Abstract
In bacteria, genes with related functions often are grouped together in operons and are cotranscribed as a single polycistronic mRNA. In eukaryotes, functionally related genes generally are scattered across the genome. Notable exceptions include gene clusters for catabolic pathways in yeast, synthesis of secondary metabolites in filamentous fungi, and the major histocompatibility complex in animals. Until quite recently it was thought that gene clusters in plants were restricted to tandem duplicates (for example, arrays of leucine-rich repeat disease-resistance genes). However, operon-like clusters of coregulated nonhomologous genes are an emerging theme in plant biology, where they may be involved in the synthesis of certain defense compounds. These clusters are unlikely to have arisen by horizontal gene transfer, and the mechanisms behind their formation are poorly understood. Previously in thale cress (Arabidopsis thaliana) we identified an operon-like gene cluster that is required for the synthesis and modification of the triterpene thalianol. Here we characterize a second operon-like triterpene cluster (the marneral cluster) from A. thaliana, compare the features of these two clusters, and investigate the evolutionary events that have led to cluster formation. We conclude that common mechanisms are likely to underlie the assembly and control of operon-like gene clusters in plants.
Keywords: genome dynamics, metabolic diversification, chromatin
Operons are a familiar feature of prokaryote genomes, where genes belonging to the same functional pathway are assembled into a single transcriptional unit. In eukaryotes, operons are rare [with a few notable exceptions such as in the genomes of nematodes (1)], and functionally related genes usually are scattered across the genome. However, eukaryotic gene order is not as random as it first appeared, and clusters of functionally related but nonhomologous genes now have been identified in the genomes of animals and fungi (2, 3). These clusters include the MHC locus in mammals (4), gene clusters for nutrient use in yeast (5–7), and numerous clusters for diverse secondary metabolic pathways in filamentous fungi (8, 9). Although the genes within these eukaryotic clusters are transcribed independently, these clusters have certain operon-like features (physical clustering and coregulation) (3).
In plants, genes for well-characterized secondary metabolic pathways such as the anthocyanin pathway are unlinked. However, the first gene cluster for a plant secondary metabolic pathway—for the synthesis of cyclic hydroxamic acids—was discovered in maize (Zea mays) in 1997 (10), and since then a secondary metabolic gene cluster for the synthesis of the triterpene avenacin has been discovered in diploid oat (Avena strigosa) (11–14), and two clusters for the synthesis of different diterpenes (momilactones and phytocassanes) have been characterized in rice (Oryza sativa) (15–18). These four clusters from cereals are all required for the synthesis of preformed or stress-induced compounds implicated in plant defense (15, 16, 19, 20). We recently identified an operon-like gene cluster in Arabidopsis, which is required for the synthesis of thalianol-derived triterpenes (the thalianol cluster) (Fig. 1A) (21). The function of the thalianol cluster is not known, but this cluster is highly conserved across different Arabidopsis accessions, suggesting that it is likely to have an important role in ecological interactions. The five plant clusters reported so far are diverse in organization and function, and all appear to have evolved independently (14, 17, 21, 22). There is good evidence indicating that these clusters are not a consequence of horizontal gene transfer from microbes because the origins of the genes within the clusters can be explained most readily by recruitment from plant primary and/or secondary metabolism (14). The clusters therefore are most likely to have formed by gene duplication, neofunctionalization, and genome reorganization. However, the underlying mechanisms are unknown. These clusters represent an emerging paradigm in plant evolutionary biology, providing tantalizing links with adaptive genome plasticity in microbes and animals.
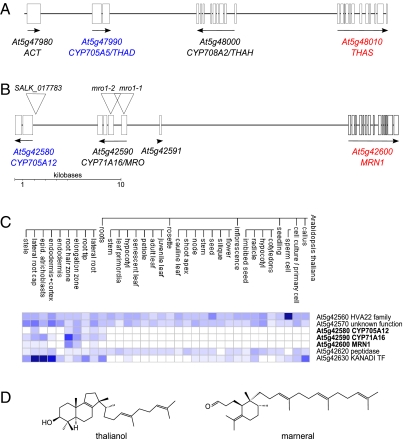
Fig. 1.
A candidate metabolic gene cluster in Arabidopsis. Maps of (A) the thalianol gene cluster (21) and (B) a candidate metabolic gene cluster on Arabidopsis chromosome 5. The boxes represent exons. Genes from the same lineage-specific clades are colored similarly, and T-DNA insertion mutants are indicated in B. At5g42591 is a nonconserved ORF that is predicted to encode a small 35-amino acid peptide of unknown function and for which there are no large-scale expression data. (C) Microarray expression profiles of the genes within the candidate cluster shown in B and four flanking genes [image adapted from Genevestigator (46)]. The putative cluster genes shown in B are indicated in bold. They are expressed only in root tissue, in a pattern similar to the thalianol cluster genes (Fig. S6).The genes flanking the candidate cluster region are not coexpressed and do not have any obvious predicted functions in secondary metabolism. Data are displayed as a heat map (blue, expressed; white, not expressed) scaled to the expression potential of each gene. (D) Structures of thalianol and marneral.
Here we report the discovery and characterization of a second operon-like gene cluster in Arabidopsis thaliana that is required for the synthesis and elaboration of a different triterpene, marneral. Comparison of the thalianol and marneral clusters indicates that although these clusters may have been founded by duplication of an ancestral gene pair, independent evolutionary events have led to the subsequent establishment of the present-day clusters. We propose a model for the sequence of events leading to cluster formation. We further show that the two clusters formed after the α whole-genome duplication event within the Brassicales and are located in dynamic chromosomal regions that are significantly enriched in transposable elements (TEs). Establishment of the coexpression patterns of the genes within the thalianol and marneral clusters appears to have been a multistep process, at least part of which is likely to have occurred after cluster assembly.
Results
The Genes for Marneral Synthesis and Modification Are Clustered.
The thalianol gene cluster in Arabidopsis consists of four contiguous coexpressed genes, At5g48010, At5g48000, At5g47990, and At5g47980, encoding enzymes that together catalyze the synthesis and elaboration of thalianol, a tricyclic triterpene that has been identified only in Arabidopsis (Fig. 1A) (21). These enzymes are the oxidosqualene cyclase (OSC) thalianol synthase (THAS), the cytochrome P450s thalianol hydroxylase (THAH) and thalian-diol desaturase (THAD) (belonging to the CYP85 and CYP71 clans, respectively), and an acyltransferase (ACT). The first committed step in the thalianol pathway is catalyzed by THAS, which belongs to a Brassicaceae-specific clade (clade II) of the OSC family (Fig. S1A). The Arabidopsis genome contains a total of 13 OSC genes, of which six members (including THAS) belong to clade II (21). We and others have noted that several of these other clade II OSC genes also are flanked by coexpressed genes in the Arabidopsis genome (21, 23). These regions therefore may represent functional gene clusters for new triterpene pathways. Here we have focused on the characterization of one of these regions, which contains the OSC gene MRN1 (also known as At5g42600). MRN1 is flanked by two coexpressed cytochrome P450 genes that belong to different P450 families (Fig. 1B). These genes have a root-specific expression pattern similar to that of the thalianol cluster (Fig. 1C) (21).
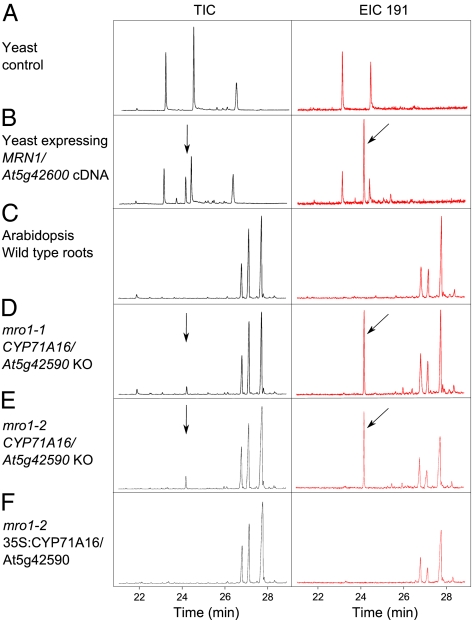
MRN1 encodes marneral synthase, an enzyme that previously had been shown to convert 2,3-oxidosqualene to marneral (an unusual monocyclic triterpene aldehyde) (Fig. 1D) when expressed in yeast (24). Marneral has not been reported in plants. Using GC-MS analysis, we detected marnerol [an alcohol spontaneously produced from marneral, as previously reported (24)] in yeast expressing MRN1 but not in extracts of roots [the tissue where MRN1 is expressed (Fig. 2)] of wild-type Arabidopsis plants. Adjacent to MRN1 is the coexpressed gene CYP71A16 (also known as At5g42590) (Fig. 1B). CYP71A16 is predicted to encode a cytochrome P450 belonging to the widespread CYP71 clan, which is greatly expanded in the Brassicaceae (Fig. S1B). We investigated whether CYP71A16 encodes an enzyme involved in the modification of marneral in planta and thus forms part of a new functional gene cluster in Arabidopsis. Marnerol was not detectable in wild-type root extracts by GC-MS (Fig. 2C). However, it was clearly detectable in the root extracts of two CYP71A16 mutants (mro1-1 and mro1-2; Fig. 2 D and E). Overexpression of CYP71A16 in mro1-2 plants restored the wild-type chemical profile (Fig. 2F). These experiments indicate that CYP71A16 is required for conversion of marneral to a downstream product or products. However, we were unable to detect any marneral derivatives in the roots of wild-type or complemented plants.
Fig. 2.
Detection of MRN1 products in yeast and Arabidopsis. Saponified extracts from yeast and Arabidopsis were analyzed for triterpene content by GC-MS. TIC, total ion chromatogram; EIC 191, extracted ion chromatogram at m/z 191. (A) Yeast empty vector control. (B) Yeast expressing the MRN1 cDNA. (C) Root extracts from wild-type Arabidopsis. (D and E) Root extracts from CYP71A16-knockout lines mro1-1 (D) and mro1-2 (E). (F) Root extracts from mro1-2 overexpressing CYP71A16. Data are representative of at least two separate experiments. The y axis (ion count) of each chromatogram is scaled to the highest peak. Arrows show peaks representing trimethylsilylated marnerol. Unlabeled peaks are trimethylsilylated sterols.
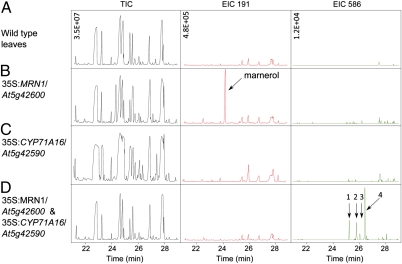
To identify the products of marneral modification by CYP71A16, we analyzed leaf extracts from plants overexpressing MRN1, CYP71A6, or both enzymes (Fig. 3). Plants overexpressing MRN1 and CYP71A16 lack the marnerol peak present in plants overexpressing MRN1 alone and accumulate seven new compounds that have ionization spectra consistent with modified forms of marneral. The four most abundant compounds are likely to represent isomers of hydroxylated desaturated marnerol (Fig. 3 and Fig. S2). These results indicate that CYP71A16 is primarily a marneral oxidase (hereafter referred to as “MRO”) and is likely to generate multiple isomers. CYP705A12 (also known as At5g42580) is a cytochrome P450 gene immediately adjacent to MRO (Fig. 1B). The predicted product of CYP705A12 belongs to a different P450 family than CYP71A16 and also is implicated in marneral metabolism, because the gene is coexpressed with MRO and MRN1 (Fig. 1C). CYP705A12 belongs to the same cytochrome P450 family as THAD, which desaturates hydroxy-thalianol (Fig. 1 A and B and Fig. S1C) (18). Together these results indicate that MRN1, MRO, and CYP705A12 are likely to constitute a functional gene cluster involved in the synthesis and elaboration of marneral. Previously we showed that accumulation of thalianol pathway intermediates has detrimental effects on plant growth and development (21). Similarly, plants overexpressing MRN1 or MRN1 and MRO1 together have pronounced dwarf phenotypes (Fig. S3).
Fig. 3.
CYP71A16 modifies the product of MRN1. Neutral extracts from Arabidopsis leaves were analyzed for triterpene content by GC-MS. TIC, total ion chromatogram; EIC 191, extracted ion chromatogram at m/z 191; EIC 586, extracted ion chromatogram at m/z 586. Leaf extracts from wild-type plants (A) or plants overexpressing MRN1 (B), CYP71A16 (C), or both MRN1 and CYP71A16 (D). Overexpression of the two enzymes resulted in the loss of the trimethylsilyl marnerol peak and the appearance of four peaks (labeled 1–4) that have ionization spectra consistent with desaturated hydroxy-marnerol. These peaks were identified using chromatogram analysis software and cannot be seen in the complex TIC. An additional three peaks of lower abundance could be detected. Ionization spectra for individual compounds are shown in Fig. S2. The plants overexpressing MRN1 and CYP71A16 presented an extreme dwarfing phenotype indicating that the products of marneral modification by CYP71A16 may inhibit plant growth and development (Fig. S3). Data are representative of at least two separate experiments. The y axis (ion count) of each column of chromatograms is to the same scale, indicated in the top left corners in A.
The Marneral and Thalianol Clusters Have both Common and Independent Features.
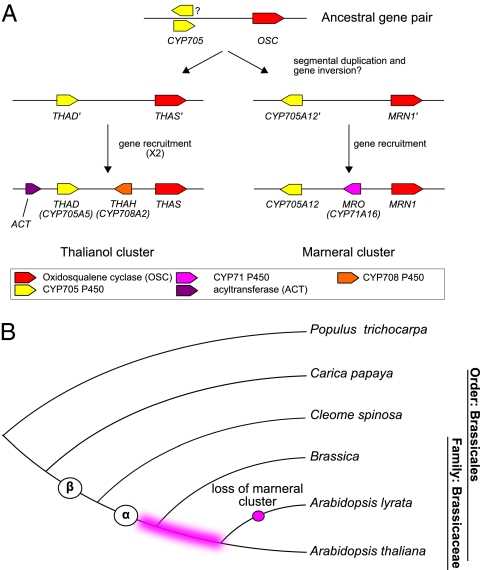
MRN1 and CYP705A12 encode enzymes belonging to the same Brassicaceae-specific clades as THAS and THAD, respectively (Fig. 1 A and B and Fig. S1). The relationship between these gene pairs suggests that the cluster regions may have formed after the segmental duplication (SD) of an ancestral cluster region. However, we note that the CYP705A12 and THAD genes are in opposite orientations, indicating that if the cluster regions formed after SD of an ancestral cluster region, a gene-inversion event must have followed the SD. Phylogenetic analysis indicates that MRN1 and CYP705A12 are both basal within their respective clades relative to their thalianol cluster counterparts THAS and THAD (Fig. S1 A and C), suggesting that these marneral biosynthetic enzymes are more ancient than THAS and THAD. However, MRO and THAH, which catalyze the second steps of the marneral and thalianol pathways, respectively (Fig. 1 A and B), are members of distantly related cytochrome P450 families and share only 27% amino acid identity. If we assume that the THAS/THAD and MRN1/CYP705A12 gene pairs share a common origin, then the most parsimonious explanation for cluster formation is that MRO and THAH were recruited independently to the marneral and thalianol cluster regions after the duplication of an ancestral OSC/ CYP705 gene-containing region (Fig. 4A). The thalianol cluster also differs from the marneral cluster in that it contains the ACT gene At5g47980 (Fig. 1A), which must have been introduced later (Fig. 4A). The sequence of events proposed in Fig. 4A assumes that the two clusters were founded by duplication of an ancestral OSC/ CYP705 gene pair. It is, of course, also possible that the THAS/THAD and MRN1/CYP705A12 gene pairs have evolved independently. In either case, it is clear that the thalianol and marneral clusters are not simply products of whole-scale cluster duplication and that independent evolutionary events have led to the establishment of the present-day clusters.
Fig. 4.
(A) Proposed scheme for formation of the thalianol and marneral clusters, based on the assumption that the two clusters were founded by duplication of an ancestral OSC/CYP705 gene pair. (B) Timing of cluster assembly. The evolutionary tree highlights the period over which the lineage-specific OSC and P450 clades arose and the thalianol and marneral clusters formed (pink shaded region). The α and β whole-genome duplication events are indicated as circles.
The Marneral and Thalianol Clusters Formed After the α Whole-Genome Duplication Event Within the Brassicales and Are Located in Dynamic Chromosomal Regions.
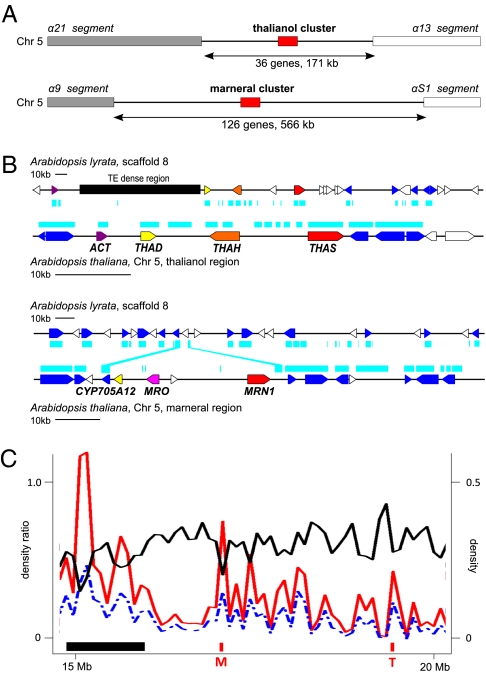
We then compared the chromosomal environments of the marneral and thalianol clusters. The Brassicales have undergone several whole-genome duplication events in their history. The β event occurred after the divergence of papaya (Carica papaya) from other Brassicales (25), whereas the α event is Brassicaceae specific and dates to ∼23–43 Mya (Fig. 4B) (26, 27). Intriguingly, the thalianol and marneral cluster each lies at the center of an island between α duplication segments (Fig. 5A). Our phylogenetic analysis indicates that the expansion of the relevant lineage-specific OSC and P450 clades occurred after the α event (Fig. S1). The thalianol cluster is represented in Arabidopsis lyrata (Fig. 5B), suggesting that clade expansion and cluster assembly already had taken place in the common ancestor of these species. Estimates of the timing of divergence of A. thaliana and A. lyrata range from 3–9 Mya (28) to more recent estimates that suggest a considerably earlier divergence date (17.9 ± 4.8 Mya) (29). However, the A. lyrata cluster differs from the A. thaliana cluster in having an insertion in the ACT gene and an intervening 100-kb TE-dense region between this gene and the next cluster gene (30) (Fig. 5B). In contrast, the marneral cluster appears to have been lost from A. lyrata (Fig. 5B). Deletion is the most likely explanation, because MRN1, MRO, and At5g42580, despite their basal position in the Brassicaceae-specific clades, lack close A. lyrata orthologs, whereas the thalianol cluster genes from A. thaliana all have paired orthologs in A. lyrata (Fig. S1).
Fig. 5.
The chromosomal context of the thalianol and marnerol gene clusters. (A) Both the thalianol and marneral clusters are located in islands between chromosomal segments that were duplicated in the α whole-genome duplication event [89% of all genes are contained within α duplication segment pairs (31)]. Apart from the gene clusters, the two junction regions do not share any other duplicated genes or extensive stretches of intergenic homology. (B) Comparative maps of the thalianol and marneral cluster regions in A. thaliana and A. lyrata. Genes are indicated by filled arrowheads; cluster genes are in red, syntenic genes in blue, and nonsyntenic genes in white. Regions of DNA with homology in both species are indicated by blue bars. (C) Distribution of gene density (black line), TE density (blue dashed line), and the TE/gene density ratio (red line) across a section of chromosome 5 that contains both the marneral (M) and thalianol (T) clusters. Centromeric and pericentromeric regions are marked by horizontal black bars. Gene and TE densities were calculated for windows of 100 kb with an overlap of 5 kb. Full chromosome plots are shown in Fig. S5.
Non-α regions are in general gene poor, contain an excess of TEs and pseudogenes, and act as acceptor sites for SDs (31, 32). TEs have properties that could contribute to the assembly of gene clusters. They promote ectopic recombination, and certain classes such as helitrons, which are present in all three clusters, can transduplicate genes (33, 34). We carried out an analysis of TE distribution in A. thaliana and found that the marneral and thalianol clusters have an unusually high TE/gene density ratio in comparison with neighboring euchromatic regions (P = 0.007 and P = 0.013) or other euchromatic intersegment islands (P = 0.027; Fig. 5C). A third region that contains the two clade II OSC genes pentacyclic triterpene synthase 1 (PEN1) and baruol synthase 1 (BARS1) (Figs. S4 and S5) (35, 36) and that may represent a further functional gene cluster also has a high TE/gene density ratio (P = 0.017). TEs typically have short half-lives (up to several million years) and, in agreement, we found that the TE composition of the thalianol cluster is markedly different A. thaliana and the closely related species A. lyrata (Dataset S1). Whatever role TEs may have played in cluster assembly, the majority of present-day TEs invaded after the lyrata-thaliana split and therefore after cluster formation.
Discussion
Previously we reported the discovery and characterization of a gene cluster in A. thaliana that is required for triterpene synthesis (the thalianol pathway) (21). Here we report on a second A. thaliana gene cluster that is required for the synthesis and elaboration of a different triterpene, marneral. The function of marneral-derived triterpenes in A. thaliana is not known, although by analogy with characterized metabolic gene clusters from other species this pathway is implicated in defense (14). Our analysis indicates that the two clusters have undergone independent evolutionary events, although they may have been founded by duplication of an ancestral OSC/CYP705 gene pair (Fig. 4A).
Both the thalianol and marneral clusters are located in dynamic chromosomal regions that are enriched in TEs. The present-day TE enrichment of the cluster regions provides important insights into cluster formation because it indicates that these regions experience a higher rate of SD turnover than neighboring euchromatic regions. The dynamic nature of these regions is highlighted further by the large differences in TE composition between the thalianol cluster in A. thaliana and A. lyrata and also by the absence of the entire marneral cluster from A. lyrata, suggesting major chromosomal rearrangements. These findings point toward a general mechanism underlying functional gene cluster assembly, because other plant gene clusters are positioned similarly in dynamic chromosomal regions. For example, the oat avenacin and the maize cyclic hydroxamic acid clusters are both subtelomeric (10, 11, 14). Subtelomeric regions are well-known hotspots for chromosomal recombination and SD (34, 37). The enhanced turnover of SDs within such regions is likely to provide an evolutionary “playground” that will accelerate the sampling of different gene combinations and thus the assembly and functional optimization of operon-like gene clusters in response to selection pressure. Preservation of secondary metabolic gene clusters within unstable chromosomal regions may be promoted by selection for the ability to produce protective compounds. Disruption of these clusters may lead not only to loss of the pathway end-products but also to accumulation of toxic/bioactive intermediates (21, 38) that may further enhance selection for clustering (14). Our demonstration that A. thaliana plants that accumulate elevated levels of marneral pathway intermediates are dwarfed (Fig. S3) is consistent with this possibility.
The marneral and thalianol cluster genes all belong to lineage-specific multigene clades that most probably were formed by a burst of tandem duplication in an ancestral Brassicaceae (Fig. S1). Many members of these clades still are arranged in tandem duplicate arrays. Genes that are prone to tandem duplication also have been shown to be more prone to ectopic transposition (39). We propose that the expansion of these lineage-specific clades is likely to have generated a supply of donor sequences that have been incorporated into nascent cluster regions. Interestingly, genes from the maize cyclic hydroxamic acid and rice diterpene metabolic gene clusters also appear to have come from clades that have expanded recently through tandem duplication and that, in at least several cases, are also lineage specific (17, 22).
Clustering of functionally related genes will facilitate the coinheritance of favorable combinations of alleles at these multigene loci (14). Clustering also may enable co-ordinate regulation of gene expression at the level of chromatin (3, 6, 8, 9, 14, 21). Consistent with this notion, we recently showed that expression of the avenacin gene cluster in oat is associated with cell type-specific chromatin decondensation (40). The genes within the marneral and thalianol clusters are coordinately expressed, and both clusters have very similar root-specific expression patterns (Fig. S6). The promoter regions of the genes within each cluster share few known regulatory motifs or regions of sequence homology (Dataset S2), and comparisons within and between the two gene clusters also failed to identify regions of significant intergenic homology (Dataset S1). There is, however, good evidence that both these clusters are likely to be regulated at the level of chromatin. As we noted previously for the thalianol cluster (21), the marneral cluster genes are strongly associated with repressive histone H3 lysine 27 trimethylation (H3K27me3), but the immediate flanking genes are not (41). Two chromatin-remodeling proteins, pickle (PKL) and pickle-related 2 (PKR2), act as transcriptional activators of H3K27me3-marked genes in roots (42). PKL binds directly to at least one gene in the thalianol cluster; the expression of three thalianol cluster genes is known to be PKL/PKR2 dependent; and expression of the marneral cluster genes appears to be partially PKL/PKR2 dependent (41).
Because THAH and MRO were recruited to each cluster independently, their H3K27me3 association must have been inherited from the parental genes or acquired after relocation. The former explanation is supported by the marked enrichment in H3K27me3 association for genes encoding members of the Brassicaceae-specific CYP71A and CYP702/708 clades. Compared with a genome average of ∼16%, 82% of the genes in the CYP71A family and 73% of the genes in the are CYP702/708 family are associated with H3K27me3. In contrast, PKL-dependent expression probably was acquired postduplication (and therefore postcluster assembly), because other members of the CYP71A and CYP702/708 clades rarely show PKL-dependent expression. Therefore, establishment of the current coexpression patterns for marneral and thalianol cluster genes appears to have been a multistep process, at least part of which is likely to have occurred after cluster assembly.
In summary, our identification of a second operon-like gene cluster for another metabolic pathway has allowed us to compare and contrast the features of two A. thaliana triterpene gene clusters, the thalianol cluster and the marneral cluster. These two clusters may have originated from a common ancestral OSC/CYP705 gene pair, although we cannot rule out the possibility that the clusters were founded independently. In either case, it is clear that independent evolutionary events have led to the establishment of the present-day clusters and that these clusters are likely to have grown by gene relocation, as has been reported for the trichothecene biosynthetic gene cluster in Fusarium (43). Both the thalianol and marneral clusters are located within dynamic chromosomal regions, contain genes from gene families prone to ectopic transposition, and are in repressive chromatin domains that are likely to facilitate coordinate regulation of gene expression. Our findings suggest that common mechanisms are likely to underlie the assembly and control of plant gene clusters. They also reveal commonalities with other eukaryotes, for example the filamentous fungi, in which gene clusters for secondary metabolism are frequently subtelomeric, TE rich, and regulated by repressive chromatin (44). As we learn more about the generic rules and mechanisms governing the formation, function, maintenance, and dissipation of such eukaryotic gene clusters, we will be able to gain insights into the forces shaping genome architecture and adaptive evolution. It also will be interesting to establish whether plant genomes contain operon-like gene clusters that have functions in processes other than secondary metabolism, because this information will shed light on our understanding of the significance of clustering for multistep processes encoded by coregulated gene ensembles.
Materials and Methods
Plant Materials and Cloning Procedures.
A. thaliana Col-0 was used as wild type for this work. The mro1-1 and mro1-2 mutants were identified from the SALK T-DNA insertion library (http://signal.salk.edu/cgi-bin/tdnaexpress/) and confirmed as knockouts by genotyping and RT-PCR (Fig. S6). A. thaliana MRN1 and MRO overexpression lines were made by transforming plants with constructs containing the full-length cDNAs under the control of the cauliflower mosaic virus 35S constitutive promoter. Full details of plant materials and cloning procedures are provided in SI Materials and Methods.
Triterpene Analysis.
The MRN1 cDNA was expressed in the yeast strain GIL77, and triterpenes were analyzed as previously described (45). Plants were grown and harvested, and the triterpenes were extracted and analyzed as described previously (21), with modifications for the identification of MRO products (details are provided in SI Materials and Methods).
Phylogenetic and Comparative Genome Analysis.
Details about phylogenetic and comparative genome analysis are provided in SI Materials and Methods.
TE Analysis.
Release 9 of the TE annotations for A. thaliana was retrieved from the Arabidopsis Information Resource (TAIR) website (http://www.arabidopsis.org). The A. thaliana interduplication segment coordinates were extracted from Bowers et al. (31). Gene-cluster regions were defined as follows: marneral cluster, Chr5v01212004 17015000–17065000; thalianol cluster, Chr5v01212004 19415000–19465000; arabidiol/baruol cluster, Chr4v01212004 8730000–8820000. For each cluster, we calculated the TE fragment and gene densities along the chromosomes. To test for enrichment, we compared the TE composition of our region of interest to random regions. To ensure a similar euchromatic genomic environment, we defined a 1-Mb window around each cluster from which we randomly extracted 10,000 regions of the same size as the given cluster. For comparisons against interduplication segments we used random windows from euchromatic interduplication segments. We then computed P values by calculating the frequency of random regions with a TE density higher than the TE density observed for the cluster. Similar results were obtained testing for enrichment in TE/gene density ratio, TE density, or TE number.
Supplementary Material
Acknowledgments
We thank Alan Jones for metabolite analysis and the John Innes Centre horticultural staff for plant care. This work was supported by the UK Biotechnological Sciences Research Council (B.F. and A.E.O.), the Engineering and Physical Sciences Research Council (A.E.O.), the Centre National de la Recherche Scientifique (B.F.), a German Research Foundation Fellowship (A.K.), and a Danish Research Agency International Studentship (K.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109273108/-/DCSupplemental.
References
- 1.Blumenthal T, Gleason KS. Caenorhabditis elegans operons: Form and function. Nat Rev Genet. 2003;4:110–118. doi: 10.1038/nrg995. [DOI] [PubMed] [Google Scholar]
- 2.Hurst LD, Pál C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 3.Field B, Osbourn A. Operons. Nat Chem Biol. 2010;6 10.1038/nchembio.359. [Google Scholar]
- 4.Horton R, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 5.Hittinger CT, Rokas A, Carroll SB. Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc Natl Acad Sci USA. 2004;101:14144–14149. doi: 10.1073/pnas.0404319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong S, Wolfe KH. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat Genet. 2005;37:777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]
- 7.Hall C, Dietrich FS. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics. 2007;177:2293–2307. doi: 10.1534/genetics.107.074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmeister D, Keller NP. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 9.Turgeon BG, Bushley KE. Secondary metabolism. In: Borkovich K, Ebbole D, editors. Cellular and Molecular Biology of Filamentous Fungi. Washington, DC: American Society of Microbiology; 2010. pp. 376–395. [Google Scholar]
- 10.Frey M, et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- 11.Qi X, et al. A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA. 2004;101:8233–8238. doi: 10.1073/pnas.0401301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi X, et al. A different function for a member of an ancient and highly conserved cytochrome P450 family: From essential sterols to plant defense. Proc Natl Acad Sci USA. 2006;103:18848–18853. doi: 10.1073/pnas.0607849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mugford ST, et al. A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell. 2009;21:2473–2484. doi: 10.1105/tpc.109.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu H-Y, Wegel E, Osbourn A. From hormones to secondary metabolism: The emergence of metabolic gene clusters in plants. Plant J. 2011;66:66–79. doi: 10.1111/j.1365-313X.2011.04503.x. [DOI] [PubMed] [Google Scholar]
- 15.Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 2004;135:2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimura K, et al. Identification of a biosynthetic gene cluster in rice for momilactones. J Biol Chem. 2007;282:34013–34018. doi: 10.1074/jbc.M703344200. [DOI] [PubMed] [Google Scholar]
- 17.Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell. 2009;21:3315–3325. doi: 10.1105/tpc.108.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Hillwig ML, Peters RJ. CYP99A3: Functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J. 2011;65:87–95. doi: 10.1111/j.1365-313X.2010.04408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE. Compromised disease resistance in saponin-deficient plants. Proc Natl Acad Sci USA. 1999;96:12923–12928. doi: 10.1073/pnas.96.22.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gierl A, Frey M. Evolution of benzoxazinone biosynthesis and indole production in maize. Planta. 2001;213:493–498. doi: 10.1007/s004250100594. [DOI] [PubMed] [Google Scholar]
- 21.Field B, Osbourn AE. Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science. 2008;320:543–547. doi: 10.1126/science.1154990. [DOI] [PubMed] [Google Scholar]
- 22.Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry. 2009;70:1645–1651. doi: 10.1016/j.phytochem.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Ehlting J, et al. An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 2008;8:47. doi: 10.1186/1471-2229-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong Q, Wilson WK, Matsuda SP. An Arabidopsis oxidosqualene cyclase catalyzes iridal skeleton formation by Grob fragmentation. Angew Chem Int Ed Engl. 2006;45:1285–1288. doi: 10.1002/anie.200503420. [DOI] [PubMed] [Google Scholar]
- 25.Ming R, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: Analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol Evol. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 29.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amoutzias G, Van de Peer Y. Together we stand: Genes cluster to coordinate regulation. Dev Cell. 2008;14:640–642. doi: 10.1016/j.devcel.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 32.Freeling M, et al. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 2008;18:1924–1937. doi: 10.1101/gr.081026.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dooner HK, Weil CF. Give-and-take: Interactions between DNA transposons and their host plant genomes. Curr Opin Genet Dev. 2007;17:486–492. doi: 10.1016/j.gde.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Fiston-Lavier AS, Anxolabehere D, Quesneville H. A model of segmental duplication formation in Drosophila melanogaster. Genome Res. 2007;17:1458–1470. doi: 10.1101/gr.6208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang T, et al. A new triterpene synthase from Arabidopsis thaliana produces a tricyclic triterpene with two hydroxyl groups. Org Lett. 2006;8:2835–2838. doi: 10.1021/ol060973p. [DOI] [PubMed] [Google Scholar]
- 36.Lodeiro S, et al. An oxidosqualene cyclase makes numerous products by diverse mechanisms: A challenge to prevailing concepts of triterpene biosynthesis. J Am Chem Soc. 2007;129:11213–11222. doi: 10.1021/ja073133u. [DOI] [PubMed] [Google Scholar]
- 37.Pryde FE, Gorham HC, Louis EJ. Chromosome ends: All the same under their caps. Curr Opin Genet Dev. 1997;7:822–828. doi: 10.1016/s0959-437x(97)80046-9. [DOI] [PubMed] [Google Scholar]
- 38.Mylona P, et al. Sad3 and sad4 are required for saponin biosynthesis and root development in oat. Plant Cell. 2008;20:201–212. doi: 10.1105/tpc.107.056531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodhouse MR, Pedersen B, Freeling M. Transposed genes in Arabidopsis are often associated with flanking repeats. PLoS Genet. 2010;6:e1000949. doi: 10.1371/journal.pgen.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wegel E, Koumproglou R, Shaw P, Osbourn A. Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell. 2009;21:3926–3936. doi: 10.1105/tpc.109.072124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aichinger E, et al. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 2009;5:e1000605. doi: 10.1371/journal.pgen.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proctor RH, McCormick SP, Alexander NJ, Desjardins AE. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol Microbiol. 2009;74:1128–1142. doi: 10.1111/j.1365-2958.2009.06927.x. [DOI] [PubMed] [Google Scholar]
- 44.Palmer JM, Keller NP. Secondary metabolism in fungi: Does chromosomal location matter? Curr Opin Microbiol. 2010;13:431–436. doi: 10.1016/j.mib.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haralampidis K, et al. A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc Natl Acad Sci USA. 2001;98:13431–13436. doi: 10.1073/pnas.231324698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hruz T, et al. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.