Abstract
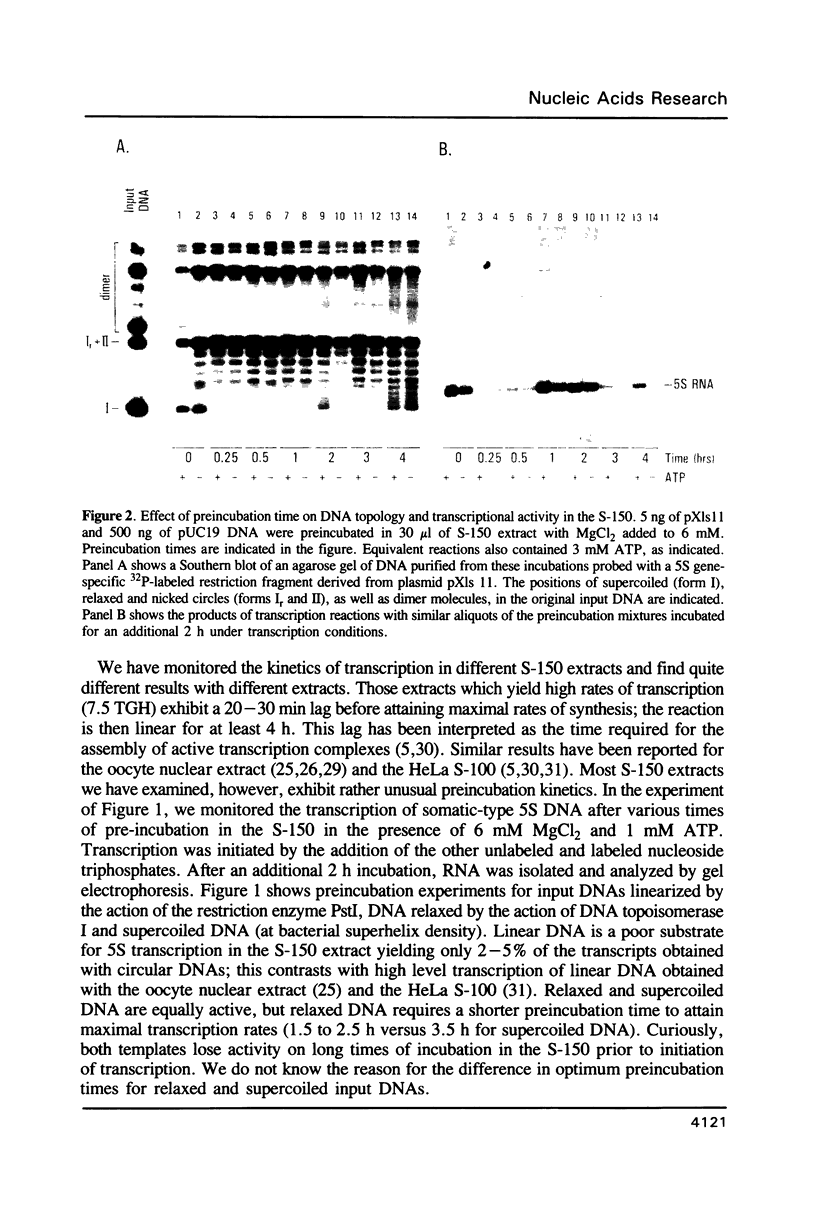
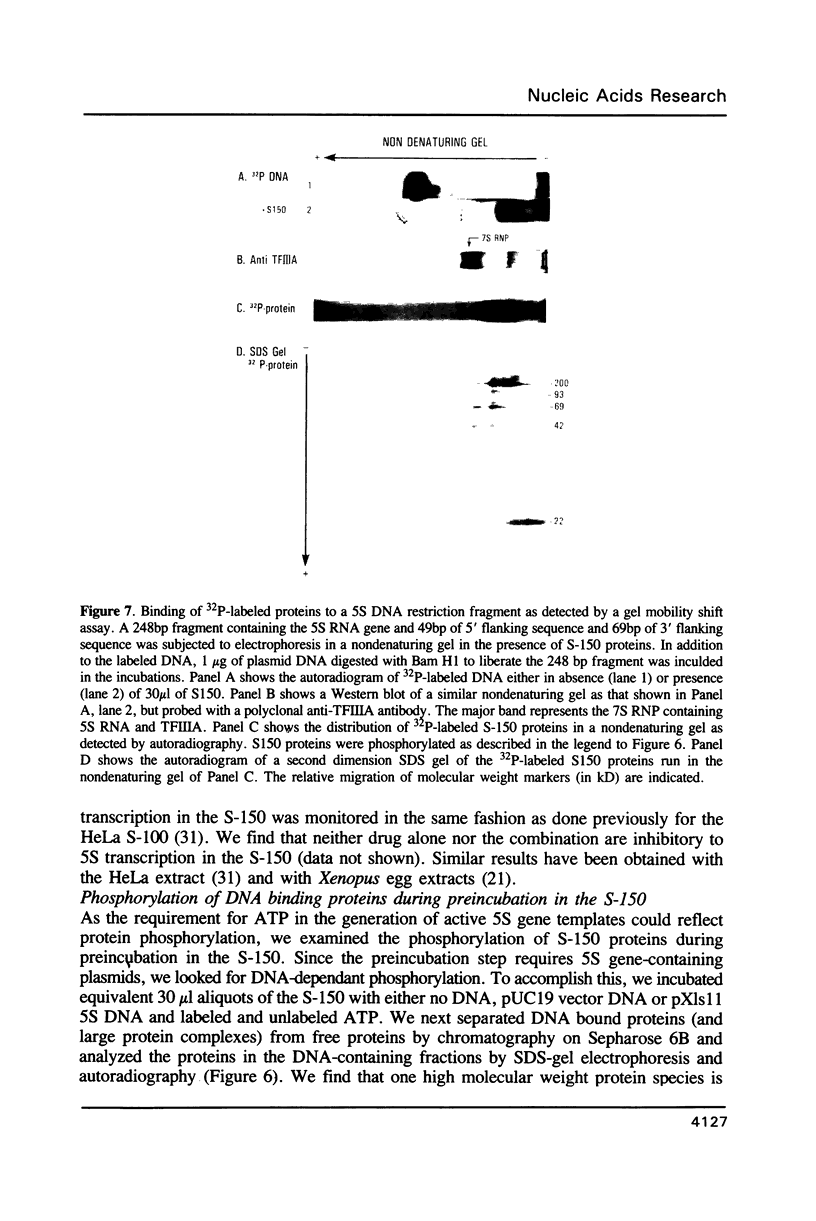
Conditions for transcription and nucleosome assembly of plasmids bearing Xenopus 5S RNA genes have been monitored in the whole oocyte S-150 extract (1). We find that the optimal conditions for transcription differ substantially from optimal conditions for nucleosome assembly. DNA molecules bearing as few as 50% of the native density of nucleosomes are transcriptionally inert. Although the 5S gene-specific transcription factor TFIIIA is in excess in this extract, these nucleosome reconstitutes do not exhibit TFIIA-like DNase footprints nor do these reconstitutes bind exogenous TFIIIA. We have also examined the nucleotide requirement for DNA supercoiling and for generation of 5S gene transcription complexes. Supercoiling associated with nucleosome assembly does not require ATP; however, nucleotide hydrolysis is required for establishment of active complexes. Phosphorylation of a 200 kdalton protein occurs in a 5S DNA-dependent manner concurrent with the generation of primed transcription complexes. Results of nondenaturing gel electrophoresis coupled with a second dimension of SDS gel electrophoresis suggest that the 200 kD protein may be a component of the 5S RNA gene transcription complex.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Bieker J. J., Roeder R. G. Characterization of the nucleotide requirement for elimination of the rate-limiting step in 5 S RNA gene transcription. J Biol Chem. 1986 Jul 25;261(21):9732–9738. [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Chen G. L., Yang L., Rowe T. C., Halligan B. D., Tewey K. M., Liu L. F. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984 Nov 10;259(21):13560–13566. [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg A. M., King B. O., Roeder R. G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984 Dec;39(3 Pt 2):479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M. DNA sequence-directed nucleosome reconstitution on 5S RNA genes of Xenopus laevis. Mol Cell Biol. 1987 May;7(5):1612–1622. doi: 10.1128/mcb.7.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M. Novobiocin inhibits RNA polymerase III transcription in vitro by a mechanism distinct from DNA topoisomerase II. Nucleic Acids Res. 1986 Mar 11;14(5):2075–2088. doi: 10.1093/nar/14.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Hoeffler W. K., Kovelman R., Roeder R. G. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988 Jun 17;53(6):907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- Hsiang Y. H., Hertzberg R., Hecht S., Liu L. F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985 Nov 25;260(27):14873–14878. [PubMed] [Google Scholar]
- Kmiec E. B., Ryoji M., Worcel A. Gyration is required for 5S RNA transcription from a chromatin template. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1305–1309. doi: 10.1073/pnas.83.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Worcel A. The positive transcription factor of the 5S RNA gene induces a 5S DNA-specific gyration in Xenopus oocyte extracts. Cell. 1985 Jul;41(3):945–953. doi: 10.1016/s0092-8674(85)80075-1. [DOI] [PubMed] [Google Scholar]
- Knezetic J. A., Luse D. S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986 Apr 11;45(1):95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Gurdon J. B. The reactivation of developmentally inert 5S genes in somatic nuclei injected into Xenopus oocytes. Nature. 1981 Feb 5;289(5797):461–465. doi: 10.1038/289461a0. [DOI] [PubMed] [Google Scholar]
- Korn L. J. Transcription of Xenopus 5S ribosomal RNA genes. Nature. 1982 Jan 14;295(5845):101–105. doi: 10.1038/295101a0. [DOI] [PubMed] [Google Scholar]
- Ladiges W. C., Raff R. F., Brown S., Deeg H. J., Storb R. The canine major histocompatibility complex. Supertypic specificities defined by the primed lymphocyte test (PLT). Immunogenetics. 1984;19(4):359–365. doi: 10.1007/BF00345410. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Matsui T. Transcription of adenovirus 2 major late and peptide IX genes under conditions of in vitro nucleosome assembly. Mol Cell Biol. 1987 Apr;7(4):1401–1408. doi: 10.1128/mcb.7.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millstein L., Eversole-Cire P., Blanco J., Gottesfeld J. M. Differential transcription of Xenopus oocyte and somatic-type 5 S genes in a Xenopus oocyte extract. J Biol Chem. 1987 Dec 15;262(35):17100–17110. [PubMed] [Google Scholar]
- Peck L. J., Millstein L., Eversole-Cire P., Gottesfeld J. M., Varshavsky A. Transcriptionally inactive oocyte-type 5S RNA genes of Xenopus laevis are complexed with TFIIIA in vitro. Mol Cell Biol. 1987 Oct;7(10):3503–3510. doi: 10.1128/mcb.7.10.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Schlissel M. S., Brown D. D. The transcriptional regulation of Xenopus 5s RNA genes in chromatin: the roles of active stable transcription complexes and histone H1. Cell. 1984 Jul;37(3):903–913. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- Setzer D. R., Brown D. D. Formation and stability of the 5 S RNA transcription complex. J Biol Chem. 1985 Feb 25;260(4):2483–2492. [PubMed] [Google Scholar]
- Shastry B. S., Ng S. Y., Roeder R. G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982 Nov 10;257(21):12979–12986. [PubMed] [Google Scholar]
- Shimamura A., Tremethick D., Worcel A. Characterization of the repressed 5S DNA minichromosomes assembled in vitro with a high-speed supernatant of Xenopus laevis oocytes. Mol Cell Biol. 1988 Oct;8(10):4257–4269. doi: 10.1128/mcb.8.10.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Tso J. Y., Van Den Berg D. J., Korn L. J. Structure of the gene for Xenopus transcription factor TFIIIA. Nucleic Acids Res. 1986 Mar 11;14(5):2187–2200. doi: 10.1093/nar/14.5.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Andrews M. T., Crawford E., Losa R., Brown D. D. Negative supercoiling is not required for 5S RNA transcription in vitro. Cell. 1987 May 8;49(3):301–303. doi: 10.1016/0092-8674(87)90279-0. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. DNA replication in vitro erases a Xenopus 5S RNA gene transcription complex. Cell. 1986 Oct 24;47(2):217–227. doi: 10.1016/0092-8674(86)90444-7. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. Differential 5S RNA gene expression in vitro. Cell. 1987 Dec 4;51(5):733–740. doi: 10.1016/0092-8674(87)90096-1. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]









