Abstract
Protein release factor 3 (RF3), a guanosine triphosphatase, binds to ribosome after release of the nascent peptide and promotes dissociation of the class I release factors during the termination of protein synthesis. Here we present the crystal structure of the 70S ribosome with RF3 in the presence of a nonhydrolyzable GTP analogue, guanosine 5′-β,γ-methylenetriphosphate (GDPCP), refined to 3.8 Å resolution. The structure shows that the subunits of the ribosome are rotated relative to each other compared to the canonical state, resulting in a P/E hybrid state for the transfer RNA. The substantial conformational rearrangements in the complex are described and suggest how RF3, by stabilizing the hybrid state of the ribosome, facilitates the dissociation of class I release factors.
Keywords: X-ray crystallography, ribosome structure, translation
Each of the major stages of translation requires specific extrinsic guanine nucleotide binding protein factors that transiently associate with the ribosome. GTP hydrolysis by these protein factors on binding to the ribosome in the appropriate state is essential for their function (1). Among these guanosine triphosphatases (GTPases), release factor 3 (RF3) is involved in the termination of protein synthesis.
Normally, protein synthesis terminates when a stop codon at the end of the coding sequence in mRNA enters the A site of the ribosome. The stop codon is recognized by a class I release factor [release factor 1 (RF1) or release factor 2 (RF2) in bacteria; eRF1 in eukaryotes], which cleaves the nascent protein chain attached to the P-site transfer RNA (tRNA) in the peptidyl transferase center of the ribosome (2). Subsequently, RF3 (eRF3 in eukaryotes), which is also known as a class II release factor, binds to the ribosome and promotes the dissociation of the class I release factor from the ribosome in a GTP-dependent manner (3).
The structures of RF1 (4) or RF2 (5, 6) bound to the 70S ribosome provided major insights into the mechanism of action of class I release factors. The role of RF3 in termination has been more complex. Although eRF3 is essential in eukaryotes, not all bacterial species have a gene for RF3 (7) and the role of GTP hydrolysis in RF3 function is not entirely clear. Cryoelectron microscopy (cryoEM) study of RF3 bound to the ribosome showed that when bound to RF3 with guanosine 5′-β,γ-imidotriphosphate (GDPNP), the ribosomal subunits had rotated relative to one another so that the P-site tRNA was in a P/E hybrid state with its acceptor arm in the E site of the 50S subunit (8).
The hybrid states of tRNA, in which the acceptor arms have moved with respect to the 50S subunit but the anticodon ends have not translocated in the 30S subunit, were shown to be an intermediate in translocation (9). CryoEM studies involving the GTPase factor elongation factor G (EF-G), which promotes translocation have shown directly that this hybrid state is characterized by 6–10 ° of counterclockwise rotation of the small ribosomal subunit with respect to the large ribosomal subunit as viewed from the solvent side of the small subunit (10, 11). Very recently, it has been shown that ribosome recycling factor (RRF) also binds to a rotated ribosome with a tRNA in the P/E hybrid state (12). These studies suggest that the conformation of the ribosome during EF-G binding in translocation, RRF binding during recycling, and RF3 binding in termination all share similarities.
A high-resolution crystal structure of the RF3-ribosome complex has the potential to reveal details of the conformational changes involved and shed light on its role in translational termination. In light of the similarities mentioned above, such a structure may provide insights into the nature of intermediate states during translocation involving EF-G.
The crystallization of GTPase factors bound to the ribosome was suggested intractable until about two years ago, when our laboratory reported the use of a mutant strain of Thermus thermophilus with ribosomes lacking ribosomal protein L9 to crystallize ribosomal complexes with elongation factor Tu (EF-Tu) and EF-G, which are involved in decoding and translocation, respectively (13, 14). We have used this L9-deletion strain of T. thermophilus to crystallize and determine the structure of the ribosome with RF3 bound to a nonhydrolyzable GTP analogue, guanosine 5′-β,γ-methylenetriphosphate (GDPCP), and describe here the structure and its implications for RF3 function. The complex represents the 70S ribosome in the state after the disassociation of class I release factors and before the hydrolysis of GTP by RF3 and its subsequent disassociation from the ribosome.
Results
Crystallization and Structure Determination of the 70S Complexed with mRNA, P-site tRNA, and RF3•GDPCP.
A complex was formed using T. thermophilus ribosomes lacking L9, fMet-tRNA, and mRNA with a start codon in the P site and a UAA stop codon in the A site, and Escherichia coli RF3•GDPCP. Because T. thermophilus lacks a gene for RF3, we used E. coli RF3 complexed with GDPCP, which binds to T. thermophilus 70S ribosome programmed with a P-site tRNA and a stop-codon UAA and RF1 in the A site of the ribosome (Fig. S1). Crystals of this complex were obtained in the space group P21 with cell dimensions of a = 204.7 Å, b = 229.3 Å, c = 307.0 Å, and β = 90.17°, with one ribosome in the asymmetric unit. The structure was solved using molecular replacement (see Materials and Methods), and refined to 3.8 Å resolution [I/σ(I) = 3.03 at 4 Å] (Fig. 1 and Table S1).
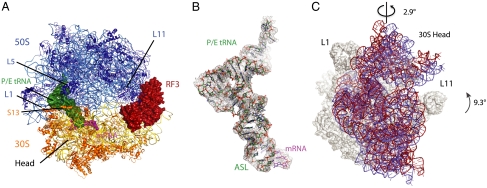
Fig. 1.
Structure of RF3•GDPCP bound to 70S ribosome in the fully rotated conformation. (A) Overview of the complex, with RF3 (firebrick), hybrid P/E tRNA (forest), mRNA (magenta) shown as surfaces, and ribosomal RNAs and proteins shown as cartoons. (B) Unbiased difference Fourier electron density maps with the refined model of P/E tRNA (forest) and mRNA (magenta). (C) Rotation of the 16S rRNA in the 30S subunit relative to the 23S rRNA in the 50S subunit from the classical state (blue) to the fully rotated state (red) showing a 9.3° 16S rRNA body domain rotation and an orthogonal 2.9° of the 16S rRNA head domain swiveling. Ribosomes in the classical and the fully rotated states were aligned using the 23S rRNA of the 50S subunit.
The tRNAfMet is clearly observed in the P/E hybrid state (Fig. 1B). The unbiased difference map also shows the global conformation of RF3 on the ribosome. The density for RF3 was relatively weaker compared to the ribosome, but it was possible to build much of it into density using the isolated crystal structure of RF3 (8) as an initial guide. In combination with the modest resolution of the structure, the relative weaker density of the protein made it difficult to be confident about the conformation of the side chains of RF3, and of the precise registry in regions that had moved relative to the isolated crystal structure (8).
The Ratcheted Form of the Ribosome.
The conformation of the ribosome bound to RF3•GDPCP is in the ratcheted state in which the 30S body is rotated by 9.3° counterclockwise relative to the 50S as viewed from the solvent side of the 30S subunit. In addition, there is an approximately 3° swiveling of the 30S head toward the E site of the 50S subunit (Fig. 1C). This ratcheted conformation of the ribosome is in contrast to structures of the ribosome solved with class I release factors in the state after peptide release for RF1 (4) or RF2 (5, 6), which are in the canonical or classical states similar to those with bound tRNA substrates (15, 16). This movement of the 30S subunit into the ratcheted state facilitates the movement of the CCA-end of the deacylated P-site tRNA into the E site of the 50S subunit. Compared to the 30S subunit, the 50S subunit shows much more localized conformational changes. Upon RF3 binding, regions in the 50S such as the L1 stalk and the L11 stalk change, and the L7/L12 stalk becomes more ordered.
Intersubunit Interactions in the Fully Ratcheted Ribosome.
Interactions between the two ribosomal subunits take place through bridges, which were first characterized by cryoEM (17) and subsequently in molecular detail by crystallography (15, 18, 19). As noted earlier, the rotation of the 30S subunit results in displacements of larger than 20 Å at the periphery of the ribosome (10, 12). Despite this large displacement, most of the bridges are maintained in the fully ratcheted ribosome, especially those that are centrally located rather than at the periphery (Fig. 2 A and B). The 30S subunit rotation is similar to that seen in the recently published RRF-bound 70S structure (12), and accordingly the intersubunit interactions in the two structures are also similar. The dominant component of the 30S subunit interface, h44, makes four critical bridge contacts with the 50S (18), of which the centrally located bridge B3 is close to the pivot point of the intersubunit rotation. The RNA minor-groove interactions in bridge B3 in the fully ratcheted state are the same as in the classical state. The other bridges adjacent to the B3 bridge are maintained through a concerted movement of the RNAs during the subunit rotation. A particularly illustrative example of such a concerted movement is the B2a bridge near the decoding center of the 30S subunit (Fig. 2C). B2a is made by the contacts between the A1913 of H69 in the 23S rRNA and the minor groove of h44 in16S rRNA. This bridge is maintained by an approximately 7 Å compression of H69 to accommodate an approximately 7 Å shift of the h44 toward the E site that results from the 30S rotation.
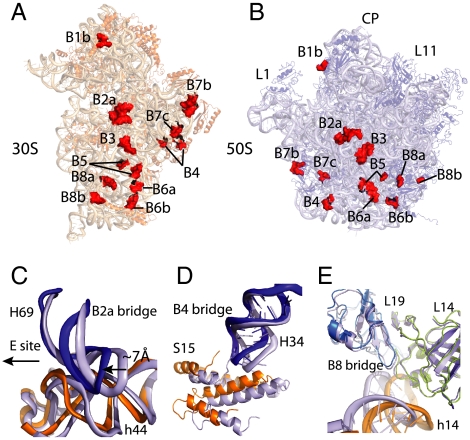
Fig. 2.
Intersubunit interactions in the fully ratcheted 70S ribosome. Interface view of the 30S (A) and the 50S (B) subunit with the intersubunit interactions or bridges (numbered as in refs. 18 and 36). Intersubunit interactions are shown as surfaces in red. The B1a bridge is not shown, as it is not well-ordered in our structure. (C) The B2a bridge. Comparison of the conformations of the B2a bridge in the ribosome between the fully rotated state (colored as 16S rRNA in orange and 23S rRNA in blue) and the classical state (light blue) showing the concerted movement of the interacting elements. (D) The B4 bridge. Comparison of the conformations of the B4 bridge in the ribosome between the fully rotated state (colored as protein S15 in orange and 23S rRNA in blue) and the classical state (light blue) showing the concerted movement of the interacting elements. (E) The B8 bridge. Comparison of the conformation of the B8 bridge in the ribosome between the fully rotated state (colored as 16S rRNA in orange, protein L19 in cyan, and protein L14 in deep olive) and the classical state (light blue) showing closer h14 and L14 interactions.
The B7a bridge, an important RNA-RNA interaction that stabilizes the 30S platform with 23S rRNA in the classical state, is broken in the hybrid state as a result of the 30S rotation. However, the nearby B4 bridge, which is closer to the pivot of the intersubunit rotation and involves hydrophobic interactions between the tip of H34 in 23S rRNA and S15 in 30S subunit, is preserved through the concerted protein-RNA movement (Fig. 2D). Proteins L14 and L19 in 50S subunit make more extensive interactions with h14 in 16S rRNA to help stabilize the rotated ribosome (Fig. 2E).
The 30S head movement in the fully rotated state is accompanied by a large lateral movement of protein S13, which places its central alpha helix directly underneath protein L5 in the 50S subunit (Fig. 1A), as was seen in earlier cryoEM (11) or crystallographic structures of the partially or fully ratcheted states (12, 20). This movement probably results in stronger protein-protein interactions in B1b bridge. The tip of H38 in 23S rRNA (known as A-site finger) that interacts with the 30S head in the classical state (B1a bridge) moves approximately 14 Å toward the 50S E site in the structure.
P/E Hybrid State tRNA.
Coupled to the 30S rotation from the classical to the hybrid state, the anticodon stem loop (ASL) and mRNA move laterally by approximately 9 Å (Fig. 3A). The tRNAfMet is in the P/E hybrid state, with the ASL in the P site of the 30S and the 3′ end of the acceptor arm in the E site of the 50S subunit. Its general conformation is similar to that seen in the recent RRF-bound 70S structure (12). The ASL remains in contact with the mRNA, as well as with the head and platform domains in the P site of the 30S subunit in essentially the same way as a P-site tRNA in the classical state (Fig. 3B) (15). However, it loses its interactions with H69 in the 23S rRNA. The acceptor arm of the P/E tRNAfMet interacts with the 50S E site in a similar way to E-site tRNA in the classical state in which the A76 stacks in between the G2421 and A2422 of 23S rRNA (Fig. 3C) (15). In addition, the G1850 to C1852 region of H68 of 23S RNA moves by approximately 2 Å to pack in the minor groove of the acceptor arm of P/E tRNA.
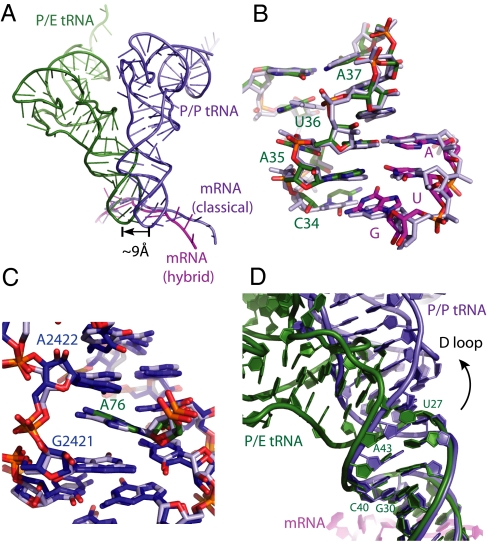
Fig. 3.
The hybrid P/E tRNA (A). Displacement of the ASL of the P/E tRNA (forest) in the hybrid state and P/P tRNA (light blue) in the classical state. The mRNA is colored in magenta in the hybrid state and in light blue in the classical state. Structural alignment is based on the 23S rRNA of the 50S subunit. (B) The conformation of the codon and ASL of the P/E tRNA (P/E tRNA ASL in green and AUG codon in magenta) and the P/P tRNA (Both P/P tRNA ASL and AUG codon are in light blue) in the P site of the 30S subunit. (C) The CCA-end of the tRNA and its interactions with 23S rRNA in the E site of the 50S subunit, showing base stacking of the A76 with residue G2421 and A2422 in the 23S rRNA for both the classical state (light blue) and the hybrid state (CCA-end of the P/E tRNA in green and the 23S rRNA in blue). (D) Superposition of the ASL of the P/P and P/E tRNA shows that (i) the two structures begin to diverge from the base pair G30:C40; and (ii) a bend starts from the base pair U27:A43 in the tRNA body toward the 50S E site. These changes allow the tRNA acceptor arm to swing toward the E site.
The ASL and acceptor arm are locally undistorted. To adopt the P/E conformation, the tRNA body needs to be twisted to allow an approximately 37° swing of the acceptor arm to enable the tRNA to simultaneously bind the 50S E site and the mRNA codon in the 30S P site. Superposing the ASL of the P/P and P/E tRNA shows that the two structures begin to diverge from the base pair G30:C40 (Fig. 3D). A sharp bend of approximately 27° toward the E site with respect to the ASL starts from the base pair U27:A43 between the ASL and the D stem in the tRNA body. These changes facilitate the distortion in the tRNA body that orients its acceptor arm toward the 50S E site.
Conformation of RF3.
Fig. 4A shows the conformation of the RF3•GDPCP in the ribosome. The highly conserved sarcin-ricin loop in the 23S rRNA is close to the nucleotide binding pocket of RF3 (Fig. 4B), as seen previously in the EF-G or EF-Tu bound ribosomal complex structures (14, 21). Protein L6 and L11 near the base of the L12 stalk are close to domain I of the RF3. Domain II of the RF3 interacts with the h5 of the shoulder domain of the 30S subunit. Among the few but important interactions, residue 309–312 in the loop between β14 and β15 in RF3 make contacts with helix h5 at the helix h5 and h15 junction formed via base pair A55:U368 in the 16S rRNA. This interaction is functionally important as a H311A mutation in this loop result in a five to sevenfold decrease of the recycling rate of RF1 and RF2 by RF3 (8).
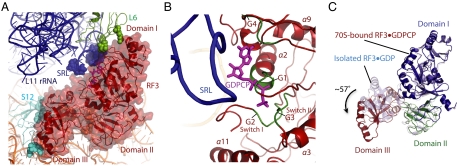
Fig. 4.
Conformation of the ribosome-bound GTPase RF3 in translation. (A) Conformation of RF3•GDPCP in the ribosome, with the interacting residues shown as spheres. (B). The sarcin-ricin loop in the 23S rRNA is in the proximity to the nucleotide binding pocket of RF3. GTP substrate analogue GDPCP is colored in magenta, and the four G motifs in the RF3 are highlighted in forest. (C). Conformational changes of RF3 upon ribosome binding. The ribosome-bound RF3•GDPCP (colored by individual domains as domain I in blue, domain II in green, and domain III in firebrick) adopts a different conformation compared to the isolated RF3•GDP structure (light blue) (8).
A rotation of approximately 57° of domain III relative to the domain I and II in RF3 compared to the isolated RF3•GDP structure (Fig. 4C) places the domain III in close contact to the 30S shoulder, where it interacts with A360 to G362 at the junction of the helix h5 and h15 in 16S rRNA and ribosomal protein S12 with the loop between β24 and α10 helix in RF3. Such a drastic domain rearrangement is very likely to be the result of conformational changes in the switch I and II regions in RF3 when a GTP substrate analogue binds.
The electron density observed for both switch I and II regions in the current structure evidences marked conformational changes underwent in these regions of RF3 going from the GDP-binding state (8) to the GTP-binding state (this study). However, because of the relatively weaker density to the rest of the protein, the A39 to E61 in the switch I region was left out in the current structure.
The overall conformation of RF3 on the ribosome in the GTP state is similar to that seen at low resolution in a cryoEM complex (8). An alignment based on the 23S rRNA backbone of our structure and the previous cryoEM structure [Protein Data Bank (PDB) ID code 2O0F and 3DG5] shows a small difference in the orientation of 70S bound RF3 (Fig. S2A). This difference can be explained by the fact that the 30S in our crystal structure is rotated by an additional approximately 4° compared to the cryoEM structure (Fig. S2 B and C).
Discussion
Comparison with Previously Observed Ratcheted States.
The ratcheting of ribosomal subunits to form hybrid states is a fundamental feature of essential steps of ribosome function such as translocation (9, 10) and recycling (12). Ratcheting involves large structural rearrangements that are made possible by the conformational flexibility of ribosomal RNA. A comparison of our structure with three other recent crystal structures of the ribosome in various stages of ratcheting (12, 20, 22) shows similar intersubunit interactions in the center of the subunit interface. Such conformation similarity suggests that the ratcheting movement in translocation, termination, and recycling share common structural features.
Not surprisingly, the centrally located bridges, which consist of both RNA-RNA and RNA-protein interactions, are maintained in the various stages of ratcheting because they do not have to undergo large relative displacements. These bridges are preserved using a concerted movement that is made possible by the flexibility of the RNA backbone. The centrally located RNA interactions are close in proximity to the binding sites of the tRNA and mRNA (15, 18). They are likely to be important in stabilizing the bound ligands in the ratcheted state. The peripherally located (RNA-protein or protein-protein) interactions, on the other hand, must endure larger conformational changes: Some of them are broken (e.g., the B7a bridge), whereas others are strengthened (e.g., B1b bridge) by newly formed interactions made possible by the 30S rotation.
It is possible, as has been suggested previously, that the swiveling of the head that accompanies the rotation of the body of the 30S subunit in forming the hybrid state of the ribosome plays an important role in translocation (12, 20, 23). In particular, it has been suggested that a preserved interaction between the tRNA anticodon stem loop and the head helps orient the tRNA into the P/E state during translocation (23). However, the precise extent of the swiveling of the 30S head in the different crystal structures varies (Fig. 1C), thus the extent to which the head swiveling contributes to the mechanism of translocation remains unclear. Aligning a 30S subunit (24) (PDB ID code 2X9R) in the classical state with the body of the ratcheted 30S subunit in the structure studied here allows us to dissect the effect of the swiveling of the head alone. It appears that the swiveling of the head allows relatively closer interactions between proteins L5 and S13, and between the A-site finger with S19. These intersubunit interactions between the central protuberance of the 50S and the 30S head domain may be functionally critical to the intersubunit rotation. Interestingly, deletions in protein S13 (25) and truncations in the A-site finger (26) cause the loss of the translocation fidelity, albeit a more rapid translocation. Thus preserving these interactions through a swiveling of the 30S head may allow for a more precise translocation, which is crucial to avoiding frameshifting during normal protein synthesis.
Comparison of the Conformation of RF3 and Other GTPases.
Translational GTPases undergo nucleotide-induced conformational changes and associate with the ribosome in either the hybrid or the classical state along the translational pathway.
Upon GTP binding, RF3 undergoes a conformational change to its GTP form. A comparison of the G-domain conformation of the GDPCP-bound RF3 from this study and the GDP-bound RF3 crystal structure (8) suggests that the switch II loop would have to move to accommodate the gamma phosphate of GTP. In addition, we see weak density indicating at least a partially ordered switch I. Such local changes are accompanied by an approximately 57° rotation of the entire domain III relative to domains I and II. The reorientation of domain III leads to interactions between this domain and the 30S subunit at helix h5 and its junction with helix h15 in 16S rRNA and ribosomal protein S12. Interestingly, mutations at P90, H92, and R452, which decrease the efficiency of RF3 in recycling RF1 or RF2 from the ribosome (8), are all located at positions whose environment would be different in the GDP form as compared to the GTP form of RF3 bound to the ribosome.
Upon GTP hydrolysis, the GDP form of RF3 (8) would not be able to make the extensive interactions between domain III and the 30S subunit. The loss of these interactions would result in lowered affinity for the ribosome and the subsequent release of RF3. Consistent with this notion, earlier biochemical studies (27) and our results (Fig. S1) show that RF3•GDP has very low affinity to the ribosome.
In addition, it is informative to compare the global conformation of the GTPase domains of the translational factors when bound to the ribosome. Two previously reported high-resolution crystal structures of ribosomal complexes with GTPase factors, namely the GDP form of EF-G in the presence fusidic acid (FA) (14), and EF-Tu in the presence of GDPCP (21) both consisted of the ribosome in the classical rather than ratcheted state.
The orientations of the GTPase domains of these factors are similar to each other, but different from that of the current RF3•GDPCP structure. Nevertheless, the regions of the ribosome that each factor makes contact with, e.g., elements of the shoulder of the 30S subunit or the sarcin-ricin loop of the 50S subunit are similar. Thus the orientation of the GTPase domain of these factors appears to depend on whether the particular factor binds to and stabilizes the ratcheted or canonical state of the ribosome. Despite the change in the orientation of the GTPase domain as a whole, each factor positions its GTP-binding pocket close to the sarcin-ricin loop when bound to the state of the ribosome appropriate for that protein factor.
Implications for the Role of RF3 in Translational Termination.
A crucial step of the translation is the termination of protein synthesis, which involves the cleavage and release of the nascent peptide chain from the P-site tRNA when the end of the coding sequence is reached, followed by the disassociation of protein release factors from the ribosome (1). The crystal structure of RF3 trapped in the GTP state on the ribosome provides an opportunity to examine the role of RF3 in accelerating the disassociation of the class I release factors in the translational termination.
Intersubunit rotation results in both a change in the conformation and a movement of over 7 Å of the decoding center. It also results in conformational changes in both the L11 region of the 50S and the head domain of the 30S. Superposing the 50S subunit of the RF2-bound 70S ribosome (24) and the RF3•GDPCP-bound 70S structure here shows steric clashes between domain II and IV of RF2 with helix h18 of the 30S body (Fig. S3) and domain I of RF2 with the L11 region of the 50S. However, there are no major backbone clashes (∼6.5 Å for the closest backbone distance) between the two release factors. The helix α13 in domain III of the RF3 is in close proximity to helix α1 in the domain I of RF2. Such structural arrangement suggests that RF2 cannot bind to the ratcheted form of the ribosome, as also suggested earlier (8). Thus by preferentially stabilizing the ratcheted form of the ribosome, RF3 probably prevents the reassociation of transiently or partially dissociated RF1/2 to the ribosome, thereby shifting the equilibrium toward dissociated RF1/2. This view would also be consistent with the fact that many species of bacteria lack RF3. Presumably, in these species, the dissociation of RF1/2 is not a limiting step. The question of why eRF3 appears to be essential in eukaryotes (7) is not clear, but perhaps it is because the protein has other roles, e.g., in mRNA decay (28).
The release of the RF3 from the ribosome sets the stage for recycling in which the ribosomal subunits are split to allow a new round of translation. Translational recycling involves the binding of RRF and subsequently EF-G to the ribosome (1). The structural similarities between the recent RRF-bound (12) and the RF3•GDPCP-bound 70S ribosome (this study) raise the possibility that RF3 may prepare the way for recycling by leaving the ribosome in the ratcheted state with a hybrid P/E tRNA, which is the conformation seen in the RRF-bound ribosomal complex.
Materials and Methods
Purification of Ribosome, tRNA and RF3 mRNA.
Ribosomes lacking ribosomal protein L9 from a mutant strain of T. thermophilus (14) were purified as previously described (15) from cells grown at the Bioexpression and Fermentation Facility at the University of Georgia. tRNAfmet was purified from E. Coli as described (15). mRNAs were purchased from Dharmacon (Thermo Scientific) with the sequence of GGCAAGGAGGAAAAAAUGUAAUACA (the start codon at the P site and stop codon in the A site are underlined).
N-terminally his-tagged RF3 from E. Coli was overexpressed using the T5 vector pQE-30 (Qiagen), purified on an Ni-nitrilotriacetate affinity column, followed by gel filtration and ion-exchange chromatography (8). The purified protein was dialyzed into a solution of 5 mM Hepes pH 7.5, 10 mM magnesium acetate, 50 mM KCl, 10 mM NH4Cl, 6 mM β-mercaptoethanol, and stored at -80 °C.
Complex Stability and Specificity Assay.
Ribosomes (final concentration 1 μM) were programmed with mRNA (final concentration 2 μM) and deacylated tRNAfMet (final concentration 4 μM) by incubation at 55 °C for 30 min in 5 mM Hepes pH 7.5, 10 mM magnesium acetate, 50 mM KCl, 10 mM NH4Cl, 6 mM β-mercaptoethanol. A second incubation step for 30 min with RF1 (final concentration 4 μM) in the same buffer was carried out for the specificity assay. The programmed ribosome complex was then cooled at a rate of 1 °C/ min to a final temperature of 37 °C. In parallel, 17.1 μM of RF3 and 10.7 mM of β, γ–methyleneguanosine 5′-triphosphate sodium salt (GDPCP, Sigma) was incubated at 37 °C for 30 min before adding to the programmed tRNA-RF1-70S mixture to a final concentration of 4 μM for RF3 and 2.5 mM for GDPCP, and the complex was further incubated at 37 °C for 10 min. For the RF3•GDP binding assay, 50 μM GDP was used for the procedure described above instead of GDPCP.
The stability and specificity of RF3•GDPCP to the 70S complexes were assayed by size-exclusion chromatography on a Superdex™ 200 (10/300) column (Amersham/GE Healthcare) and SDS-PAGE. The presence of a cognate stop codon UAA in the A site was a requirement for RF1 and RF3•GDPCP binding to the ribosomes as shown by Coomassie-stained gels or Sypro ruby-stained gels obtained from SDS-PAGE (Fig. S1).
Complex Formation and Crystallization.
Ribosomes and mRNA at the final concentration of 4.0 μM and 8.0 μM were incubated at 55 °C for 6 min in a buffer containing 5 mM Hepes pH 7.5, 10 mM magnesium acetate, 50 mM KCl, 10 mM NH4Cl, 6 mM β-mercaptoethanol; tRNAfMet was then added into the mixture to a final concentration of 16 μM and the complex was incubated at 55 °C for 30 min. The programmed ribosome complex was then subjected to the decrease of temperature at 1 °C/ min to the final temperature of 37 °C. Separately, RF3 (68.5 μM) and GDPCP (42.8 mM) were incubated together at 37 °C for 30 min before adding to the programmed ribosome mixture to a final concentration of 16 μM for RF3 and 10 mM for GDPCP. The complex was incubated at 37 °C for another 10 min, and left at 19 °C for 20 min before crystallization.
Following the addition of the detergent HEGA-9 to a concentration of 78 mM to the 70S-tRNAfMet-RF3•GDPCP complex, crystals were grown via vapor diffusion in sitting drop trays by adding 3.3 μL reservoir [100 mM MES-KOH pH 6.7, 20 mM KCl, 7.4% (wt/vol) PEG 20 K] to 3 μL of the sample. Crystals with stick or plate morphology grew to full size in two weeks. Crystals were cryoprotected stepwise until reaching a final solution containing 100 mM MES-KOH pH 6.7, 20 mM KCl, 8.1% (wt/vol) PEG 20 K and 25% (wt/vol) PEG 400, and then harvested and frozen by plunging into liquid nitrogen.
Data Collection, Structure Determination, and Refinement.
Data were obtained at 100 K at beamline X06SA of the Swiss Light Source, Villigen. Data were collected in 20–30° wedges on nine separate crystals because of high radiation sensitivity of the crystals, and were integrated and scaled using XDS (29). The crystals are in the space group P21, with unit cell dimensions a = 204.7 Å, b = 229.3 Å, c = 307.0 Å, and β = 90.17°. A molecular replacement solution using the 50S subunit of the 70S ribosome (15) as the first search model followed by the 30S subunit as the second search model was done using Phaser (30). The solution showed one copy of a ribosome in the asymmetric unit.
Refinement was carried out using Phenix (31) and CNS (32) with the following general scheme: rigid body refinement of the 50S and 30S ribosomal subunits; followed by an additional rigid body refinement in which the ribosomal RNA domains, ribosomal proteins, were defined as separate rigid groups. Regions in the 70S ribosome that had changed as a result of the 30S rotation and the ligand binding were rebuilt, followed by the building of mRNA, tRNA, and RF3. Further rigid body refinement that defined mRNA, tRNA, and individual domains in the RF3 as separate rigid groups were carried out, followed by reiterative buildings and refinements using rounds of energy minimization and B-factor refinement. Electron density maps were generated from the CNS program.
Least-square superposition for the structural comparison of the ribosome and the bound ligands in the classical and fully rotated state were carried out in the superpose program in CCP4 suite (33). RNA structures were analyzed using Curves+ (34). All model building was done using Coot (35) and figures were created in PyMOL (www.pymol.org).
Supplementary Material
Acknowledgments.
We thank Dr. Meitian Wang and Dr. Takashi Tomizaki at Swiss Light Source X06SA for assistance with data collection. We thank Dr. Raphael Gasper-Schönenbrücher for a critical discussion on the G-protein structure and function, and Dr. Jade Li for structural refinement. We thank Dr. Kate Godin for the help with the Curve+ program on RNA structure analysis, and Dr. Israel Sanchez and Dr. Cajetan Neubauer for helpful discussions. This work is supported by the Medical Research Council, the Wellcome Trust, the Agouron Institute, and the Louis-Jeantet Foundation. H.J. is a recipient of a Ruth L. Kirschstein postdoctoral fellowship (award number F32GM087083) from the National Institute of General Medical Sciences, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3ZVO and 3ZVP).
See Profile on page 15676.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112185108/-/DCSupplemental.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Korostelev AA. Structural aspects of translation termination on the ribosome. RNA. 2011;17:1409–1421. doi: 10.1261/rna.2733411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. Release factor RF3 in E. coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 5.Weixlbaumer A, et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kisselev LL, Buckingham RH. Translational termination comes of age. Trends Biochem Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 8.Gao H, et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 9.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 10.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 11.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 12.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 16.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank J, et al. A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem Cell Biol. 1995;73:757–765. doi: 10.1139/o95-084. [DOI] [PubMed] [Google Scholar]
- 18.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 19.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 23.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin H, Kelley AC, Loakes D, Ramakrishnan V. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci USA. 2010;107:8593–8598. doi: 10.1073/pnas.1003995107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cukras AR, Green R. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J Mol Biol. 2005;349:47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komoda T, et al. The A-site finger in 23S rRNA acts as a functional attenuator for translocation. J Biol Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 27.Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor rf3. Cell. 2001;107:115–124. doi: 10.1016/s0092-8674(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–200. [Google Scholar]
- 30.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr Sect D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Gabashvili IS, et al. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.