes that include initiation factor TFIID and the Mediator coactivator, thus raising the possibility of dynamic interactions of MLL partners in transcriptional regulation. Direct functions of these complexes in transcription elongation, dependent upon the nature of the associated factors, have been established. Significantly, our analysis of reconstituted, biochemically defined minimal complexes has allowed us to detail direct protein-protein interactions and functions without the possible influence of other factors that are invariably associated with complexes purified from mammalian cells. These more rigorous analyses have established the formation of distinct, well-defined complexes, with overlapping subunits, that may interact dynamically to regulate different steps of transcriptional activation. Finally, using MLL–AF9 as an example, we have established the relevance of defined protein interactions to MLL fusion protein function in target gene expression and transformation in vivo.
Functionally Distinct Complexes Purified Through Common MLL Fusion Partner Proteins.
Our identification of overlapping and unique associations of fusion partners with natural AF4-, AF9- and ELL-containing protein complexes does not support the general view that these factors necessarily function through a single, static large macromolecular complex. All these complexes exhibited Pol II CTD serine 2 kinase activities, consistent with common presence of P-TEFb, but establishment of their possible roles in transcriptional elongation await appropriate assays with DSIF and NELF. In contrast, the AF4- and ELL-purified complexes, but not the AF9 complex, displayed potent elongation activities that correlated with the presence of ELL and ELL-associated factors (EAFs). These results indicate that the fusion partner complexes indeed do act, minimally, at the stage of elongation—as has been presumed but not heretofore shown. Also of note, the presence of components of general initiation factor TFIID in the AF9-purified complex raises the possibility of a functional interaction of a special AF9- and AF4-containing complex with TFIID at an early (initiation) step in transcription (and possible exclusion of ELL-EAF1/2 association by TFIID). Similarly, the presence of Mediator components in an elongation-active AF4 complex may reflect a functional interaction of these components at a later stage in transcription—possibly in an initiation to elongation transition—and is consistent with previous reports of postinitiation Mediator functions (25).
Common MLL Fusion Partners form Distinct, Well-Defined Complexes with Overlapping Subunits.
The isolation of natural complexes by us and others (12–17) has guided functional tests of associated proteins in cell-based assays and, as reported here, allowed specific in vitro assays where direct effects can be unequivocally established. Of note, however, the combined molecular masses of earlier reported interacting proteins (AF4/AFF4, AF9/ENL, P-TEFb and, in some cases, ELL•EAF1/2) in the presumed single complexes are considerably smaller than the reported sizes of the purified complexes (15, 17), thus indicative of possible aggregation and/or the presence of other identified/unidentified proteins. Based on these considerations, as well as compositional and functional differences in the natural fusion partner complexes analyzed here, we undertook the reconstitution of defined higher order complexes to identify the intrinsic interactions and associated functions of minimal sets of factors.
These analyses have clearly shown that defined subsets of these fusion partners indeed form distinct complexes with overlapping subunits, and that stable minimal complexes can directly associate with other factors to form higher order complexes. Thus, our analyses show, first, that AF4 and AFF4 (individually) interact directly with P-TEFb, through its cyclinT1 subunit, to form active complexes that in turn, through AF4 or AFF4, interact directly with AF9 to form corresponding higher order complexes. Importantly, AF4 and AFF4 were shown to provide the critical links for association of AF9 with P-TEFb and, consistent with an inability to incorporate both into a single complex, to form distinct AF9•AF4•P-TEFb and AF9•AFF4•P-TEFb complexes. These latter results also suggest partially redundant functions between AF4 and AFF4, consistent with the results of an analysis of AF4 null mice (26). However, they are seemingly inconsistent with the report (17) of a minimal heterodimeric complex containing both AF4 and AFF4 in association with P-TEFb and AF9/ENL, unless other factors facilitate such an association and allow coimmunoprecipitation.
Our reconstitution analyses show, second, that P-TEFb provides the linkage for association of the ELL•EAF1/2 complex with the AF4•P-TEFb complex, through an interaction with cyclinT1. A possible functional synergy between these two complexes at different steps in transcription awaits further investigation. Third, and consistent with earlier studies of ENL, our analyses have shown a direct AF9–DOT1 interaction that mediates formation of a defined distinct AF9•DOT1 complex. Importantly, again consistent with other studies (15, 17, 27), the AF9•DOT1 complex failed to show an association with the minimal reconstituted AF4•P-TEFb complex, because both AF4 and DOT1 interact at the same region of AF9 (Fig. 5).
Thus, our reconstitution analyses represent an important first step in elucidating the functions of individual fusion partners and, mechanistically, interactions among defined minimal complexes that may be important for disease outcome.
Role of Fusion Partner Interactions in MLL–AF9-Mediated Transcriptional Up-Regulation and Leukemogenesis.
Our results also provide direct mechanistic evidence for the role(s) of fusion partners in MLL fusion-mediated higher order complex assembly. Thus, we showed that the minimal 90-amino acid AF9 fragment in MLL–AF9 (i) retains an ability to form higher order complexes both with AF4•P-TEFb and with DOT1 and (ii) facilitates assembly of higher order MLL–AF9-containing complexes through direct associations with AF4•P-TEFb and with DOT1. Notably, the role for AF4 family proteins, through direct AF9 interactions, in assembly of an MLL–AF9•AF4•P-TEFb complex was also established. Importantly, however, MLL–AF9-mediated target gene (HOXA9) expression and cell transformation were abolished not only by AF9 mutations that abolished direct interactions with DOT1 and with AF4•P-TEFb but also by AF9 mutations that selectively reduced the AF4•P-TEFb interaction without affecting the DOT1 interaction. Overall, these results establish the importance of an AF9–AF4 interaction both for higher order complex formation and for MLL–AF9-mediated target gene activation and cell transformation. They also indicate that the AF9 domain-mediated DOT1 interaction alone is not sufficient for these MLL–AF9-mediated processes.
A Model for Fusion Partner Protein Interactions Important for Transcriptional Regulation.
Our analyses of both natural (Fig. 1) and reconstituted (Fig. 5D) MLL fusion partner complexes point to dynamic associations of these factors during different stages of transcription.
We speculate, as mentioned in part above that (i) the ELL-deficient, TFIID-containing, AF9-purified complex may reflect a role for AF9 in recruitment of an AF4•P-TEFb complex at or close to the point of initiation to overcome a DSIF/NELF-mediated inhibition of the transition to productive elongation, (ii) the Mediator-associated AF4-purified complex may reflect recruitment/stabilization of an AF4•P-TEFb•ELL•EAF1/2 complex through an interaction with Mediator, and (iii) the AF9-deficient, ELL-purified complex containing AF4/AFF4 and P-TEFb may reflect the loss of AF9 during the transition to a fully productive elongation phase. These models emphasize dynamic (metastable) interactions between more stable subcomplexes, and also raise the possibility that transitions between these different complexes (during different stages of transcription) might necessarily involve transient formation of larger transition complexes. The conservation of MLL fusion partner protein interactions in MLL fusion protein complexes (this and earlier studies) suggests the likely relevance of corresponding complexes to MLL fusion-mediated leukemogenesis.
Materials and Methods
See SI Materials and Methods for details of standard experimental methods for baculovirus-based reconstitution of protein complexes, affinity-purification of FLAG-HA protein complexes, coimmunoprecipitation and mass spectrometric analyses, and transcription elongation, CTD kinase, histone methyltransferase, HoxA9 expression, ChIP and hematopoietic transformation assays.
Supplementary Material
Acknowledgments.
We thank Dr. David Price (University of Iowa) for advice in establishing transcription elongation assays and for P-TEFb expressing baculoviruses. D.B. was supported by a Mary Jose and Henry Kravis Fellowship from The Rockefeller University and by a Career Development Fellowship from the Leukemia and Lymphoma Society. Work in this study was supported by Leukemia and Lymphoma Society Specialized Centers of Research (SCOR) Grant 7132-08, Starr Cancer Consortium Grant I4-A430, and National Institutes of Health Grants CA113872 and CA129325.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111498108/-/DCSupplemental.
References
- 1.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 2.Marschalek R. Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol. 2011;152:141–154. doi: 10.1111/j.1365-2141.2010.08459.x. [DOI] [PubMed] [Google Scholar]
- 3.Huret JL, Dessen P, Bernheim A. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia. 2001;15:987–989. doi: 10.1038/sj.leu.2402135. [DOI] [PubMed] [Google Scholar]
- 4.Krivtsov AV, et al. H3K79 methylation profiles define murine and human MLL–AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy N. Leukaemia: MLL makes friends and influences. Nat Rev Cancer. 2010;10:529. doi: 10.1038/nrc2904. [DOI] [PubMed] [Google Scholar]
- 7.Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 8.Estable MC, et al. MCEF, the newest member of the AF4 family of transcription factors involved in leukemia, is a positive transcription elongation factor-b-associated protein. J Biomed Sci. 2002;9:234–245. doi: 10.1007/BF02256070. [DOI] [PubMed] [Google Scholar]
- 9.Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okada Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 13.Mueller D, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He N, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobhian B, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Perales S, Cano F, Lobato MN, Rabbitts TH. MLL gene fusions in human leukaemias: In vivo modelling to recapitulate these primary tumourigenic events. Int J Hematol. 2008;87:3–9. doi: 10.1007/s12185-007-0001-3. [DOI] [PubMed] [Google Scholar]
- 19.Kong SE, Banks CA, Shilatifard A, Conaway JW, Conaway RC. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc Natl Acad Sci USA. 2005;102:10094–10098. doi: 10.1073/pnas.0503017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 21.Erfurth F, Hemenway CS, de Erkenez AC, Domer PH. MLL fusion partners AF4 and AF9 interact at subnuclear foci. Leukemia. 2004;18:92–102. doi: 10.1038/sj.leu.2403200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a–AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corral J, et al. An Mll–AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: A method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 24.Dobson CL, et al. The mll–AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 1999;18:3564–3574. doi: 10.1093/emboj/18.13.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isnard P, Core N, Naquet P, Djabali M. Altered lymphoid development in mice deficient for the mAF4 proto-oncogene. Blood. 2000;96:705–710. [PubMed] [Google Scholar]
- 27.Mohan M, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.