Abstract
Acyl-homoserine lactone (AHL) quorum sensing controls gene expression in hundreds of Proteobacteria including a number of plant and animal pathogens. Generally, the AHL receptors are members of a family of related transcription factors, and although they have been targets for development of antivirulence therapeutics there is very little structural information about this class of bacterial receptors. We have determined the structure of the transcription factor, QscR, bound to N-3-oxo-dodecanoyl-homoserine lactone from the opportunistic human pathogen Pseudomonas aeruginosa at a resolution of 2.55 Å. The ligand-bound QscR is a dimer with a unique symmetric “cross-subunit” arrangement containing multiple dimerization interfaces involving both domains of each subunit. The QscR dimer appears poised to bind DNA. Predictions about signal binding and dimerization contacts were supported by studies of mutant QscR proteins in vivo. The acyl chain of the AHL is in close proximity to the dimerization interfaces. Our data are consistent with an allosteric mechanism of signal transmission in the regulation of DNA binding and thus virulence gene expression.
Keywords: , , bacterial communication, X-ray crystallography
Quorum sensing enables bacteria to sense cell-population density through the synthesis, release, and subsequent response to signaling molecules (reviewed in refs. 1 and 2). In Proteobacteria, the quorum sensing signals are often acyl-homoserine lactones (AHLs), which consist of a homoserine lactone ring with an acyl chain (reviewed in refs. 3 and 4). The AHLs can diffuse out of and into cells, and at a threshold concentration they bind to transcription factors that activate specific sets of genes. Generally, the signal synthases are members of the LuxI protein family, and the transcription factors, which respond to AHLs, are members of the LuxR protein family. Signal specificity resides in the variable acyl side chain (reviewed in refs. 5 and 6).
AHL-dependent quorum sensing activates virulence in many plant and animal pathogens, and the LuxR family of AHL receptors has been a target for therapeutic drug development (6–12). Unfortunately, there is a paucity of structural data for members of the LuxR family of AHL-responsive transcription factors (13). Crystal structures of TraR from the plant pathogen Agrobacterium tumefaciens bound to the cognate AHL 3-oxo-octanoyl-homoserine lactone (3OC8-HSL) and DNA-binding site (14, 15), as well as CviR from a human pathogen Chromobacterium violaceum bound to antagonist have been reported (16). Aside from these, the structures of the N-terminal AHL-binding domains of LasR from the opportunistic human pathogen Pseudomonas aeruginosa (11, 17) and SdiA from Escherichia coli (18) have been reported. In the latter two cases, it has not proven possible to obtain structures for the full-length proteins (11, 18).
The structures of TraR bound to 3OC8-HSL (14, 15) confirmed the view from genetic dissection that LuxR homologs are homodimers of two-domain polypeptides with N-terminal ligand-binding domains (LBDs) and a C-terminal DNA-binding domain (DBD) (19, 20). The DBD is a classical helix-turn-helix motif, where each subunit binds to one half of the palindromic 18-bp binding site. Despite the dyad symmetry of the DNA, TraR binds as an asymmetric dimer because of a 90° rotation of the AHL-binding domains relative to the DNA (14, 15). In contrast, the structure of CviR bound to strong or weak antagonists formed a nearly symmetric cross-subunit configuration with the DBD sequestered in a DNA-binding incompetent conformation (16). Although the AHL-binding site in the structure of the LBD of SdiA bound to octanoyl-HSL (C8-HSL) (18) is similar to TraR and CviR (14–16), structures of the LasR LBD bound to 3-oxo-dodecanoyl-homoserine lactone (3OC12-HSL) (11, 17) dimerize and bind AHL differently compared to full-length TraR and CviR (14–16).
We were interested in the LuxR homolog QscR from P. aeruginosa because it has been studied in vitro in some depth and is stable at high concentrations (21, 22). P. aeruginosa has two complete quorum sensing regulatory circuits, LasI and LasR, which produce and respond to 3OC12-HSL, and RhlR and RhlI, which produce and respond to butyryl-homoserine lactone (C4-HSL) (5, 23, 24). QscR is an additional LuxR homolog in P. aeruginosa, which responds to the LasI-generated 3OC12-HSL (21, 22).
Despite the existence of hundreds of LuxR family members and their importance in virulence, there is little information on how the AHL signals are transduced. It has been generally difficult to study full-length receptors in vitro because of their lack of stability in the absence of a ligand and insolubility at concentrations required for structural characterization (11, 25, 26). We have determined the crystal structure of QscR bound to 3OC12-HSL and analyzed its unique features in vivo. Because the subunit architectures of the full-length TraR bound to AHL and DNA and CviR bound to antagonists are vastly different, the structure of QscR bound to an activating ligand in the absence of DNA reveals a unique step in the activation process.
Results
QscR–3OC12-HSL Forms a Symmetric Homodimer with a Cross-Subunit Architecture.
We determined the structure of full-length QscR bound to an AHL using X-ray crystallography. QscR was expressed, purified, and crystallized in the presence of 3OC12-HSL, which is a strong activating ligand (22) with a high binding affinity of 3.1 nM (21). The structure was determined in spacegroup P3121 at a resolution of 2.55 Å using molecular replacement with a model based on TraR (SI Appendix, Table S1) (14, 15). The QscR model was continuous in chain A from Arg5 to the C-terminal amino acid residue Asn237. Chain B had discontinuous electron density, and the overall B factors were elevated relative to chain A. QscR is an approximately symmetrical dimer with a unique configuration of the two domains in each chain (Fig. 1A). The N-terminal LBD is connected by a short linker (aa 165–174) to the DBD. The LBD forms an α-β-α sandwich from beta strands β1–β5 and α-helices α1–α5 with highly conserved residues that are predicted to be required for binding to AHLs. A canonical helix-turn-helix motif forms the DBD. The AHL binds in a pocket formed between α-helices α3–α5 and beta-sheets β1–β5 (Fig. 1B). Although QscR polypeptides have the general structure of the LuxR family consisting of two functional domains, an N-terminal LBD and a C-terminal DBD, the dimer has unique features that have not been seen in any of the LuxR family structures determined to date (13).
Fig. 1.
Structure of QscR bound to 3OC12-HSL. (A) Ribbon representation shows chain A (cyan), chain B (green), 3OC12-HSL (orange), with oxygen atoms in red and the nitrogen atom in blue. The LBD, the DBD, the first visible amino acid, and the C terminus are indicted for each chain. (B) Electron density map surrounding 3OC12-HSL in chain A. The σa-weighted 2Fo-Fc map contoured at 1.5σ is shown in gray, with the underlying model of QscR (cyan).
The QscR homodimer forms a unique nearly symmetric cross-subunit conformation. This configuration has extensive dimerization interfaces, where the LBD of one subunit interacts with the LBD and the DBD of the other subunit (Fig. 1A and SI Appendix, Fig. S1). As a result, both intermolecular and intramolecular contacts have potential functional implications. Symmetrical contacts between the LBDs of chains A and B (LBD-A and LBD-B) about the twofold axis of the structure (Fig. 2A) occur between A-Glu84 and B-Lys121 (2.6 Å), A-Ser147 and B-Ser147 (3.3 Å), and between A-Lys121 and B-Glu84 (2.5 Å) (SI Appendix, Fig. S1). Additional intermolecular interactions include the hydrogen-bonding interactions present between both Arg42 and Arg79 of the LBD of one monomer with Asn237 of the DBD of the opposite monomer (Fig. 2B). These have the potential to affect dimerization and the ability of 3OC12-HSL–QscR to bind DNA.
Fig. 2.
QscR dimerization interactions. (A) Subset of the predicted intermolecular contacts between the LBDs of chain A (cyan) and B (green). Ionic interactions are shown between both Glu84 with Lys121, and Tyr85 forms van der Waal’s contacts with the opposite LBD. This interface is near the acyl-chain region of the AHL-binding pocket (AHL is in tan). (B) Intermolecular interactions at the interface of LBD-A with DBD-B and LBD-B. Each chain is colored as in A. Hydrogen-bonding contacts are shown between Arg42 and Arg79 with the carboxylate group of Asn237. This interface lies near the acyl-chain region of the AHL-binding pocket. (C) Activity assays in E. coli containing the QscR expression vector pJN105Q and the PA1897-lacZ vector pJL101 comparing the wild-type QscR (WT) to four of the substitutions of key residues noted in A and B. Each assay was conducted at three different concentrations of AHL (10, 50, and 250 nM for 3OC12-, C12-, and 3OC14-HSLs and 10, 50, and 250 μM for 3OC6-HSL). Activity of WT QscR in the presence of 250 nM 3OC12-HSL was set as 100%. Values are means of three independent experiments and bars show standard deviations.
To assess whether the unique dimerization interface formed by the QscR LBDs is functionally relevant in vivo, we performed activity assays with substitution mutants that were designed to disrupt the interface. Substitution of Glu84 or Lys121 to alanine results in a dramatic reduction of QscR activity (Fig. 2C). Furthermore, the relative response to different AHLs is similar to wild-type QscR. This observation suggests that the substitutions decrease the overall activity of QscR. It is thought that the dimer form of QscR is more stable relative to the monomer, and that the ability to dimerize is important for the response of QscR to AHLs (21).
The interactions observed between the LBD and the DBD in opposite subunits are also different than those observed in the other LuxR family crystal structures. For functional validation, we performed activity assays in vivo with substitution mutants that were designed to disrupt the interactions of arginine residues in the LBD with the C-terminus of QscR. Fig. 2C shows that substitution of Arg42 or Arg79 to alanine results in a dramatic decrease in QscR activity. These results validate the overall structure observed for the QscR–3OC12-HSL complex. Again, the relative response to different AHLs is similar to the wild-type QscR, which suggests that the specific length of the acyl chain does not influence this interaction significantly. Because of the proximity of both Arg42 and Arg79 to the AHL-binding pocket, the binding of AHLs in the conformation observed in this structure undoubtedly influences this region of the dimerization interface.
The Acyl Chain of 3OC12-HSL Is Buried Within the LBD of QscR.
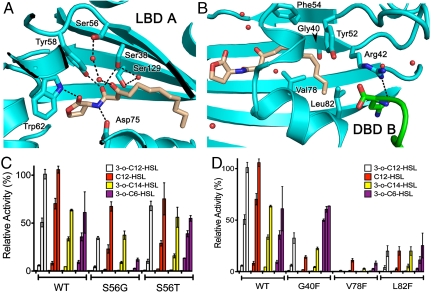
3OC12-HSL makes numerous hydrogen-bonding contacts with QscR (Fig. 3A). These include the direct interactions between the 2-amino group of Trp62 and the carbonyl group of the lactone ring (3.0 Å), between the phenolic hydroxyl group of Tyr58 and the 1-oxo group of the acyl chain (2.5 Å), and between the carboxylate group of Asp75 and the amino group of the AHL (2.6 Å). These interactions are generally conserved among AHL receptors (SI Appendix, Fig. S1). In addition, the nonconserved residue Ser38 forms hydrogen bonds in the AHL-binding pocket with the 1-oxo group and a water molecule. Two other water molecules mediate the interaction between Ser56 and the 3-oxo position of the AHL.
Fig. 3.
AHL-binding pocket interactions. (A) Hydrogen-bonding contacts between 3OC12-HSL and QscR. Water molecules are shown as red spheres. Dashed lines indicate presumed hydrogen bonds. (B) Hydrophobic interactions formed between the aliphatic tail of 3OC12-HSL and QscR. (C and D) Activity assays comparing the wild-type QscR (WT) to five of the substitutions of key residues noted in A and B. Each assay was conducted at the same three different concentrations of AHL shown in Fig. 2. Values are means of three independent experiments, and bars show standard deviations.
AHL-contacting residues of QscR were assessed for their effect on the specificity of AHL recognition by testing QscR with single amino acid substitutions (Fig. 3 A and B). We expected a Ser56 to glycine (S56G) substitution would disturb the water mediated hydrogen bonding with the 3-oxo position of the acyl chain and lessen the preference of QscR for 3-oxo-HSLs. Indeed, the response of the QscR S56G mutant to all 3-oxo-HSL species [3OC12-HSL, 3-oxo-tetradecanoyl-HSL (3OC14-HSL), and 3-oxo-hexanoyl-HSL (3OC6-HSL)] was decreased more than twofold, whereas the response to the unsubstituted AHL C12-HSL was decreased only 50%, and represented the strongest in vivo response for this mutant (Fig. 3C). A serine to threonine substitution (S56T) on the other hand, did not have much influence on specificity, as was expected, because it should still be able to form hydrogen bonds with water molecules in the binding pocket. These results suggest that Ser56 influences selection of 3-oxo versus unsubstituted AHLs.
The acyl chain of the AHL makes numerous hydrophobic and van der Waals interactions with QscR. Contacts include Tyr52, Val78, Leu82, Ile125, and Arg42 (Fig. 3B and SI Appendix, Fig. S1). These residues lie at the distal end of the AHL-binding pocket near the site of the chain A-Arg42-chain B-DBD interaction (Fig. 2B). We suggest that these could influence acyl-chain length recognition. To test this hypothesis, we produced QscR mutants predicted to decrease the space within the acyl-chain–binding pocket and thus increase the response of QscR to AHLs with shorter acyl-chain lengths. Residues Gly40, Val78, and Leu82 point into the acyl-chain–binding pocket, and substitutions with the larger phenylalanine residue had varied effects on the activity and specificity of QscR. The V78F substitution decreased the overall in vivo activity of QscR. In contrast, G40F and L82F substitutions either increased or only slightly decreased the activity of QscR with 3OC6-HSL, whereas the response to AHLs with longer acyl chains was more greatly decreased (Fig. 3D). Strikingly, QscR G40F responded much better to lower concentrations of 3OC6-HSL than the wild-type QscR, which indicates that the specificity of QscR was altered by the addition of a large side chain in the acyl-chain–binding pocket. Thus, residues in the distal portion of the binding pocket are important determinants for QscR signal specificity.
AHL Recognition by QscR and LasR Is Distinct from TraR, SdiA, and CviR.
Next, we examined the AHLs and their corresponding binding pockets in TraR, SdiA, CviR, LasR, and QscR. In contrast to the HSL portion of the AHL-binding pocket, the acyl-chain–binding region is poorly conserved among the AHL receptors. Fig. 4A and SI Appendix, Fig. S2A show that the overall structure of the AHL is similar in the QscR and LasR complexes, but in the SdiA, TraR, and CviR complexes, the AHL adopts a dramatically different conformation. The acyl chain of 3OC12-HSL is similarly embedded in a cavity for QscR and LasR (11, 17) near the region that forms the LBD–DBD dimer interface in QscR (Fig. 2B). In contrast, the acyl chain of AHLs bound to TraR (14, 15), SdiA (18), and CviR (16) extend toward the solvent (SI Appendix, Fig. S2B).
Fig. 4.
Ligand binding and dimerization of QscR compared to other AHL receptors. (A) Ligands in the superimposed QscR (cyan), LasR (purple), TraR (gold), SdiA (blue), and CviR (green). (B) The LBD dimer of QscR (cyan) superimposed on LasR (purple), TraR (gold), and CviR (green). AHLs are not shown. The orientation of the LBDs in QscR and LasR are similar, but for TraR one of the LBDs is rotated nearly 150° relative to QscR, and for CviR, one LBD is rotated 43° in the opposite direction and is shifted by nearly 30 Å.
The AHL-binding pockets differ between the AHL receptors that bind 3OC12-HSL and those that bind AHLs with shorter acyl chains. The shape of the QscR AHL-binding pocket closely resembles that of LasR, but is distinct from TraR, SdiA, and CviR (SI Appendix, Fig. S2B). Measurements of the cavity or pocket volumes indicate that the AHL-binding site of QscR is approximately the same size as LasR (SI Appendix, Table S2). However, the analyses are less reliable for the other AHL receptors, because the AHL extends into the solvent region. To determine how tightly packed the AHL is within the QscR signal-binding pocket, we analyzed the atomic packing density for each AHL–AHL receptor interaction (SI Appendix, Table S2). We found that LasR and QscR have comparable binding surfaces and tighter packing densities than TraR, SdiA, or CviR, which is consistent with the modes of AHL binding by the different LuxR homologs.
The LBD–LBD Dimerization Interfaces of QscR and LasR Are Distinct from Those of TraR, SdiA, and CviR.
Structural comparisons revealed differences between the dimerization interactions of the LBDs of the AHL receptors. The root-mean-square deviations (rmsd) in SI Appendix, Table S3, based on the superposition of the most amino acid residues between each AHL receptor and QscR, gave values between 1.57 Å for LasR and 1.81 Å for SdiA for the majority of the residues in the LBD. However, the CviR (bound to an antagonist) and CviR′ [bound to hexanoyl-HSL (C6-HSL)] matched fewer residues, indicating that CviR and CviR′ are the most structurally diverged from QscR. Despite the similarities of the overall LBD fold, there are major differences in the details of these structures that can be seen in diagrams of their superposition (Fig. 4B and SI Appendix, Fig. S2A). QscR and LasR LBD dimers superimpose quite well with an rmsd of 1.97 Å, and as such have the same general mode of dimerization. In contrast, when one monomer of the TraR or CviR dimer is superimposed on QscR, the other monomer is displaced from the position of the other monomer of QscR. For TraR the monomer is rotated by nearly 150°, and in CviR it is rotated in the opposite direction and translated. Thus, the TraR and CviR LBD–LBD dimerization interfaces are dramatically different from both QscR and LasR and they are also unlike each other.
QscR Appears to Be Poised for DNA Binding.
In the QscR structure the individual DBDs form a dimer similar to that observed for TraR. The QscR and TraR DBDs are highly similar as seen by the rmsd value of 0.73 Å (SI Appendix, Table S3). This is only slightly greater than the expected coordinate error for the comparison of these two structures and stands in contrast to the significant differences between the structures of LBDs. Perhaps the most striking observation in the comparison of these full-length receptors is that the dimerization interface and structures of the DBD dimers of QscR and TraR are nearly superimposable, with an rmsd of 1.5 Å (SI Appendix, Table S3). This is in contrast to the differences observed in LBD–LBD dimerization (Fig. 4B) and orientations of LBDs to the DBDs of TraR relative to QscR, which were too different to measure accurately (SI Appendix, Fig. S3). The similarity of the DBD dimer of QscR to TraR enabled the superposition of the QscR and TraR DBDs to produce a model of a QscR–DNA complex (Fig. 5A). Although the QscR structure was determined in the absence of DNA, little structural arrangement of the protein or DNA would be required for QscR to recognize DNA.
Fig. 5.
Model of DNA recognition by QscR. (A) Superposition of the QscR-AHL complex (cyan) onto the DBD and DNA from the TraR–AHL–DNA complex (in gold). (B) CD spectra of QscR with 3OC12-HSL and QscR with 3OC12-HSL and DNA. (C) Thermal melting monitored by CD of QscR in the presence of 3OC12-HSL, with and without DNA.
We next assessed the effect of DNA binding on the QscR–3OC12-HSL complex. Circular dichroism (CD) revealed the secondary structure and stability of QscR in the absence and presence of DNA. QscR–3OC12-HSL has a largely α-helical spectrum (Fig. 5B), and DNA binding increases the α-helical content by a few percent. DNA also dramatically increased the thermal stability of QscR–3OC12-HSL, which denatured at 50.4 °C without DNA and 64 °C with DNA (Fig. 5C). Limited proteolysis of QscR bound to 3OC12-HSL (SI Appendix, Fig. S4) showed proteolytic susceptibility of the linker between the LBD and DBD. However, addition of DNA protected QscR from this cleavage. The cooperativity observed for QscR binding to DNA (21) is consistent with QscR existing primarily as a monomer at nanomolar concentrations, but at higher concentrations, such as the 1–3 μM used here, QscR exists in a monomer–dimer equilibrium. Therefore, exposure of the linker to the protease would be greater when QscR is in the monomer form and less in the dimer form and bound to the DNA.
Finally, we compared the potential interactions of QscR with TraR with DNA in an effort to gain insights into the molecular basis of sequence-specific DNA recognition. The TraR-binding site is palindrome composed of two 7-bp half sites separated by a 4-bp spacer (14, 15, 27). The TraR- and QscR-binding sites have only two base pairs in common (SI Appendix, Fig. S5) (22). One is at position 6 of the half site. TraR has R206 at the position to contact this base pair, and in QscR the residue is also a basic residue (K208). The first base pair in the spacer region is also the same, and this has no base-specific interactions in the TraR ternary complex structure. Otherwise, the base sequence and the residues that are poised to interact with the DNA are completely different between QscR and TraR.
Discussion
There is very little structural information on members of the large family of LuxR-type proteins involved in AHL quorum sensing (13). Our structure of full-length P. aeruginosa QscR bound to its activating ligand 3OC12-HSL shows unique features as well as those that have been seen in structures reported for these LuxR family members. QscR–3OC12-HSL has a nearly symmetric cross-subunit architecture that is poised to bind to DNA. The LBD dimerization and the AHL-binding pocket are most similar to LasR (11, 17), whereas the dimerization of the DBDs is similar to TraR (14, 15). Of the three available full-length structures, QscR and CviR have cross-subunit structures and TraR does not. Because full-length CviR structures were determined with bound antagonists, it was possible that the cross-subunit structure might be a consequence of antagonist binding. However, this is not the case for QscR and our modeling of QscR–DNA interactions suggests that the DBDs in the cross-subunit structure are poised for DNA binding.
QscR recognizes 3OC12-HSL in almost exactly the same way as LasR (11, 17). However, QscR shows greater promiscuity in its response to AHLs than does LasR in P. aeruginosa (22). Although the binding pocket surface areas, packing densities, and pocket volumes are nearly identical in QscR and LasR, different interactions involving the 3-oxo-position of the acyl chain may be responsible for the more relaxed specificity of QscR relative to LasR. Ser56 participates in a loose network of water-mediated hydrogen bonds with the 3-oxo group. It is equivalent to G54 in LasR, Thr51 in TraR, Val78 in CviR, and Thr61 in SdiA, but none of these residues interact either directly or through water molecules with the 3-oxo position of the AHL. Instead, Arg61 in LasR forms a specific water-mediated interaction with the 3-oxo-position, which would be expected to increase the specificity of LasR for 3-oxo-substituted AHLs. Thus, Ser56 of QscR influences the specificity of QscR for AHLs by allowing both 3-oxo-substituted as well as unsubstituted AHLs to bind to the receptor. The equivalent residues in TraR, SdiA, and CviR pack against the aliphatic portion of the AHL as it leaves the AHL-binding cavity and would not be expected to influence AHL selectivity. Studies of TraR or LuxR AHL specificity mutants also have not revealed a specificity determinant at this position (28, 29). As we continue to learn about determinants of signal specificity from structural studies, it should become possible to predictably alter specific residues of LuxR family members to obtain proteins with desired changes in signal specificity.
Interestingly, the conformation of the AHL in the binding pocket is not conserved among AHL receptors. An “internalized” conformation of the AHL was observed in 3OC12-HSL–bound QscR and LasR LBD (11, 17), but not in AHLs or antagonists with short acyl chains (Fig. 4). The more severely bent conformation results in the potential for interactions at the surface of the AHL receptor, such as those seen in the CviR and CviR′ structures (14–16). In contrast, the internalized conformation allows for the stabilization of the LBD in the regions required for interactions with the DBDs that give rise to the active conformation of the receptor. The residue at the position equivalent to Gly40 may contribute to the mode of AHL binding. Bulkier residues at this position would block access to the far end of this pocket near the dimerization interface and allow AHLs with shorter acyl chains to participate in stabilizing cooperative interactions normally only available to the bulkier ligands. Indeed, this hypothesis is supported by the change in selectivity of QscR observed for the G40F substitution mutant (Fig. 3). Analysis of other AHL receptors will be needed to generalize how much the internalized or surface-exposed modes of AHL binding are determined by acyl-chain length or some other function of the receptor.
The LBD dimerization interface is also not conserved among AHL receptors. QscR and LasR have nearly identical LBD–LBD interactions, which is in contrast to the other AHL receptors (Fig. 4). The conformation of the AHL in the QscR- and LasR-binding pockets may promote the dimerization and binding of QscR to DNA because of allosteric transmission as a result of binding the AHL. Indeed, both Arg42 and Arg79 are in very close proximity to the AHL-binding pocket near the acyl chain of 3OC12-HSL. This may contribute to stabilization of the residues involved in the dimerization interfaces and a conformation that is competent to bind to DNA.
QscR–3OC12-HSL has a nearly symmetric architecture. The TraR–3OC8-HSL–DNA complex is in an active form, but the subunit architecture is asymmetric, with far fewer interactions between the LBD and DBDs, and even these occur within the same subunit. TraR also has a unique mechanism of antiactivation. The antiactivator protein, TraM, inhibits the DNA binding of TraR (30, 31). Structures of the TraR–3OC8-HSL–TraM complex showed that TraM binds between the LBD and DBD in a symmetric configuration, making numerous contacts with DBD, LBD, and the linker between them, which places the DBDs in a position that is incompetent to bind to DNA (13, 32–34). Because TraM can form a complex with TraR while it is simultaneously bound to DNA (34), it is quite unlikely that TraR adopts a cross-subunit architecture, and indeed the linker of TraR probably evolved to bind to TraM and not necessarily to make the extensive interactions with the LBD or DBDs that have been observed in the QscR or CviR structures (16). Recently, a protein similar in function to TraM, QslA, was shown to be an antiactivator of LasR, but the molecular details of the LasR–QslA interaction are unknown (35). Thus, the different architectures observed for full-length LuxR family members potentially reflect specialized features of their physiological activity, such as direct regulation by other factors.
The structures of CviR and CviR′ bound to antagonist signals have different but nearly symmetric cross-subunit architectures; however, the DBDs are sequestered in a conformation that precludes DNA binding (16). It is unclear how crystal packing influences the subunit arrangements, but the structures have been validated, and they likely represent valid states of the R proteins. As such, the cross-subunit architecture provides a convenient way for AHL receptors to switch readily from an inactive to active form. QscR is poised to bind to the DNA and as such represents an active form of AHL receptor. The CviR structures represent antagonized or inactive receptors, with the DBD dimerization helices sequestered at the LBD albeit with a similarly crossed configuration (16). For activation to occur, the DBDs would only have to swing around to permit the C-terminal dimerization helices to interact and give rise to a structure such as QscR. Without a ternary complex structure for either QscR or CviR, it is not possible to know if this crossed-subunit configuration persists when the proteins are bound to DNA. Although it is interesting that the LuxR-like AHL receptors have such great structural variability, our ability to generalize a mechanism for the function of AHL receptors will be limited without further structural studies.
Materials and Methods
QscR Purification.
The QscR–3OC12-HSL complex was expressed and purified from recombinant E. coli as in ref. 21 with modifications described in SI Appendix.
Structure Determination and Analysis.
The details of QscR–3OC12-HSL crystallization are in SI Appendix. The structure was solved at a resolution of 2.5 Å using molecular replacement. Individual domains of TraR [Protein Data Bank (PDB) ID codes 1H0M and 1L3L] and LasR [PDB ID code 3IX3 (17)], which had been trimmed and modified with SCRWL software (36), were used as search models in the PHASER module of the CCP4 software suite (37). The model was built using Coot (38), and refinement was conducted with Refmac in the CCP4 program suite and CNS (39). Group translation/libration/screw refinement was used in the final round, as there were large regions of chain B with higher than average mobility and B factors. Several small sections of the chain and several side chains in chain B were not modeled because of this disorder.
The structure was analyzed for stereochemical and geometrical quality with PROCHECK (40). The rmsd values were calculated by using LSQMAN (41) as an alignment of the regions of QscR chain A, as listed in SI Appendix, Table S3. The values obtained were calculated using the default parameters by imposing a fast force alignment of each molecule followed by an improved fit and global sequence positioning of each fragment; this yielded the best-fit rsmd for Method 2. Models compared to QscR were LasR [PDB ID codes 3IX3 (17) and 2UV0 (11)], TraR [PDB ID codes 1L3L (14) and 1HM0 (15)], SdiA [PDB ID code 2AVX (18)], CviR [PDB ID code 3QP5 (16)], and CviR′ [PDB ID code 3QP1 (16)]. Pocket finder (42) and CASTp (43) were used for cavity volume analyses. Contacts were identified by using CNS (39), and interfacial packing densities were calculated using FADE (44). Figures were made using PyMol, VMD (45), and Photoshop (Adobe).
Assessing QscR Activity in Vivo.
Cultures of E. coli carrying pJL101, which has the QscR-responsive PA1897 promoter fused to lacZ (22), and the constructed mutant pJN105Q plasmids (see SI Appendix) were grown in 50 mM MOPS-Na (pH 7.0)-buffered Luria–Bertani broth containing ampicillin (100 μg/mL) and gentamicin (15 μg/mL) at 37 °C with shaking. At an optical density of 0.4 to 0.5 at 600 nm, L-arabinose was added to 0.2%, and 500 μL of the cultures were transferred to 2-mL wells of 96-well deep-well plates with the indicated amounts of AHLs. The plates were sealed, vortexed, and incubated for 2 h at 37 °C with shaking. To monitor lacZ transcription, β-galactosidase activity was measured using a Galacto-Light Plus kit (Tropix) as described elsewhere (46).
Circular Dichroism.
CD analyses were performed similarly to those described previously (47). QscR was diluted from 293.5 μM to 8 μM in 5 mM Tris-HCl pH 7.8 and 150 mM NaCl. Data were recorded on a Jasco-J815 spectropolarimeter with Peltier temperature control. Spectra were collected at 4 °C. The thermal melting data collection was measured at 222 nm using a temperature range of 4 to 90 °C with a ramp rate of 1.5 °C/ min.
Supplementary Material
Acknowledgments.
We thank Leah Feeley and Dr. Jake Herman for help with crystallization of QscR. We also thank Dr. Jay Nix and the staff at beamline 4.2.2 at the Advanced Light Source, Lawrence Berkeley National Laboratory. The University of Colorado Denver Biomolecular X-ray Crystallography Center was supported in part by funding from the Howard Hughes Medical Institute, the University of Colorado Cancer Center, and National Institutes of Health. This work was supported by National Science Foundation Grant MCB0821220 and National Institutes of Health Grant R56AI081872 (to M.E.A.C.) and National Institutes of Health Grant R01GM59026 (to E.P.G.). K.-I.O. received a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3SZT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112398108/-/DCSupplemental.
References
- 1.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: The LuxR–LuxI family of cell density–responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 3.Salmond GP, Bycroft BW, Stewart GS, Williams P. The bacterial ‘enigma’: Cracking the code of cell–cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 4.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 5.Schuster M, Greenberg EP. LuxR-type proteins in Pseudomonas aeruginosa quorum sensing: Distinct mechanisms with global implications. In: Winans SC, Bassler BL, editors. Chemical Communication Among Bacteria. Washington, DC: AMS Press; 2008. pp. 133–144. [Google Scholar]
- 6.Stevens AM, Queneau Y, Soulere L, von Bodman S, Doutheau A. Mechanisms and synthetic modulators of AHL-dependent gene regulation. Chem Rev. 2011;111:4–27. doi: 10.1021/cr100064s. [DOI] [PubMed] [Google Scholar]
- 7.Mattmann ME, Shipway PM, Heth NJ, Blackwell HE. Potent and selective synthetic modulators of a quorum sensing repressor in Pseudomonas aeruginosa identified from second-generation libraries of N-acylated L-homoserine lactones. Chembiochem. 2011;12:942–949. doi: 10.1002/cbic.201000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu HB, Lee JH, Kim JS, Park S. Inhibitors of the Pseudomonas aeruginosa quorum-sensing regulator, QscR. Biotechnol Bioeng. 2010;106:119–126. doi: 10.1002/bit.22672. [DOI] [PubMed] [Google Scholar]
- 9.Borlee BR, Geske GD, Blackwell HE, Handelsman J. Identification of synthetic inducers and inhibitors of the quorum-sensing regulator LasR in Pseudomonas aeruginosa by high-throughput screening. Appl Environ Microbiol. 2010;76:8255–8258. doi: 10.1128/AEM.00499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattmann ME, et al. Synthetic ligands that activate and inhibit a quorum-sensing regulator in Pseudomonas aeruginosa. Bioorg Med Chem Lett. 2008;18:3072–3075. doi: 10.1016/j.bmcl.2007.11.095. [DOI] [PubMed] [Google Scholar]
- 11.Bottomley MJ, Muraglia E, Bazzo R, Carfi A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem. 2007;282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 12.Muh U, et al. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc Natl Acad Sci USA. 2006;103:16948–16952. doi: 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Churchill ME, Chen L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem Rev. 2011;111:68–85. doi: 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang RG, et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 15.Vannini A, et al. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen G, et al. A strategy for antagonizing quorum sensing. Mol Cell. 2011;42:199–209. doi: 10.1016/j.molcel.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou Y, Nair SK. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem Biol. 2009;16:961–970. doi: 10.1016/j.chembiol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao Y, et al. Structure of the Escherichia coli quorum sensing protein SdiA: Activation of the folding switch by acyl homoserine lactones. J Mol Biol. 2006;355:262–273. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 19.Choi SH, Greenberg EP. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc Natl Acad Sci USA. 1991;88:11115–11119. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanzelka BL, Greenberg EP. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J Bacteriol. 1995;177:815–817. doi: 10.1128/jb.177.3.815-817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oinuma K, Greenberg EP. Acyl-homoserine lactone binding to and stability of the orphan Pseudomonas aeruginosa quorum-sensing signal receptor QscR. J Bacteriol. 2011;193:421–428. doi: 10.1128/JB.01041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Lequette Y, Greenberg EP. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 23.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urbanowski ML, Lostroh CP, Greenberg EP. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J Bacteriol. 2004;186:631–637. doi: 10.1128/JB.186.3.631-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch M, et al. N-acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 2000;19:631–641. doi: 10.1093/emboj/19.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White CE, Winans SC. The quorum-sensing transcription factor TraR decodes its DNA binding site by direct contacts with DNA bases and by detection of DNA flexibility. Mol Microbiol. 2007;64:245–256. doi: 10.1111/j.1365-2958.2007.05647.x. [DOI] [PubMed] [Google Scholar]
- 28.Chai Y, Winans SC. Site-directed mutagenesis of a LuxR-type quorum-sensing transcription factor: Alteration of autoinducer specificity. Mol Microbiol. 2004;51:765–776. doi: 10.1046/j.1365-2958.2003.03857.x. [DOI] [PubMed] [Google Scholar]
- 29.Collins CH, Leadbetter JR, Arnold FH. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat Biotechnol. 2006;24:708–712. doi: 10.1038/nbt1209. [DOI] [PubMed] [Google Scholar]
- 30.Luo ZQ, Qin Y, Farrand SK. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J Biol Chem. 2000;275:7713–7722. doi: 10.1074/jbc.275.11.7713. [DOI] [PubMed] [Google Scholar]
- 31.Fuqua C, Burbea M, Winans SC. Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J Bacteriol. 1995;177:1367–1373. doi: 10.1128/jb.177.5.1367-1373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vannini A, Volpari C, Di Marco S. Crystal structure of the quorum-sensing protein TraM and its interaction with the transcriptional regulator TraR. J Biol Chem. 2004;279:24291–24296. doi: 10.1074/jbc.M401855200. [DOI] [PubMed] [Google Scholar]
- 33.Chen G, Jeffrey PD, Fuqua C, Shi Y, Chen L. Structural basis for antiactivation in bacterial quorum sensing. Proc Natl Acad Sci USA. 2007;104:16474–16479. doi: 10.1073/pnas.0704843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin Y, Su S, Farrand SK. Molecular basis of transcriptional antiactivation. TraM disrupts the TraR-DNA complex through stepwise interactions. J Biol Chem. 2007;282:19979–19991. doi: 10.1074/jbc.M703332200. [DOI] [PubMed] [Google Scholar]
- 35.Seet Q, Zhang LH. Anti-activator QslA defines the quorum sensing threshold and response in Pseudomonas aeruginosa. Mol Microbiol. 2011;80:951–965. doi: 10.1111/j.1365-2958.2011.07622.x. [DOI] [PubMed] [Google Scholar]
- 36.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey S. The CCP4 suite—Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski RA. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 41.Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
- 42.Laurie AT, Jackson RM. Methods for the prediction of protein-ligand binding sites for structure-based drug design and virtual ligand screening. Curr Protein Pept Sci. 2006;7:395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- 43.Dundas J, et al. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell JC, Kerr R, Ten Eyck LF. Rapid atomic density methods for molecular shape characterization. J Mol Graph Model. 2001;19:325–330. doi: 10.1016/s1093-3263(00)00079-6. [DOI] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VMD—Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klass J, et al. The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity. Nucleic Acids Res. 2003;31:2852–2864. doi: 10.1093/nar/gkg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.