Abstract
Polyploidy has been a common process during the evolution of eukaryotes, especially plants, leading to speciation and the evolution of new gene functions. Gene expression levels and patterns can change, and gene silencing can occur in allopolyploids—phenomena sometimes referred to as “transcriptome shock.” Alternative splicing (AS) creates multiple mature mRNAs from a single type of precursor mRNA. Here we examined the evolution of AS patterns after polyploidy, with natural and two resynthesized allotetraploid Brassica napus lines, using RT-PCR and sequencing assays of 82 AS events in duplicated gene pairs (homeologs). Comparing the AS patterns between the two homeologs in natural B. napus revealed that many of the gene pairs show different AS patterns, with a few showing variation that was organ specific or induced by abiotic stress treatments. In the resynthesized allotetraploids, 26–30% of the duplicated genes showed changes in AS compared with the parents, including many cases of AS event loss after polyploidy. Parallel losses of many AS events after allopolyploidy were detected in the two independently resynthesized lines. More changes occurred in parallel between the two lines than changes specific to each line. The PASTICCINO gene showed partitioning of two AS events between the two homeologs in the resynthesized allopolyploids. AS changes after allopolyploidy were much more common than homeolog silencing. Our findings indicate that AS patterns can change rapidly after polyploidy, that many genes are affected, and that AS changes are an important component of the transcriptome shock experienced by new allopolyploids.
Keywords: gene duplication, whole genome duplication, molecular evolution
Polyploidy is ubiquitous in plants, with many plants being evolutionarily recent polyploids, and all angiosperms having one or more ancient polyploidy events in their lineage (1–3). Polyploids can display novel phenotypes that may contribute to their evolutionary success (4). Gene expression levels and patterns in natural and synthetic polyploids have been widely studied to investigate the immediate and long-term effects of polyploidy. Some of the phenomena revealed include nonadditive patterns of gene expression in polyploids compared with their progenitors, divergence in expression levels and patterns between the homeologs, and gene silencing, with the effects being variable in different organ types, in different developmental stages, and in response to abiotic stresses (reviewed in refs. 5–8).
Alternative splicing (AS) is a posttranscriptional mechanism that can regulate gene expression. AS also allows multiple proteins to be produced from the same gene. In addition, AS can cause transcript degradation by including premature stop codons within the coding region that target the transcripts for nonsense-mediated decay, and potentially lower levels of gene expression (9, 10). Premature stop codons introduced by AS can sometimes result in truncated proteins that are functional (11). AS can result in phenotypic changes and impact many important physiological processes, such as photosynthesis and defense responses (9). In plants, it is estimated that ∼33% of genes are alternatively spliced, with intron retention being the most common type of AS (12, 13).
Changes in AS patterns after polyploidy may be an important aspect of the evolution of duplicated genes in polyploid plants. A recent study showed extensive divergence in AS patterns between duplicated genes generated by an ancient polyploidy event in the Arabidopsis lineage (14). However, there has been only one report of differential AS patterns in duplicated genes in natural polyploids: some populations of polyploid Capsella bursa-pastoris show AS in a FLOWERING LOCUS C (FLC) homeolog, and it is associated with flowering time variation (15). Little is known about AS changes at the onset of allopolyploidy and upon interspecific hybridization. New AS events after interspecific hybridization have been shown in the S locus in an interspecific hybrid of Arabidopsis and its derived allopolyploid (16), and for two SR splicing factors in an interspecific Populus hybrid (17).
Here, we studied AS patterns in a set of homeologous genes in a natural allopolyploid, Brassica napus (canola), compared with its progenitor diploid species—B. rapa and B. oleracea—and with resynthesized Brassica allotetraploid S5 lines derived from the same parental species as the natural allopolyploid (18, 19). We assayed AS events by RT-PCR and sequencing a sizable number of genes to (i) evaluate AS patterns between the resynthesized allopolyploids and their parents, (ii) compare AS patterns between natural B. napus and its diploid progenitor species as well as identify differences between the homeologs in B. napus, (iii) detect possible organ and stress-specific AS patterns in B. napus, (iv) compare the patterns of AS between natural and resynthesized B. napus lines, and (v) compare AS patterns between two independently resynthesized allopolyploid lines.
Results
Analysis of Alternative Splicing Events in Homeologs of Natural B. napus.
Allotetraploid B. napus, as well as accessions of its diploid parental species, were used to study the evolution of AS events in homeologous genes. AS was analyzed by performing RT-PCR and resolving the products on agarose gels (see examples in Fig. 1A). Seventy-four genes, including 82 AS events, were assayed (Table 1 and Table S1). In eight genes, two AS events were assayed. An AS event is a single case of AS, such as retention of a single intron. The most common type of AS event studied was intron retention (IR), which is the most frequent type of AS in plants (9). In a polyploid plant, both homeologs could show an AS event, or one of the homeologs might show the AS event in some of its transcripts. To determine if one or both homeologs in B. napus showed a particular AS event, the AS bands from the RT-PCR gels were sequenced and SNPs between the homeologs were evaluated (Fig. 1B). Two different organ types, cotyledons and leaves from seedlings, and two abiotic stress treatments, heat and cold, were used because abiotic stresses are known to affect AS patterns in some genes, and sometimes AS is organ specific (20). Two biological replicates were assayed for each organ and stress condition (see Materials and Methods for details); we did not find any presence/absence variation between the replicates.
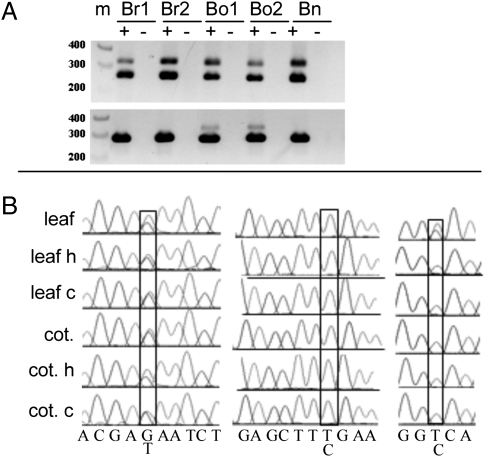
Fig. 1.
RT-PCR and sequencing assays of AS. (A) RT-PCR assays of AS in three Brassica species. Examples of RT-PCR gels showing AS events in B. napus (Bn) compared with two accessions of the diploid progenitor species B. rapa (Br1 and Br2) and B. oleracea (Bo1 and Bo2) in cotyledons. (Upper) Gene 22. (Lower) Gene 16. Plus and minus signs indicate presence or absence of reverse transcriptase. Numbers indicate size markers (M). (B) Sequencing chromatograms showing AS of one or both homeologs in B. napus in two different organ types and under two abiotic stress conditions. Shown are short regions of the chromatograms from direct sequencing of the AS bands from the agarose gels from three example genes. C, cold stress; H, heat stress; cot, cotyledons. (Left) Gene 1 showed two peaks at an SNP site, in each case indicating AS in both homeologs. (Center) Gene 17 showed one peak at an SNP site, in each case indicating AS in only the homeolog derived from B. rapa. (Right) Gene 3 showed two peaks at an SNP site in leaf under normal conditions, indicating AS in both homeologs; however, there was only one peak at the SNP site in all other cases, indicating AS only in the B. rapa homeolog. Multiple SNPs were evaluated for each gene.
Table 1.
AS patterns in homeologs of natural B. napus and the resynthesized allopolyploids
| No. | Gene function/putative function | B.n. | Syn 1 | Syn 2 | No. | Gene function/putative function | B.n. | Syn 1 | Syn 2 |
| 1 | Unknown function | AC | A♦ | AC | 40 | Tetrahydrofolate dehydrogenase | C♦ | C♦ | C♦ |
| 2 | Vacuolar protein | AC | C* | AC | 41 | Homolog of mammalian Bax | AC | AC | AC |
| 3 | Translation initiation factor | A♦ | A♦ | AC | 42 | Ascorbate peroxidase | AC | AC | AC |
| 4 | Vesicle membrane protein | AC | C♦ | C♦ | 43 | Subunit of dehydrogenase | A* | AC | AC |
| 5 | Unknown function | AC | AC | AC | 44 | Disulfade isomerase-like | NIa♦ | A | A |
| 6-1 | Protein TYR phosphatase-like | C♦ | A♦ | A♦ | 45-1 | Unknown function | AC | AC | AC |
| 6-2 | Protein TYR phosphatase-like | C♦ | C♦ | C♦ | 45-2 | Unknown function | AC | AC | AC |
| 7 | Beta carbonic anhydrase 6 | AC | A♦ | AC | 46 | Unknown function | AC | AC | AC |
| 8 | Ascorbate peroxidase | AC | AC | AC | 47 | Nuclear cap binding protein | C♦ | C♦ | C♦ |
| 9 | 20S proteasome subunit | A* | AC | AC | 48 | Glycine-rich protein | AC | AC | AC |
| 10 | Plasma membrane protein | AC | A | AC | 49 | Microsomal signal peptidase | A♦ | A♦ | A♦ |
| 11 | Unknown function | NIa♦ | AC | AC | 50 | DEAD/DEAH box helicase | AC | AC | AC |
| 12 | Unknown function | AC | AC | AC | 51 | DNA damage responses | A* | nd | nd |
| 13 | Unknown function | AC | AC | AC | 52 | Haloacid dehalogenase-like | AC | AC | AC |
| 14 | Ubiquitin-conjugating enzyme | AC | A♦ | A♦ | 53 | Unknown function | AC | AC | A♦ |
| 15 | Unknown function | AC | AC | AC | 54 | Prohibitin 2 | AC | AC | AC |
| 16 | Cold acclimation protein | A | A | A | 55 | Nudix hydrolase homolog | AC | AC | AC |
| 17 | Dormancy-associated protein | A♦ | AC | A♦ | 56 | ERF subfamily | AC | AC | A♦ |
| 18 | CDC2 kinase subfamily | AC | AC | AC | 57 | Dehydrogenase reductase | C* | C♦ | C♦ |
| 19 | Protein kinase AME3 | AC | AC | AC | 58-1 | Inorganic pyrophosphatase | AC | NI | NI |
| 20 | Lammer-type protein kinase | AC | AC | AC | 58-2 | Inorganic pyrophosphatase | AC | NI | NI |
| 21 | Ferritin | AC | A♦ | A♦ | 59 | Chromatin remodeling factor | AC | C♦ | C♦ |
| 22 | Ferredoxin-like superfamily | AC | AC | AC | 60 | Pantothenate kinase | AC | C♦ | C♦ |
| 23 | Poly(A) polymerase | AC | AC | C♦ | 61 | Vacuolar protein sorting | AC | A♦ | A♦ |
| 24 | SR splicing factor | AC | C* | AC | 62-1 | Unknown function | AC | C* | C* |
| 25 | DNA repair | AC | AC | AC | 62-2 | Unknown function | AC | C* | C* |
| 26 | DNA repair | A− | C* | C* | 63 | Peroxiredoxin Q | AC | AC | AC |
| 27 | Glycine-rich protein | AC | AC | AC | 64-1 | Unknown function | AC | AC | AC |
| 28-1 | Splicing factor RSZ33 | A− | A− | A− | 64-2 | Unknown function | C♦ | C♦ | C♦ |
| 28-2 | Splicing factor RSZ33 | A− | A− | A− | 65 | Thioredoxin-like protein | A | A* | A* |
| 29 | Unknown function | A | A | A | 66 | Unknown function | A♦ | C* | AC |
| 30 | Transferase activity | A | AC | AC | 67-1 | Cold acclimation protein | AC | AC | AC |
| 31 | Dihydropyrimidine | C− | C♦ | AC | 67-2 | Cold acclimation protein | AC | AC | AC |
| 32 | Homeodomain-leucine zipper | C♦ | AC | C♦ | 68 | GTP-binding family | C♦ | C♦ | C♦ |
| 33 | Kinase | A | A | A | 69 | 26S proteasome | C | C | C |
| 34 | Ubiquitin related | AC | AC | AC | 70 | Rab GTPase homolog H1e | C♦ | C♦ | C♦ |
| 35 | d-cysteine desulfhydrase | AC | C* | C* | 71 | Isopropylmalate dehydrogenase | AC | AC | AC |
| 36 | 20S proteasome beta subunit | AC | AC | AC | 72 | Chlorophyll b reductase | AC | C♦ | C♦ |
| 37 | Ankyrin repeat family | AC | AC | AC | 73 | Calmodulin | AC | AC | AC |
| 38 | Mitochondrial translocase | A | A | A | 74 | Transmembrane protein | A♦ | A♦ | A♦ |
| 39-1 | Cytochrome b561 | AC | NI | NI | |||||
| 39-2 | Cytochrome b561 | C | C | NIc♦ |
AS status in cotyledons of B. napus (B.n.), resynthesized line 1 (Syn 1), and resynthesized line 2 (Syn 2). A is the homeolog derived from B. rapa, and C is the homeolog derived from B. oleracea. No., gene pair number. AC, AS in A and C homeologs; A, AS in A homeolog only; C, AS in C homeolog only; A♦, change in AS after allopolyploidy/loss of AS in C homeolog; C♦, change in AS after allopolyploidy/loss of AS in A homeolog, AC, change in AS after allopolyploidy/gain of AS in C homeolog; AC, change in AS after allopolyploidy/gain of AS in A and C homeologs; A*, no expression of the C homeolog; C*, no expression of the A homeolog; A−, loss of the C homeolog; C−, loss of the A homeolog; NI, no AS isoform present; NIa♦, no AS isoform, loss of AS in A homeolog; NIc♦, no AS isoform, loss of AS in C homeolog; nd, not determined. Underlining indicates differences in AS or expression among organs or stresses (Table S2).
Forty-five of the AS events (of 82) occurred in the transcripts of both homeologs in a gene pair of B. napus under all growing conditions and in both organ types (Table 1 and Table S2). In contrast, 18 AS events were present in only one homeolog in at least one organ type or under one stress treatment; those included 10 events with only the A homeolog, derived from B. rapa, and eight events with only the C homeolog, derived from B. oleracea. In six cases, AS was present in both of the homeologs in some organ types or stress conditions and in only one of the homeologous genes in other organ types or stress conditions. For example, gene pair 3, a translation initiation factor, showed AS of both homeologous genes in leaves under normal growing conditions; however, only the homeolog derived from B. rapa showed AS in leaves of B. napus under heat and cold stresses and in cotyledons under normal, heat, and cold stress conditions (Fig. 2C). Thus, for a few cases, AS in a particular homeolog was organ specific or stress specific. For genes that showed AS in only one homeolog, the result was further verified by doing RT-PCR with a primer located in the retained intron (Table S3). In this assay only the AS form was amplified and not the fully spliced form, to potentially detect low levels of the AS form in one homeolog that might have been missed by coamplifying the major and minor forms by RT-PCR. Sequencing of those RT-PCR products confirmed the presence of one homeolog in the AS band in each case.
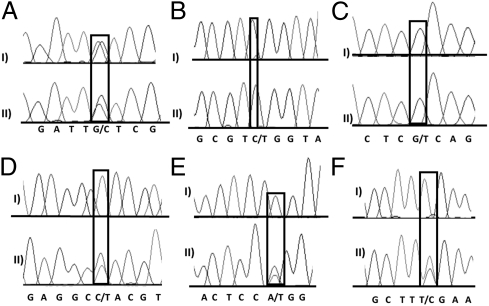
Fig. 2.
Sequencing chromatograms of AS in cotyledons of the two resynthesized allopolyploid lines. Shown are short regions of the chromatograms from direct sequencing of the AS bands from the agarose gels from six example genes. (A) gene 13, (B) gene 29, (C) gene 6-2, (D) gene 10, (E) gene 31, and (F) gene 3. Scoring was done in the same way as in Fig. 1. Multiple SNPs were evaluated for each gene.
In the cases where only one of the homeologs had AS, the fully spliced form was also sequenced to determine whether homeolog silencing occurred. In most cases there was expression of the fully spliced form from both homeologs, indicating that homeolog silencing did not occur. However, in five cases there was homeolog silencing in one or more of the organ types or stress conditions (Table 1, Table S2, and Fig. S1). If only one of the homeologous genes was expressed, then PCR and sequencing of the gene from genomic DNA was done to determine if there was evidence for gene loss or rearrangement that disrupted the gene. One homeolog in each of three gene pairs was not detected (Table 1); therefore, homeolog loss or chromosome rearrangement occurred such that the gene is not present or intact. To evaluate the possibility of recombination between the homeologs, we evaluated SNP sites in the exons surrounding the AS events. In each case, both homeologs were present without recombination between them in the region evaluated for AS. We did not examine homeologous recombination elsewhere in the genes, which therefore could exist in some cases.
Gene pairs with AS in only one of the two homeologs may represent changes in AS patterns after polyploidy. To compare AS patterns in B. napus with models of its diploid progenitor species, B. rapa and B. oleracea, two accessions of each diploid species were used. Among 80 AS events, 69 showed conservation among the two accessions each of B. rapa and B. oleracea (Table S4). In contrast, 11 AS events showed varied presence/absence among the diploid lines. Eight of those events were present in only one homeolog in the polyploid, strongly suggesting that the difference in AS between the homeologs was due to AS absence in the corresponding diploid and inheritance of this pattern in the polyploid. In two cases we found no AS isoform in B. napus (Table 1); these were interpreted as loss of AS in the A homeolog because both accessions of B. rapa showed the AS event. Another case, gene 39-1, may have AS gain in both homeologs in B. napus, because none of the diploid lines showed the event.
When accounting for gene silencing, loss or recombination, and likely inheritance of AS from the diploid progenitors, a total of 16 events (20%) showed likely AS change in the polyploid compared with its progenitors. Those changes could have taken place early in allopolyploid evolution or later over evolutionary time. It is notable that loss of AS from one homeolog was more common than silencing of one homeolog.
The vast majority of the events assayed are IR, which is the most common type of AS in plants, with many of those causing premature stop codons that could result in production of truncated proteins or in transcript degradation by nonsense-mediated decay that may lower the total level of expression of the genes. Evolutionary conservation can provide some clues as to the possibility of the AS isoforms being functional. An analysis of the AS events indicated that most show evolutionary conservation between species (SI Text and Table S5).
Analysis of Alternative Splicing Events in Two Resynthesized B. napus Allotetraploid Lines.
To determine if extensive AS changes in homeologs might occur within a few generations after polyploid formation, we assayed AS in two resynthesized B. napus allotetraploid lines in the S5 generation, derived by hybridization between B. rapa and B. oleracea and chromosome doubling (18, 19). We determined whether one or both homeologs showed AS in the resynthesized allopolyploids for 81 AS events in 73 genes using leaves and cotyledons of seedlings. AS was assayed by RT-PCR and sequencing of the RT-PCR products (Fig. 2). No differences in AS patterns were found between the two organ types. In resynthesized line 1, 39 events were present in both homeologs, 15 events were found in only the A homeolog derived from B. rapa, and 14 events were present in only the C homeolog derived from B. oleracea (Fig. S2 and Table 1). In resynthesized line 2, 42 events were present in both homeologs, 14 events were found only in the A homeolog, and 14 events were present only in the C homeolog. Thirty-six of the AS events were found in both homeologs in both of the synthetic allopolyploid lines (Table 1). In contrast, 23 of the AS events were present in only one homeolog in both resynthesized lines, and surprisingly in each case it was the same homeolog (Table 1). There were no examples of AS in only the A homeolog of one line and in only the C homeolog of the other line. Ten of the AS events were found in both homeologs of one line but only one homeolog of the other line. For cases that showed AS in only one homeolog, the result was further verified by RT-PCR with a primer located in the intron; sequencing of those RT-PCR products confirmed the presence of one homeolog in the AS band.
In each case where only one homeolog showed AS, we also determined if one or both homeologs showed the fully spliced form or if there was homeolog silencing, and if both homeologous genes were present, as discussed previously with natural B. napus. Three genes showed homeolog silencing in one resynthesized line or the other, whereas three other genes showed homeolog silencing in both resynthesized lines (Table 1). One gene showed evidence of homeolog loss or recombination. The possibility of homeologous nonreciprocal recombination (HNR) accounting for AS in one homeolog was also evaluated. HNR has been shown to occasionally occur between homeologs in the synthetic Brassica allopolyploid lines (19). PCR was performed with genomic DNA, and the products were then sequenced. In each case, both homeologs were present in genomic DNA, without recombination, in the region evaluated for AS, and thus HNR has not occurred in those regions of the genes.
Overall, the synthetic allopolyploids showed a surprisingly high number of cases where AS was present in only one homeolog. Two possibilities could account for this observation: there were changes in AS after allopolyploidy, or the diploid parents differed in their AS patterns. To distinguish between those two possibilities, the AS patterns in the two diploid parents were assayed by RT-PCR. There were five events that were not present in the B. rapa parent and 10 events that were not present in the B. oleracea parent; three events were not present in either parent or the polyploids (Table S4). Overall, there was a change in AS from the diploid parents for 21 events in resynthesized line 1 (26%) and 24 events in resynthesized line 2 (30%), in most cases loss of a parental AS event from one homeolog. Seventeen of the AS events showed changes from the diploid parents in both lines, whereas there were four events that showed AS change only in resynthesized line 1 and seven events that showed AS change only in resynthesized line 2. Gene 10, a plasma membrane protein, showed the AS event in both homeologs of resynthesized line 2 even though the event was absent from transcripts of the B. oleracea parental line, indicating an AS gain after allopolyploidy.
A particularly interesting case of AS change in the resynthesized allopolyploids was the PASTICCINO2 (PAS2) gene, encoding a tyrosine phosphatase-like protein involved in cell division and differentiation, in which two AS events were assayed in different regions of the gene. The RT-PCR reactions yielded three products: the fully spliced form, retention of intron 2, and retention of intron 8 (Fig. 3). The RT-PCR products were sequenced to determine which homeologs were present in the resynthesized allopolyploids. Whereas intron 2 was only retained in the transcripts from the A homeolog, intron 8 was only retained in the transcripts from the C homeolog (Fig. 3) from both resynthesized lines. Both diploid parents showed both AS events. Thus, the two AS events were partitioned between the homeologs after polyploidy. Each of the retained introns creates a premature stop codon, which could result in production of truncated proteins or in transcript degradation by nonsense-mediated decay that may lower the total level of expression from the genes. In natural B. napus, only the C homeolog shows both events; thus there has been parallel loss of AS from one homeolog, compared with the synthetic lines, but not partitioning of AS events between homeologs.
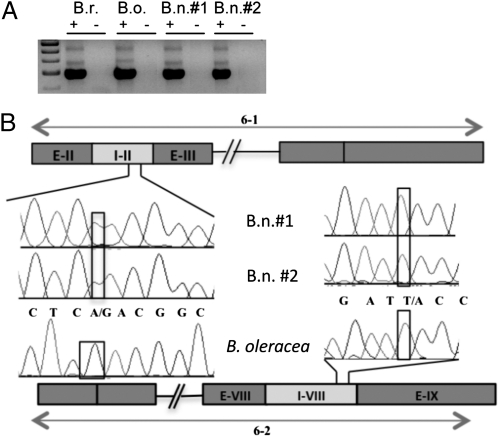
Fig. 3.
Partitioning of AS events in the PAS2 gene (gene 6) between homeologs in the resynthesized allopolyploids. (A) RT-PCR gel: The 6-2 isoform is the Top band, the 6-1 isoform is the Middle band, and the fully spliced form is the Bottom band. B.r. indicates B. rapa, O indicates B. oleracea, and B.n. indicates resynthesized B. napus. Plus and minus signs indicate presence or absence of reverse transcriptase. (B) Chromatograms from direct sequencing of the AS bands in the resynthesized allopolyploid: SNP sites are boxed. Multiple SNPs were evaluated for each region. Only the A homeolog was present in the 6-1 isoform that retains intron 2, and only the C homeolog was present in the 6-2 isoform that retains intron 8. Chromatograms from B. oleracea are shown for comparison.
Comparisons Between Natural and Resynthesized B. napus Lines.
The resynthesized allopolyploids showed more cases of changes in AS patterns, compared with the diploids, than did natural B. napus. This result was unexpected because we predicted that the resynthesized allopolyploids would show fewer AS changes than a natural allopolyploid species. The two resynthesized lines are more similar to each other in AS patterns than either is to natural B. napus (Fig. S2), indicating more divergence between the resynthesized lines and the natural line than between the two resynthesized lines. There were many genes where the AS patterns were the same in all three lines, mostly cases of AS in both homeologs. In contrast, there are eight events in eight genes (about 10% of the assayed AS events) that showed parallel changes in AS patterns, compared with the diploids, among both resynthesized lines and natural B. napus. For example, gene 47 encoding a subunit of the nuclear cap-binding protein complex (CBP20) involved in AS (21) showed AS of only the C homeolog in natural B. napus and both resynthesized lines. There were three genes that showed parallel changes in AS in one resynthesized line and the natural B. napus line. There were no parallel cases of homeolog silencing among the resynthesized lines and the B. napus line. Overall the resynthesized lines showed AS patterns in the homeologs that were largely different from the natural B. napus line, although a few genes showed parallel homeolog-specific losses of AS.
Discussion
Evolution of Alternative Splicing Patterns in a Natural Allopolyploid.
AS is a fundamental aspect of gene expression that can affect gene function by creating new protein isoforms and lowering the total level of gene expression by premature stop codon formation followed by transcript degradation (9). The evolutionary patterns of AS changes in natural polyploid species compared with their progenitor diploid species are unknown. We found that 20% of the gene pairs examined showed AS changes in B. napus compared with the parental diploid species. This value was higher than the number of gene pairs showing silencing of one homeolog, 6% of the gene pairs examined. Several gene pairs showed organ-specific AS of homeologs, indicating that AS in homeologs of a polyploid can be regulated in an organ-specific manner. Organ-specific homeolog silencing and biased expression are well documented in allopolyploids (reviewed in ref. 6), but there have not been previous reports of organ-specific AS of homeologs. A few gene pairs showed effects of heat and cold treatments on homeolog-specific AS patterns. Effects of abiotic stress treatments on homeologous gene expression have been investigated in cotton allopolyploids, where considerable quantitative changes to transcription levels in response to different abiotic stresses were shown for many genes, and regulatory subfunctionalization was shown for an alcohol dehydrogenase gene (22, 23).
Surprisingly Extensive and Parallel Changes in Alternative Splicing Patterns in the Resynthesized Allopolyploids.
The resynthesized Brassica allopolyploids show changes in homeolog-specific AS events in 26–30% of the gene pairs examined compared with their parental lines. A surprisingly high number (21%) of the genes showed parallel changes in AS events in the two resynthesized lines. This result shows that changes in AS in some genes after allopolyploidy are repeatable, and suggests that the phenomenon is not entirely due to random events. If the changes were entirely or mostly random, one would expect to find cases where AS in the A homeolog was lost in one line and AS in the C homeolog was lost in the other line. We did not find any cases of that type. Most of the changes were homeolog-specific losses of AS in one homeolog, but in one case there was a homeolog-specific gain of an AS event in one line. Our study assayed known events identified from expressed sequence tags in B. oleracea and thus we would preferentially detect AS losses over gains of novel AS events. Therefore, there were likely more changes in AS patterns in the assayed genes than revealed in this study. Overall, changes in AS patterns after polyploidy appear to be a common occurrence in resynthesized Brassica allopolyploids. We evaluated the presence and absence of AS events, and levels of AS transcripts were not quantified, although the levels of AS transcripts could vary between the homeologs in some cases. Thus, changes in AS patterns in the resynthesized allopolyploids are likely more extensive if AS transcript levels are considered.
Not only was the amount of AS change in the synthetic Brassica allopolyploid surprisingly high, but there were considerably more cases of AS changes than in natural B. napus (20% of the homeologous gene pairs). Why would the synthetic allopolyploids show more AS changes after polyploidy than natural B. napus? One possibility is that natural selection has acted in the natural polyploid such that some AS changes within the first few generations after allopolyploidy have been eliminated. Another possibility is that the variety of molecular events that accompany the merger of two divergent genomes in a common nucleus during allopolyploidy and in the first few generations afterward result in variable and unstable AS patterns during the first few generations. For example, there may be misregulation of genes whose products function in AS, such as SR-splicing factors. Stochastic expression patterns have been observed in a few genes among selfing generations during the first five generations after allopolyploidy in Arabidopsis (24). Following the initial stochastic sorting out of AS patterns, stability may ensue, perhaps resulting in AS patterns becoming more like the parental patterns over time. Although a hypothesis based on stochastic AS changes in the first few generations may sound attractive, our finding of a relatively high frequency of parallel AS changes in two independently synthesized lines is not supportive of it. A third possibility is that different parental populations of B. rapa and B. oleracea created the two allopolyploids (natural vs. resynthesized), and the amount of genetic changes in allopolyploids might vary when different diploid populations of each species form an allopolyploid.
Partitioning of Parental AS Patterns Between Duplicates in the Resynthesized Allopolyploids.
PASTICCINO2 (PAS2) showed partitioning of parental AS pattern between homeologs in both resynthesized allopolyploids. There was retention of intron 2 only in the homeolog derived from B. rapa, and retention of intron 8 only in the homeolog derived from B. oleracea. Partitioning of AS forms between duplicated genes is indicative of subfunctionalization of AS. If each splice form has a different function then it could lead to retention of both genes. In PAS2 the function(s) of the AS forms are unknown. Considering the presence of premature stop codons in each retained intron, the transcripts may get degraded by nonsense-mediated RNA decay (9), with the resulting effect of potentially lowering the total level of expression from the genes. Regulation of expression level might be an important function of the AS in PAS2. In Arabidopsis thaliana there is a PAS2 mutant with defective splicing of the final intron, resulting in that intron being retained in a majority of the transcripts (25). The total level of transcript accumulation is lowered, presumably because of transcript decay. The mutant shows impaired embryo and seedling development as well as ectopic cell proliferation. In contrast, overexpression of PAS2 slowed down cell division and inhibited seedling growth (26). Thus, too little or too much expression of PAS2 is detrimental, and AS may be a way of regulating the expression level.
There were only eight genes in this study where two AS events were assayed. Considering the large number of genes that contain multiple introns with the potential for AS, there could be a sizable number of genes in the synthetic allopolyploid that show partitioning of AS. Previously in allopolyploids, subfunctionalization of organ-specific expression patterns was shown in allotetraploid cotton and Tragopogon (27, 28). However, there have been no previous reports of partitioning of AS patterns in a synthetic or natural allopolyploid. Partitioning of AS patterns between duplicated genes has been seen when comparing gene structures in different plant species (29, 30).
Changes in AS Are Part of the Transcriptome Shock Experienced by New Allopolyploids.
Previous studies of several different synthetic allopolyploids showed that nonadditive gene expression, biased expression of homeologs, and homeologous gene silencing are phenomena that occur after polyploidization events (reviewed in refs. 5–8). Those events have been referred to as transcriptome shock (28, 31). AS changes represent a new form of transcriptome shock that affects many genes during the first few generations after allopolyploid formation. Our results show that AS changes in homeologs are considerably more common than homeolog silencing, which has been estimated as affecting ∼1–9% of homeologs in other studies (24, 32–34). How common are changes in AS forms after allopolyploidization? The findings here show changes in AS occurred in 26% of the genes that were examined in the resynthesized allopolyploids. In A. thaliana and rice, about 33% of genes have AS (12, 13), and the frequency of AS in genes in Brassica is probably equivalent. The B. napus genome is estimated to contain ∼100,000 genes (35). If ∼26% of the homeologous pairs show changes in AS in synthetic B. napus compared with their diploid parents, then AS in ∼4,300 gene pairs may be affected after polyploidy.
This study of AS patterns in more than one gene in a synthetic or natural allopolyploid is unique. Thus, at this point it is unknown if extensive changes in AS patterns occur after polyploidy in other plants or if the synthetic Brassica allopolyploids are unusual in this regard. Also unknown is whether interspecific hybridization, chromosome doubling, or molecular events in the first generations resulted in most of the AS alterations in the resynthesized allopolyploids. Two previous studies showed that AS patterns can change after interspecific hybridization. New AS variants were present in two SR splicing factor genes after interspecific hybridization in Populus (17) and in the S locus in an interspecific Arabidopsis hybrid (16). Two studies of synthetic allopolyploids showed splicing changes within a few generations after allopolyploidy: a different single (not alternative) spliced product appeared in the por gene of a cucumber allopolyploid (36), and the levels of the AS form of one gene changed in a synthetic wheat allopolyploid (37). Future studies of other allopolyploids, autopolyploids, and interspecific F1 hybrids will be needed to determine the extent of AS changes and their timing after interspecific hybridization and chromosome doubling.
What molecular mechanisms might cause changes in AS patterns in resynthesized allopolyploids? One possibility is that the combination of diverged AS factors from both parental species could cause different interactions with the target genes, resulting in changes in AS patterns. This hypothesis is based on proposals regarding the combination of diverged regulatory factors in an allopolyploid, causing changes in gene expression levels and patterns (38). Another possible mechanism is that epigenetic changes that occur in some allopolyploids, including histone modifications and cytosine methylation changes, might play a role in the AS changes. In contrast to the resynthesized Brassica allopolyploids, divergence in AS patterns between homeologs in natural B. napus may have been caused by divergence in sequences that are important for AS, such as sequences where SR proteins bind. There is a relatively low level of sequence divergence (about 1–2% SNPs and 1–4% insertions/deletions) between homeologous gene sequences of B. napus compared with its progenitor species (39).
What implications might AS changes in allopolyploids have for allopolyploid evolution? Differentially regulating AS between homeologs might allow more regulatory flexibility in duplicated genes, particularly in regards to regulating gene expression levels by AS, as shown for some genes (9). New AS forms in allopolyploids that create new protein isoforms, including truncated proteins, could result in new gene functions. AS changes in allopolyploids could be responsible for some of the phenotypic differences from their diploid parents, although this study was not designed to examine AS in candidate genes for phenotypes. In addition, AS changes in homeologs might lead to duplicate gene retention if AS forms are partitioned between duplicates, and both forms are essential for function.
Materials and Methods
Detailed information can be found in SI Materials and Methods. Briefly, the synthetic allotetraploids were created by crossing doubled-haploid B. rapa and B. oleracea followed by spontaneous chromosome doubling or colchicine doubling (18). The lines were propagated to the fifth generation by single-seed descent (19). Plants were grown under constant conditions in growth chambers for 15 d. Cold and heat treatments were done for 24 h at 4 °C and 38 °C, respectively. Nucleic acids were extracted using standard procedures. Genes were chosen randomly among those that showed AS in B. oleracea from publicly available ESTs. By comparing ESTs of B. napus, B. rapa, and B. oleracea, SNP sites in B. napus ESTs were identified. Phylogenetic analyses were performed to identify homeologs and paralogs. RT-PCR was performed on cDNAs derived from DNase-treated RNAs, using oligo(dT) for reverse transcription, with PCR primers that amplified both homeologs and the genes from the diploids. Products were resolved by agarose gel electrophoresis, followed by band cutting and Sanger sequencing. SNP sites on the chromatograms were scored to infer whether one or both homeologous genes were present in the sequenced AS bands.
Supplementary Material
Acknowledgments
We thank J. Wendel and the K. Adams laboratory for comments on the manuscript, and P. McVetty from the University of Manitoba for his gift of the B. napus seeds. This research was supported by the Natural Science and Engineering Research Council of Canada. R.Z. was partially supported by a fellowship from the China Scholarship Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109551108/-/DCSupplemental.
References
- 1.Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 2.Soltis DE, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- 3.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 4.Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZJ, Ni ZF. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays. 2006;28:240–252. doi: 10.1002/bies.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams KL. Evolution of duplicate gene expression in polyploid and hybrid plants. J Hered. 2007;98:136–141. doi: 10.1093/jhered/esl061. [DOI] [PubMed] [Google Scholar]
- 7.Doyle JJ, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 8.Hegarty MJ, Hiscock SJ. Genomic clues to the evolutionary success of polyploid plants. Curr Biol. 2008;18:R435–R444. doi: 10.1016/j.cub.2008.03.043. [DOI] [PubMed] [Google Scholar]
- 9.Reddy ASN. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 10.Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 11.Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filichkin SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G, et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010;20:646–654. doi: 10.1101/gr.100677.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang PG, Huang SZ, Pin AL, Adams KL. Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis. Mol Biol Evol. 2010;27:1686–1697. doi: 10.1093/molbev/msq054. [DOI] [PubMed] [Google Scholar]
- 15.Slotte T, et al. Splicing variation at a FLOWERING LOCUS C homeolog is associated with flowering time variation in the tetraploid Capsella bursa-pastoris. Genetics. 2009;183:337–345. doi: 10.1534/genetics.109.103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nasrallah JB, Liu P, Sherman-Broyles S, Schmidt R, Nasrallah ME. Epigenetic mechanisms for breakdown of self-incompatibility in interspecific hybrids. Genetics. 2007;175:1965–1973. doi: 10.1534/genetics.106.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scascitelli M, Cognet M, Adams KL. An interspecific plant hybrid shows novel changes in parental splice forms of genes for splicing factors. Genetics. 2010;184:975–983. doi: 10.1534/genetics.109.112557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukens LN, et al. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 2006;140:336–348. doi: 10.1104/pp.105.066308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palusa SG, Ali GS, Reddy ASN. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- 21.Raczynska KD, et al. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010;38:265–278. doi: 10.1093/nar/gkp869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu ZL, Adams KL. Expression partitioning between genes duplicated by polyploidy under abiotic stress and during organ development. Curr Biol. 2007;17:1669–1674. doi: 10.1016/j.cub.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 23.Dong S, Adams KL. Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytol. 2011;190:1045–1057. doi: 10.1111/j.1469-8137.2011.03650.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, et al. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellec Y, et al. Pasticcino2 is a protein tyrosine phosphatase-like involved in cell proliferation and differentiation in Arabidopsis. Plant J. 2002;32:713–722. doi: 10.1046/j.1365-313x.2002.01456.x. [DOI] [PubMed] [Google Scholar]
- 26.Da Costa M, et al. Arabidopsis PASTICCINO2 is an antiphosphatase involved in regulation of cyclin-dependent kinase A. Plant Cell. 2006;18:1426–1437. doi: 10.1105/tpc.105.040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buggs RJ, et al. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr Biol. 2011;21:551–556. doi: 10.1016/j.cub.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Cusack BP, Wolfe KH. When gene marriages don't work out: Divorce by subfunctionalization. Trends Genet. 2007;23:270–272. doi: 10.1016/j.tig.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Rösti S, Denyer K. Two paralogous genes encoding small subunits of ADP-glucose pyrophosphorylase in maize, Bt2 and L2, replace the single alternatively spliced gene found in other cereal species. J Mol Evol. 2007;65:316–327. doi: 10.1007/s00239-007-9013-0. [DOI] [PubMed] [Google Scholar]
- 31.Hegarty MJ, et al. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 32.Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams KL, Percifield R, Wendel JF. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics. 2004;168:2217–2226. doi: 10.1534/genetics.104.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buggs RJ, et al. Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol Ecol. 2010;19(Suppl 1):132–146. doi: 10.1111/j.1365-294X.2009.04469.x. [DOI] [PubMed] [Google Scholar]
- 35.Mun JH, et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009 doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuang Y, Chen JF, Jahn M. Expression and sequence variation of the cucumber Por gene in the synthesized allotetraploid Cucumis × hytivus. Mol Biol Rep. 2009;36:1725–1731. doi: 10.1007/s11033-008-9374-5. [DOI] [PubMed] [Google Scholar]
- 37.Terashima A, Takumi S. Allopolyploidization reduces alternative splicing efficiency for transcripts of the wheat DREB2 homolog, WDREB2. Genome. 2009;52:100–105. doi: 10.1139/G08-101. [DOI] [PubMed] [Google Scholar]
- 38.Riddle NC, Birchler JA. Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 2003;19:597–600. doi: 10.1016/j.tig.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Cheung F, et al. Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell. 2009;21:1912–1928. doi: 10.1105/tpc.108.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.