Abstract
Transcription factors TFIIB and TFIIF are both required for RNA polymerase II preinitiation complex (PIC) assembly, but their roles at and downstream of initiation are not clear. We now show that TFIIF phosphorylated by casein kinase 2 remains competent to support PIC assembly but is not stably retained in the PIC. PICs completely lacking TFIIF are not defective in initiation or subsequent promoter clearance, demonstrating that TFIIF is not required for initiation or clearance. Lack of TFIIF in the PIC reduces transcription levels at some promoters, coincident with reduced retention of TFIIB. TFIIB is normally associated with the early elongation complex and is only destabilized at +12 to +13. However, if TFIIF is not retained in the PIC, TFIIB can be lost immediately after initiation. TFIIF therefore has an important role in stabilizing TFIIB within the PIC and after transcription initiates.
Keywords: general transcription factors, transcript initiation
Promoter-directed transcript initiation by RNA polymerase II (pol II) on double-stranded templates requires at minimum the general transcription factors TBP (TATA box-binding protein), TFIIB, TFIIF, TFIIE, and TFIIH. Genetic and biochemical evidence indicates that TFIIB can direct transcription start site selection (1–4) with the assistance of TFIIF (5–8). When TFIIB is in complex with pol II, a TFIIB domain referred to as the finger (9) or reader (10) enters pol II and approaches the catalytic center. Structural investigations of the yeast preinitiation complex (PIC) revealed a segment of TFIIF localized near the TFIIB domain that presumably directs transcription start site selection (7, 11, 12). It was suggested that this interaction with TFIIF is important in stabilizing TFIIB within the PIC (12). Release of TFIIB from the elongation complex (EC) is reported to begin 7 to 16 bases downstream of transcription start (13). TFIIF should remain associated with the EC to facilitate rapid and effective transcript elongation (14, 15), but this factor may be transiently lost after initiation (16).
The functional roles of TFIIB and TFIIF within the PIC are not fully understood. Transcription from a preformed transcription bubble required only TFIIB and TBP, although TFIIF was strongly stimulatory in that system (17). RNA synthesis from bubble templates in the presence of the complete set of general factors was also very strongly dependent on TFIIF (18). These results raise an important question: Do TFIIB and TFIIF participate in the formation of initial bonds, or are they simply structural components that direct pol II to the correct start site? An important related issue concerns the continued residence of these factors within the EC: Is the proposed loss of TFIIF after initiation linked to the expected release of TFIIB?
In the present paper we take advantage of our recent observation that phosphorylation of TFIIF with casein kinase 2 (CK2) abolishes the ability of TFIIF to associate with pol II in ECs and stimulate elongation effectively (19), while leaving TFIIF’s support of transcript initiation intact. We found to our considerable surprise that although phosphorylated TFIIF (P-TFIIF) is fully capable of supporting PIC formation, the resulting complexes do not retain TFIIF. Initiation and promoter clearance proceeds normally in the complete absence of TFIIF, showing that TFIIF is not a required factor for either of these events. On some promoters, a reduction in transcription was seen with P-TFIIF, which correlated with reduced loading of TFIIB in those PICs. We also show that transcription complexes normally retain TFIIB well into the promoter clearance process; i.e., +12 to +13. However, when PICs lack TFIIF, TFIIB is apparently lost immediately from the transcription complex at initiation. Our results suggest that a major function of TFIIF is to assist and stabilize TFIIB.
Results
TFIIF Is Not Required for Initiation of Transcription, but It Is Essential for Effective Recruitment and Retention of TFIIB.
Our laboratory recently showed that phosphorylation of TFIIF by CK2 reduces or eliminates the functional interaction of TFIIF with the pol II EC without blocking the ability of TFIIF to support initiation (19). Given the link between TFIIF and TFIIB, we were interested in determining whether CK2 modification of TFIIF affects the loading of TFIIB into the PIC or the retention of TFIIB after initiation. To test this, PICs were assembled on bead-attached templates using recombinant human TBP, TFIIB, and TFIIE along with pol II and TFIIH purified from HeLa cells (18, 20). Recombinant TFIIF was either phosphorylated with CK2 (P-TFIIF) or unmodified (U-TFIIF). Complexes were assembled in DNA excess and extensively washed before analysis to minimize nonspecific association of factors with the templates. The promoters were variants of the TATA box containing adenovirus major late (AdML) promoter, modified such that transcription in the absence of one NTP stalls pol II at a particular downstream location. For example, on the pML-8g templates, the first G residue on the nontemplate strand appears at position +8. Two variants of the pML-8g template were used, in which the spacing from the TATA box to transcription start was either wild type (8gW) or 2 bp shorter (8g2D) (18). As measured by synthesis of a 9-mer transcript, P-TFIIF was almost fully active in PIC assembly on the 8gW template; however, transcription was reduced by about 2-fold when P-TFIIF was used instead of U-TFIIF on the 8g2D promoter (Fig. 1 A and B). No RNA was made if any one of the transcription factors was absent from the reaction (Fig. S1A).
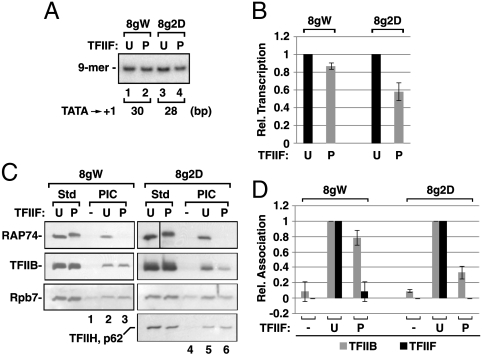
Fig. 1.
PICs assembled with P-TFIIF support initiation but fail to retain TFIIF and may recruit TFIIB inefficiently. (A) PICs assembled with U- or P-TFIIF on the 8gW or 8g2D promoters were washed and incubated with CpA, dATP, [α-32P]CTP, UTP, and 3′-deoxy-GTP for 5 min at 30 °C to generate 9-mer RNAs. (B) RNA levels in A were quantified and plotted with the U-TFIIF values for each promoter set to 1. The values shown are averages of three independent experiments, +/- SD. (C) PIC assembly reactions were performed on 8g2D or 8gW as in A (lanes 2, 3 and 5, 6), except no TFIIF was added in lanes 1 and 4. Retention of TFIIB, the RAP74 subunit of TFIIF, the Rpb7 subunit of pol II, and the p62 subunit of TFIIH in the PICs were analyzed by Western blotting as described in Materials and Methods. The two left-hand lanes (Std) in each panel contained 2 ng of U- or P-TFIIF, 0.75 ng of TFIIB, 0.5 ng of Rpb7, and 1 μL of TFIIH, respectively. (D) Average amounts of TFIIB and TFIIF from two (8gW) or three (8g2D) experiments are plotted with the level in the U-TFIIF reactions set to 1. Error bars indicate the SD (8g2D) or the range of values (8gW).
To determine the basis for the P-TFIIF dependent transcription defect on the 8g2D template, we used immunoblotting to assay the loading of TFIIB into 8g2D and 8gW PICs assembled with P- or U-TFIIF. As shown in Fig. 1 C and D, the reduction in transcription from 8g2D with P-TFIIF was paralleled by a corresponding reduction in recruitment of TFIIB to the 8g2D PICs. Probing for other PIC components showed that for the 8g2D template, assembly with P-TFIIF leads to a drop in pol II recruitment roughly proportional to the reduction in both RNA synthesis and TFIIB recruitment; however, TFIIH recruitment was not affected by the modification of TFIIF (Fig. 1C). To confirm the specificity of factor association with the template, we assembled PICs with or without the TBP promoter recognition factor. Neither TFIIB nor TFIIH were template-bound in the absence of TBP (Fig. S2C); some binding of pol II was detected in TBP-less reactions, possibly reflecting polymerase associating with template ends (Fig. S2C).
We also tested for the presence of TFIIF in PICs assembled with P-TFIIF, using an antibody to the RAP74 subunit. We were very surprised to discover that P-TFIIF was nearly (8gW) or completely (8g2D) absent from complexes made with P-TFIIF (Fig. 1 C and D). Tests with antibody to RAP30 confirmed that neither P-TFIIF subunit was retained in PICs assembled with the modified factor (Fig. S2A). RAP74 was present in PICs obtained with U-TFIIF (Fig. 1C). This was the strongly predicted result from earlier studies with both mammalian (21) and yeast (12) systems. However, we were unable to demonstrate promoter-dependent association of U-TFIIF in our PICs. Unlike the case for TFIIB and TFIIH, U-TFIIF was template-bound when TBP was absent from the assembly reactions (Fig. S2C). This association is puzzling because it does not reflect nonspecific binding of U-TFIIF alone to template DNA (Fig. S2B). We would emphasize that regardless of the unexpected TBP-independent binding of U-TFIIF to the template, we can firmly conclude that although TFIIF is absolutely required for assembly of the promoter-directed PIC, TFIIF need not remain in the PIC to support initiation. Further, the deficit in transcriptional activity seen with P-TFIIF on the 8g2D promoter is paralleled by a deficit in TFIIB recruitment. It is important to stress that PICs assembled with P-TFIIF are not generally defective, because there is only a minor reduction in RNA synthesis and TFIIB content when P-TFIIF was used to assemble complexes on the 8gW promoter.
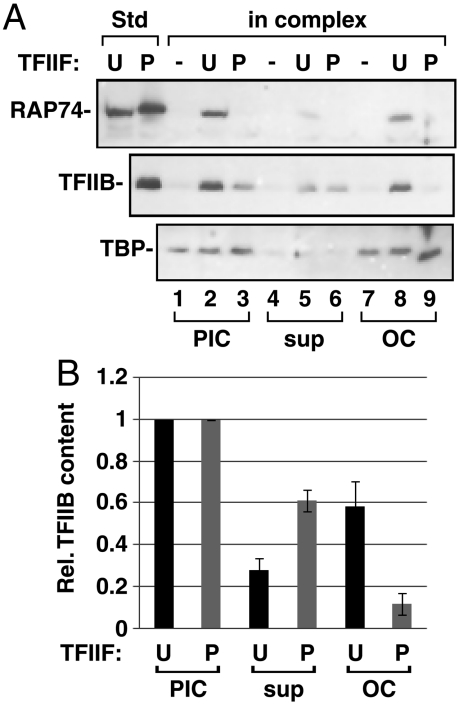
Because TFIIB recruitment or retention can be defective when PICs do not retain TFIIF, we next asked if TFIIB is retained during open complex formation in PICs made with P-TFIIF. ATP was added to PICs assembled with U- or P-TFIIF on the 8g2D template and after 5 min, the template-bound and released fractions were tested (Fig. 2). Complexes assembled with U-TFIIF mostly retained TFIIB in the open complex after ATP addition (compare lanes 2, 5, and 8 of Fig. 2A), whereas the opposite was found when PICs were formed with P-TFIIF (compare lanes 3, 6, and 9 of Fig. 2A). Quantitative results are shown in Fig. 2B. TBP was present equally regardless of ATP exposure or the state of TFIIF used for PIC assembly.
Fig. 2.
In PICs which lack TFIIF, TFIIB is destabilized upon open complex formation. PIC assembly reactions on 8g2D contained U-TFIIF, P-TFIIF, or no TFIIF; PICs were subsequently washed. (A) PICs were either untreated (lanes 1–3) or incubated with ATP for 5 min at 30 °C and then separated into template-bound (OC) and released (sup) fractions. Proteins were resolved and TFIIF (RAP74), TFIIB, and TBP were detected by Western blotting as described in Materials and Methods. (B) The results from A are plotted with the level of TFIIB set to 1 for each PIC. Values are the average of two experiments, +/- the range.
PICs That Lack TFIIF Are Fully Functional in First-Bond Formation and in Promoter Clearance.
The failure to retain TFIIB efficiently in open complexes assembled with P-TFIIF raises the question of whether multiple rounds of transcript initiation could occur with such PICs, for example, the repeated abortive synthesis of trimer RNAs using dinucleotide primers (22). This seemed an important question in light of earlier studies with premelted templates (8, 23), which reported that TFIIF stimulates first-bond formation. The results of an abortive initiation assay using CpA primer and [α-32P] CTP are shown in Fig. 3. As was the case with longer transcripts, CpApC production was completely dependent on both TFIIF (Fig. 3A) and TBP (Fig. S1B). The relative efficiencies of trimer production with complexes formed with U- or P-TFIIF on the 8gW and 8g2D templates (Fig. 3B) paralleled 9-mer synthesis levels with these promoters (Fig. 1 A and B). Comparison of trimer and 8-mer levels made from the same PIC preparation showed a 13.3-fold molar excess of abortive to productive transcripts (Fig. S1C). This is consistent with multiple rounds of trimer production from each PIC, as predicted from our earlier studies of abortive initiation at the AdML promoter (22). Thus, there is no apparent defect in repetitive first-bond formation in transcription complexes that lack TFIIF.
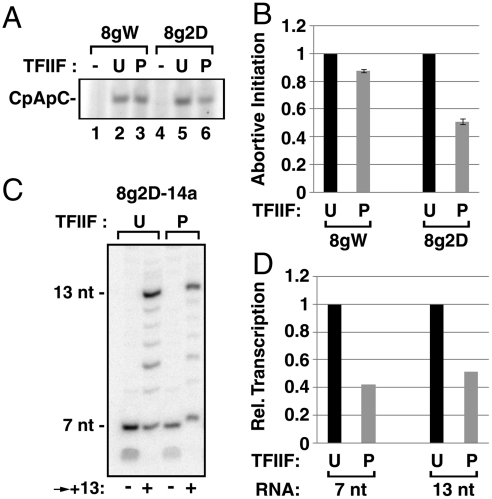
Fig. 3.
The absence of TFIIF from PICs does not compromise abortive initiation or promoter clearance. (A) PIC assembly reactions on 8gW and 8g2D contained U-TFIIF, P-TFIIF, or no TFIIF. PICs were washed and then incubated for 5 min with CpA and labeled CTP to generate CpApC. (B) Levels of CpApC from three independent experiments were plotted, +/- SD; the values obtained with U-TFIIF were set to 1. (C) ATP was used to initiate U- or P-TFIIF-mediated transcription on 8g2D-14a. ECs were advanced to the G stop at +8, washed, and then chased with GTP, UTP, and CTP to the A stop for 2 min at 30 °C. (D) RNAs of the indicated lengths were quantified, with the U-TFIIF value set to 1 for each transcript length. In the experiment shown, transcription was initiated with 500 μM ATP. In two other tests, results of which are not included in this figure, transcription was initiated with 50 μM ATP or 1 mM ApC. In those cases as well, we observed that the fraction of 7-mers that chased to 13 was the same regardless of the modification state of TFIIF.
Based on previous studies (23, 24), it might be expected that some aspect of promoter escape or clearance would be defective when the early stages of transcript elongation occur in the absence of TFIIF. We took advantage of our earlier observation that complexes halted at +7 on the 8g2D promoter have not achieved promoter clearance, as judged by the presence of extended transcription bubbles in these complexes and a continued requirement for TFIIH helicase activity for effective continuation of RNA synthesis (18). We also showed that once pol II has advanced to +13 on this template, clearance has occurred as judged by collapse of the upstream segment of the initial transcription bubble and lack of continued dependence on TFIIH (18). As expected (Figs. 1A and 3 A and B), P-TFIIF-dependent transcription from the 8g2D promoter showed a 2-fold reduction in 7-nt RNA production compared to transcription with U-TFIIF. Significantly, the same fraction of transcription complexes that restarted from +7 reached +13 successfully regardless of the continued presence of TFIIF in the initial PICs (Fig. 3 C and D). Our data, therefore, do not indicate any defect in promoter clearance for early elongation complexes that lack TFIIF.
The Onset of Significant Release of TFIIB Occurs as the Nascent RNA Is Extended from 12 to 13 nt.
Early studies on the fate of TFIIB during transcription indicated that TFIIB is released from the EC shortly after initiation (16). Subsequent work placed the onset of TFIIB release between positions +7 and +16, with most TFIIB lost during full elongation (13). These results left open the question of the mechanistic relationship between TFIIB release and critical structural changes in the early EC, which should influence TFIIB retention. These include occupancy of the pol II RNA exit channel by the nascent RNA, which should displace a segment of TFIIB (10, 25), and the collapse of the initial, overly long transcription bubble, which should remove TFIIB contacts with the upstream, nontemplate strand (10, 18, 26). Transcription complexes for some TFIIB release studies were assembled in nuclear extracts (13), which contain numerous kinases including CK2. This could be significant because TFIIB may be lost from the transcription complex immediately if TFIIF is phosphorylated by CK2 (Fig. 2). These considerations led us to test for retention of TFIIB in ECs in our system. Preliminary studies had indicated that when PICs are assembled with U-TFIIF, TFIIB is relatively stable in ECs halted at +7. We therefore established a two-step TFIIB release assay (Fig. 4 and Fig. S3) in which ECs stalled at +7 were advanced to one of four different downstream locations. At these positions the EC was upstream of clearance (+8), just prior to clearance (+10), just beyond clearance (+13), and fully committed to elongation (+74—see ref. 18). At 1, 2, and 4 min after nucleotide addition, the templates were separated from supernatants and both fractions were probed for TFIIB and pol II (Rpb7 subunit). Under the transcription conditions used, pol II advanced from +7 to +8, +10, or +13 in a few seconds; pol II reached +74 in approximately 15 s and runoff in approximately 20 s.
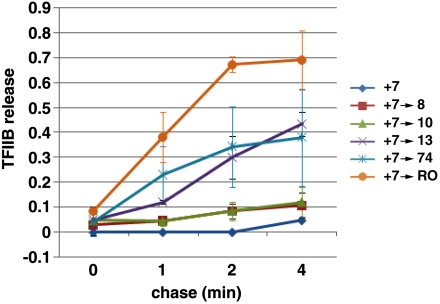
Fig. 4.
TFIIB becomes destabilized within the EC when the transcript is extended from 7 to 13 nt. Quantitative analysis of two or three (+7 → 10) independent experiments on TFIIB release over a 4-min time course. The fraction of TFIIB lost is shown for complexes advanced from +7 to the indicated positions, with the TFIIB level in the 7-mer complex set to 1. The values plotted show the averages +/- the range, or +/- SD for the +7 → 10 test. Examples of the primary data are shown in Fig. S1.
TFIIB remained relatively stably associated with the complex (Fig. 4 and Fig. S3B) when pol II advanced to +8 or +10, whereas advancing only 3 bp more (+13) caused instability equivalent to a complex that had advanced much farther (+74; Fig. 4 and Fig. S3A). Each release/retention pattern was compared with an NTP treatment that does not support elongation to ensure that release did not simply result from the incubation itself. The rates of TFIIB release from the various ECs are summarized in Fig. 4, with losses given relative to the amount of TFIIB present in the initial 7-mer ECs. Approximately 40% release within 4 min was observed when ECs were advanced to +13 or to +74. In contrast, less than 10% loss of TFIIB was observed for translocations to +8 and +10. Measurement of the level of RNA synthesis obtained with our PICs showed that during the 5-min reactions that generated the 7-mer complexes, only 35–40% of the PICs had produced a transcript (as judged by comparison of TFIIB levels on the templates versus amounts of RNA synthesized—see detailed SI Materials and Methods). It is possible that some of our PICs were defective in initiation. Alternatively, initiation in these PICs may be slow, consistent with our earlier observation that human PICs have not completed single-round initiation even after a 20-min incubation with NTP substrates (22). In any event, the fraction of TFIIB lost by the 13- or 74-mer complexes is fully consistent with the portion of PICs that were active in RNA synthesis. These results point to a very narrow window in the late promoter clearance process, during extension of the RNA from 10 to 13 nt, in which TFIIB’s interaction with the EC is destabilized.
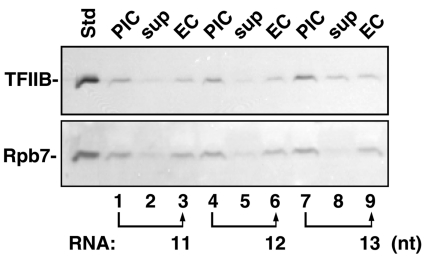
The comparison of TFIIB release between positions +10 and +13 was particularly interesting, because the 10-mer and 13-mer complexes are immediately prior to and just downstream of the bubble collapse transition, which is a major hallmark of promoter clearance (10, 18). To determine if collapse of the upstream segment of the initial transcription bubble is the exact boundary of TFIIB destabilization, we prepared a set of three ECs halted with 11-, 12-, and 13-nt RNAs. Our earlier studies on similar complexes indicated that bubble collapse should have occurred in all three of these ECs (18). As expected from the results in Fig. 4, TFIIB began to dissociate from ECs stalled at RNA length of 13 nt. However, no significant amount of TFIIB was released by ECs stalled at the RNA lengths of 11 and 12 nt (Fig. 5, compare lane 8 with lanes 2 and 5, respectively). The retention of pol II (Rpb7) in the ECs shows that there was not extensive nonspecific dissociation of any of these complexes (Fig. 5).
Fig. 5.
Extension of the nascent transcript from 12 to 13 nt marks the threshold of efficient TFIIB release. PICs were assembled with unmodified TFIIF on 11g or 8g2D-14a templates and transcribed for 5 min at 30 °C. PICs on 11g were incubated with ApC (for 11-nt RNA) or CpA (for 12-nt RNA), dATP, CTP, UTP, and 3′-deoxy-GTP. PICs on 8g2D-14a were incubated with ApC, dATP, CTP, GTP, and UTP to give 13-nt RNAs. Levels of TFIIB and pol II (as Rpb7) in the initial PICs, retained in the ECs and released to the supernates after transcription, were analyzed by immunoblotting as described in Materials and Methods. Std lane contained 0.75 ng TFIIB or 0.5 ng Rpb7.
Advancing Pol II from +6 to +7 Can Initiate the Destabilization of TFIIB in Early ECs.
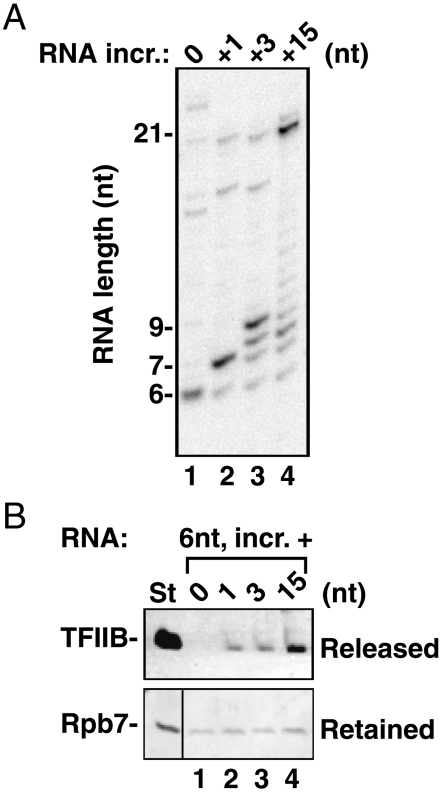
The results in Fig. 5 suggest that the loss of potential upstream DNA contacts at bubble collapse is not sufficient to drive release of TFIIB. The other event thought to influence loss of TFIIB early in elongation is the developing clash of the TFIIB reader/finger segment with the advancing nascent RNA. It was speculated that this interaction should begin with nascent RNAs of 6 or 7 nt (9, 10, 25). We therefore generated ECs containing only 6-nt RNAs and assayed for loss of TFIIB as these complexes advanced by 1, 3, or 15 nt (Fig. 6A). TFIIB is relatively stable in 7-mer complexes containing TFIIF (Fig. 4 and Fig. S3), but remarkably, addition of only one nucleotide to a 6-mer caused a slight destabilization of TFIIB in a 2-min incubation (Fig. 6B). ECs with 6-mer RNAs released no TFIIB during the same treatment (lane 1). The extent of the release remained the same when polymerase advanced by 3 nt (lane 3) but strongly increased with extension by 15 nt in the control reaction (lane 4). The stabilities of the ECs were verified by Rpb7 retention (Fig. 6B, lower row).
Fig. 6.
Advancing from 6 to 7 nt can initiate the destabilization of TFIIB in early ECs. PICs were formed with U-TFIIF on the 6gW template. (A) Initial complexes were advanced to the G stop with CpA, dATP, UTP, and [α-32P] CTP for 5 min at 30 °C. After washing, the 6-mer complexes were incubated for 2 min with ATP only (no advance), GTP (for 7-mers), GTP and UTP (for 9-mers), or GTP, UTP, and CTP (for 21-mers). (B) ECs were generated as in A, except that reaction volume was scaled up threefold and nonlabled NTPs were used. TFIIB or pol II (as Rpb7) was detected by immunoblotting in the supernatant fraction (TFIIB) or the retained fraction (Rpb7) after the advance of the 6-mer complexes. The left-hand lane (St) contained 2.5 ng TFIIB or 0.5 ng Rpb7.
Discussion
General transcription factors TFIIF and TFIIB are both required, along with TBP, TFIIE, and TFIIH, to assemble a pol II PIC on a double-stranded template (27, 28). On a premelted template, TFIIB (along with TBP) is sufficient to obtain a functional PIC, but the activity of such complexes is very strongly stimulated by TFIIF (17, 18). Thompson et al. (29) showed that TFIIF rescues the transcriptional activity of otherwise-inactive PICs assembled with mutant forms of TFIIB. These findings, plus recent structural studies of yeast pol II PICs (12, 30), support the generally accepted idea that TFIIF (or at least RAP30—see ref. 24) is required for transcript initiation and not simply for assembly. We show here that this is not necessarily the case. PICs assembled on the natural AdML promoter (8gW) with a phosphorylated form of TFIIF do not retain TFIIF but are nevertheless fully competent for normal transcript initiation, as well as multiround abortive initiation. PICs that lack TFIIF give rise to ECs that have no detectable deficit in passage through the bubble collapse transition to promoter clearance. The defect in complexes that lack TFIIF centers on TFIIB, which is not stably maintained after initiation in those complexes.
Early studies of pol II PIC assembly stressed the importance of the TBP/TFIIB/DNA interaction in recruiting TFIIB to the template (reviewed in ref. 28). However, we find that in the absence of stable association of TFIIF with the PIC, the presence of TBP and a canonical TATA element are not sufficient to guarantee effective recruitment (8g2D template) and retention of TFIIB. Thus, the TFIIB-TFIIF interaction (12) is a critical part of establishing and maintaining TFIIB in the PIC, a point emphasized by the loss of TFIIB from open complexes in the absence of TFIIF. Earlier studies (8, 23) that suggested an important role for TFIIF in the initial stages of transcription relied on premelted templates in order to generate PICs with or without TFIIF. Our results emphasize both the importance of the TFIIF-TFIIB interaction and the promoter dependence of this interaction. It is interesting that RNA polymerases I (31) and III (32) both contain structural motifs that resemble segments of TFIIF. The archaeal RNA polymerases, in contrast, do not require a factor analogous to TFIIF for initiation, even though initiation for those polymerases does depend on a TFIIB analog (see, for example, ref. 33).
Structural studies predicted that the reader/finger domain of TFIIB would be displaced from the RNA exit channel by the advancing nascent RNA, beginning with 6- or 7-nt transcripts (10, 25). It was also predicted that the linker region of TFIIB should lose its contact with the free nontemplate strand at the upstream end of the transcription bubble when this DNA reanneals during promoter clearance (10). We did see a slight loss of TFIIB as pol II advanced from +6 to +7, but TFIIB remained relatively stably associated with the EC until the nascent RNA was 13 nt long. This is well past the predicted initial collision point of the reader/finger with the nascent RNA, and it is also 2 bp downstream of the point at which the upstream segment of the transcription bubble reanneals on the promoter we used (18). Thus, full destabilization of the association of TFIIB and the EC may require the loss of both TFIIB’s nontemplate strand contacts and nearly all of TFIIB’s association with the RNA exit channel.
Particularly in the context of the mechanism of TFIIB release, it is important to note that our modified AdML template (8g2D) has a TATA box to transcription start spacing of only 28 bp. A recent study showed that 45% of mammalian TATA-containing promoters have a TATA to +1 spacing of either 30 bp (as in the 8gW promoter from which 8g2D is derived) or 31 bp, whereas about 4% have the 28-bp spacing present in 8g2D and about 6% have a spacing 2 bp longer than the major class (34). The somewhat unusual (but nonetheless natural) spacing in 8g2D could influence both the RNA length for initial collision between TFIIB and the nascent RNA and the interactions of the TFIIB linker with the transcription bubble. We also note that our TFIIB release studies involved initiation with CpA; these transcripts have a slightly different effective length than RNAs initiated with ATP (see ref. 35). Thus, the exact transcript length at which TFIIB becomes destabilized may vary with both promoter sequence and transcription substrate. Regardless of these considerations, the destabilization of TFIIB appears to occur well downstream of major predicted structural transitions in the nascent transcript elongation complex for the promoter that we used. We emphasize that 8g2D is not a significantly defective promoter, because it supports levels of PIC formation equivalent to those observed with 8gW when U-TFIIF is used in assembly.
It is useful to place our findings on TFIIF within the larger context of gene regulation. TFIIF is phosphorylated in vivo (36, 37), and major sites for TFIIF modification by HeLa extracts are CK2 consensus sites (38). CK2 is required for the activity of some pol II promoters in vitro (39), and CK2 associates with both promoters (39) and active genes (40) in vivo. The reduced transcriptional activity of the 8g2D promoter (relative to the 8gW promoter) with P-TFIIF shows that even closely related promoters could be differentially controlled by CK2 modification of TFIIF. The failure of P-TFIIF to remain in the PIC is potentially significant because it is now appreciated that pol II pauses at roughly 50 bases within the transcription unit on many mammalian genes. Release from this pause is an important aspect of control of gene expression (41–45). Biochemical studies suggest that TFIIF is essential for the paused EC to advance into productive transcript elongation (15). CK2 modification of TFIIF, which drives TFIIF out of the PIC and inhibits its reassociation with pol II during elongation (19), could play a role in the regulated release of pol II from the +50 pause region. The importance of the TFIIF-TFIIB interaction in this regulatory event was highlighted by a report that deletions within TFIIB can lead to failure to retain TFIIF during transcript elongation (46). Finally, termination of transcription by pol II in yeast involves a TFIIB-mediated interaction between the terminator and the promoter-proximal region (47). Given the importance of TFIIF in stabilizing TFIIB interaction with the transcription complex, it is tempting to speculate that TFIIF should also participate in the process of termination.
Materials and Methods
Detailed descriptions are given in SI Text for template construction, preparation of proteins, and analysis of factor retention and release during transcription. Preinitiation complexes were assembled and transcription reactions were performed on immobilized template as described earlier (18, 20) and in detail in SI Text.
Supplementary Material
Acknowledgments.
We thank Erica Golemis (Fox Chase Cancer Center, Philadelphia, PA) for the antibody to Rpb7. This work was supported by National Institutes of Health Grant GM 29487 (to D.S.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104591108/-/DCSupplemental.
References
- 1.Pinto I, Wu W-H, Na JG, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 2.Pardee TS, Bangur CS, Ponticelli AS. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J Biol Chem. 1998;273:17859–17864. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- 3.Hawkes NA, Roberts SG. The role of human TFIIB in transcription start site selection in vitro and in vivo. J Biol Chem. 1999;274:14337–14343. doi: 10.1074/jbc.274.20.14337. [DOI] [PubMed] [Google Scholar]
- 4.Cho EJ, Buratowski S. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J Biol Chem. 1999;274:25807–25813. doi: 10.1074/jbc.274.36.25807. [DOI] [PubMed] [Google Scholar]
- 5.Sun Z-W, Hampsey M. Identification of the gene (SSU71/TFG1) encoding the largest subunit of transcription factor TFIIF as a suppressor of a TFIIB mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:3127–3131. doi: 10.1073/pnas.92.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghazy MA, Brodie SA, Ammerman ML, Ziegler LM, Ponticelli AS. Amino acid substitutions in yeast TFIIF confer upstream shifts in transcription initiation and altered interaction with RNA polymerase II. Mol Cell Biol. 2004;24:10975–10985. doi: 10.1128/MCB.24.24.10975-10985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freire-Picos MA, Krishnamurthy S, Sun ZW, Hampsey M. Evidence that the Tfg1/Tfg2 dimer interface of TFIIF lies near the active center of the RNA polymerase II initiation complex. Nucleic Acids Res. 2005;33:5045–5052. doi: 10.1093/nar/gki825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khaperskyy DA, Ammerman ML, Majovski RC, Ponticelli AS. Functions of Saccharomyces cerevisiae TFIIF during transcription start site utilization. Mol Cell Biol. 2008;28:3757–3766. doi: 10.1128/MCB.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostrewa D, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 11.Majovski RC, Khaperskyy DA, Ghazy MA, Ponticelli AS. A functional role for the switch 2 region of yeast RNA polymerase II in transcription start site utilization and abortive initiation. J Biol Chem. 2005;280:34917–34923. doi: 10.1074/jbc.M502932200. [DOI] [PubMed] [Google Scholar]
- 12.Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 2010;29:706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran K, Gralla JD. Control of the timing of promoter escape and RNA catalysis by the transcription factor IIB fingertip. J Biol Chem. 2008;283:15665–15671. doi: 10.1074/jbc.M801439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luse DS, Spangler LC, Újvári A. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J Biol Chem. 2011;286:6040–6048. doi: 10.1074/jbc.M110.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 16.Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 17.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 18.Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 19.Újvári A, Pal M, Luse DS. The functions of TFIIF during initiation and transcript elongation are differentially affected by phosphorylation by casein kinase 2. J Biol Chem. 2011;286:23160–23167. doi: 10.1074/jbc.M110.205658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Újvári A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat Struct Mol Biol. 2006;13:49–54. doi: 10.1038/nsmb1026. [DOI] [PubMed] [Google Scholar]
- 21.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1421. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 22.Luse DS, Jacob GA. Abortive initiation by RNA polymerase II in vitro at the Adenovirus 2 major late promoter. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 23.Yan Q, Moreland RJ, Conaway JW, Conaway RC. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J Biol Chem. 1999;274:35668–35675. doi: 10.1074/jbc.274.50.35668. [DOI] [PubMed] [Google Scholar]
- 24.Chang C, Kostrub CF, Burton ZF. RAP30/74 (transcription factor IIF) is required for promoter escape by RNA polymerase II. J Biol Chem. 1993;268:20482–20489. [PubMed] [Google Scholar]
- 25.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 26.Fiedler U, Timmers HTM. Analysis of the open region of RNA polymerase II transcription complexes in the early phase of elongation. Nucleic Acids Res. 2001;29:2706–2714. doi: 10.1093/nar/29.13.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 29.Thompson NE, Glaser BT, Foley KM, Burton ZF, Burgess RR. Minimal promoter systems reveal the importance of conserved residues in the B-finger of human transcription factor IIB. J Biol Chem. 2009;284:24754–24766. doi: 10.1074/jbc.M109.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: A model for the structure of PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Geiger SR, et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell. 2010;39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 32.Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem. 2010;285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werner F, Weinzierl ROJ. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol Cell Biol. 2005;25:8344–8355. doi: 10.1128/MCB.25.18.8344-8355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponjavic J, et al. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pal M, Luse DS. The initiation-elongation transition: Lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23. Proc Natl Acad Sci USA. 2003;100:5700–5705. doi: 10.1073/pnas.1037057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sopta M, Carthew RW, Greenblatt J. Isolation of three proteins that bind to mammalian RNA polymerase II. J Biol Chem. 1985;260:10353–10360. [PubMed] [Google Scholar]
- 37.Kitajima S, Chibazakura T, Yonaha M, Yasukochi Y. Regulation of the human general transcription initiation factor TFIIF by phosphorylation. J Biol Chem. 1994;269:29970–29977. [PubMed] [Google Scholar]
- 38.Rossignol M, Keriel A, Staub A, Egly JM. Kinase activity and phosphorylation of the largest subunit of TFIIF transcription factor. J Biol Chem. 1999;274:22387–22392. doi: 10.1074/jbc.274.32.22387. [DOI] [PubMed] [Google Scholar]
- 39.Lewis BA, Sims RJ, Lane WS, Reinberg D. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol Cell. 2005;18:471–481. doi: 10.1016/j.molcel.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Egyházi E, Ossoinak A, Filhol-Cochet O, Cochet C, Pigon A. The binding of the α subunit of protein kinase CK2 and RAP74 subunit of TFIIF to protein-coding genes in living cells is DRB sensitive. Mol Cell Biochem. 1999;191:149–159. [PubMed] [Google Scholar]
- 41.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price DH. Poised polymerases: On your mark…get set…go! Mol Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Nechaev S, Adelman K. Promoter-proximal pol II: When stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- 44.Margaritis T, Holstege FCP. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 45.Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- 46.Tran K, Gralla JD. The TFIIB tip domain couples transcription initiation to events involved in RNA processing. J Biol Chem. 2010;285:39580–39587. doi: 10.1074/jbc.M110.171850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.